Abstract
Clostridium perfringens type E isolates produce iota-toxin, which is encoded by iap and ibp genes. Using Southern blot analyses, the current study identified iap/ibp plasmids of ∼97 or ∼135 kb among eight type E isolates. For most of these isolates, their iap/ibp plasmid also encoded urease and lambda-toxin. However, the beta2-toxin gene, if present, was on a different plasmid from the iap/ibp plasmid. For all isolates, the iap/ibp plasmid carried a tcp locus, strongly suggesting that these plasmids are conjugative. Overlapping PCR analyses demonstrated some similarity between the iap/ibp plasmids and enterotoxin-encoding plasmids of type A isolates. Additional PCR analyses demonstrated that the iap/ibp locus is located near dcm sequences, an apparent plasmid hot spot for toxin gene insertion, and that two IS1151-related sequences are present in the iap/ibp locus. To begin testing whether those IS1151-like sequences can mobilize iap/ibp genes, a PCR assay was performed that amplifies a product only from circular DNA forms that could represent transposition intermediates. This PCR assay detected circular forms containing iap/ibp genes and silent enterotoxin gene sequences, with or without an IS1151-like sequence. Collectively, these results suggest that a mobile genetic element carrying iap/ibp has inserted onto a tcp-carrying enterotoxin plasmid in a type A isolate, creating a progenitor iap/ibp plasmid. That plasmid then spread via conjugation to other isolates, converting them to type E. Further iap/ibp plasmid diversity occurred when either the iap/ibp genes later remobilized and inserted onto other conjugative plasmids or some iap/ibp plasmids acquired additional DNA sequences.
Clostridium perfringens isolates are classified into one of five types (A to E) (19, 25), based upon production of four typing toxins (alpha, beta, epsilon, and iota). Each C. perfringens type is associated with certain human or animal diseases (19, 25). Type E isolates cause enteritis or enterotoxemias in rabbits, lambs, cattle, and dogs (19).
By definition, type E isolates must produce both alpha- and iota-toxin (19, 25). Alpha-toxin is a 42.5-kDa single polypeptide with phospholipase C, sphingomyelinase, and hemolytic and lethal properties (23). Iota-toxin, a binary toxin comprised of two noncovalently lined components (named ιa and ιb), catalyzes ADP-ribosylation of host cell actin at Arg-177 (2). Beyond alpha- and iota-toxin, at least a few type E isolates have been shown to carry additional potential virulence genes encoding beta2-toxin, urease, or lambda-toxin (7, 12, 14, 16-18).
While the plc gene encoding alpha-toxin is chromosomal (22, 28), plasmids are also important for the virulence of Clostridium perfringens type E isolates. For example, it has been shown elsewhere (3, 12, 18) that many (if not all) type E isolates carry their iap and ibp genes (encoding, respectively, the ιa activity or ιb binding components of iota-toxin) on a large plasmid(s). While the iap/ibp plasmid remains poorly characterized, it was found that highly conserved (but silent) cpe sequences are located immediately upstream of the plasmid-borne iap/ibp genes in many, if not all, type E isolates (3, 26).
The presence of highly conserved, silent cpe sequences in the iap/ibp locus of many or all type E isolates is interesting considering that iap/ibp homologues can also be carried by other pathogenic clostridial species, including C. difficile and C. spiroforme (15). The presence of similar binary toxin genes in other pathogenic clostridia led to the proposal (3) that a foreign genetic element carrying iap/ibp may have inserted into the promoter region of a plasmid-borne cpe gene in a type A isolate, creating a progenitor iap/ibp plasmid that subsequently transferred to other C. perfringens isolates, converting those plasmid recipients to type E. This hypothesis later received indirect support from a study (5) demonstrating that the cpe plasmids of type A isolates can transfer via conjugation, probably due to their carriage of a Tn916-related tcp locus similar to that mediating conjugative transfer of the C. perfringens tetracycline resistance plasmid pCW3 (1). Further supporting this hypothesis was the detection of tcp sequences in one type E isolate (21), although it was not determined whether tcp sequences are located on the iap/ibp plasmid in that or other type E isolates.
Support is also emerging for a close association between insertion sequences (ISs) and many C. perfringens toxin genes. For example, an IS1469 sequence is located immediately upstream of the cpe gene in most or all type A isolates (6, 20). Furthermore, either IS1151 or IS1470-like sequences are present downstream of most plasmid-borne cpe genes in type A isolates (20, 21). When chromosomal, the IS1469-cpe sequences are flanked by both upstream and downstream IS1470 sequences that may represent an integrated transposon (6). It has been proposed that those flanking ISs can mobilize the chromosomal cpe gene since IS- and cpe-containing circular forms, which may represent transposition intermediates, have been detected in lysates of chromosomal cpe type A isolates (4). IS elements can also be associated with toxin genes in other C. perfringens types: e.g., an IS1151 element is located upstream of the plasmid-borne etx gene of some type D isolates (11, 12). An IS1151-like sequence has also been identified downstream of the silent cpe sequences in many type E isolates (3), but it remains unknown whether any IS element is located downstream of the iap/ibp sequence in these isolates or whether IS elements can mediate movement of iap/ibp-containing DNA into circular intermediate forms.
Regardless of whether a downstream IS1151 or IS1470-like sequence is present, the plasmid-borne cpe gene in type A isolates is always located near an upstream dcm gene (13, 20). That finding has suggested that the dcm region may represent a plasmid hot spot for insertion of mobile genetic elements carrying the cpe gene (21). Interestingly, similar dcm sequences have also been detected in some isolates of non-type A C. perfringens, including type E isolate 853 (20). However, it is unknown whether the dcm sequences of isolate 853 are located on the iap/ibp plasmid or whether dcm sequences are present in most type E isolates.
Because type E virulence plasmids have received such limited research attention to date, our current study sought to characterize their size, diversity, and virulence gene carriage using a previously studied collection of type E isolates mainly originating from diseased animals. In addition, to gain possible insights into C. perfringens virulence plasmid evolution, we further studied the organization of the iap/ibp locus in type E isolates in order to evaluate the possible presence in this locus of additional IS elements and to evaluate whether this locus is near dcm sequences.
MATERIALS AND METHODS
Bacterial strains.
With the exception of type E reference strain NCIB10748 and isolate 576 (an NCIB10748 subculture obtained from J. Glenn Songer), the other six type E isolates used in this study, i.e., isolates 51, 294, 572, 853, 1987, and B2085, originated from neonatal calves that were diagnosed in the mid-1990s with hemorrhagic enteritis in the states of KS, MO, CO, and WY (3). Previous multiplex PCR analyses and iota-toxin activity assays (3) had classified these eight isolates as type E. By pulsed-field gel electrophoresis (PFGE), isolates NCIB10748, 51, 294, 572, 853, 1987, and B2085 did not appear to share a clonal relationship (3). All type E isolates were maintained as stock cultures in 15% glycerol and were stored at −80°C.
Isolates ATCC 3624, strain 13, F5603, and F4969 are C. perfringens type A isolates. F5603 and F4969 carry, respectively, the previously sequenced cpe plasmid pCPF5603 or pCPF4969 (21). All of these type A isolates were maintained as stock cultures in cooked meat medium (Oxoid) and were stored at −20°C. Escherichia coli strain DH5α was grown in LB medium and used for transformation experiments.
Preparation of DIG-labeled probes.
Digoxigenin (DIG)-labeled DNA probes were constructed using the PCR DIG probe synthesis kit (Roche). Primers for amplifying internal iap, cpb2, urease gene (ure), cpe, tcpH, tcpF, IS1151, comEC, and lambda-toxin gene (lam) sequences, as well as reaction 6 in the overlapping PCR assay for the pCPF4969 variable region (21), are listed in Table S1 in the supplemental material.
PFGE and Southern hybridizations.
DNA plugs for PFGE were prepared as previously described (27). Briefly, overnight cultures of C. perfringens were grown in TGY (3% proteose peptone [Difco], 2% d-glucose [Fisher], 1% yeast extract [Difco], and 0.1% sodium thioglycolate [Sigma]). Those cultures were centrifuged, and the bacterial pellets were resuspended in 2% PFGE-certified agarose (Bio-Rad), for a final agarose concentration of 1%. Those plugs were then electrophoresed in a CHEF-DR II PFGE system (Bio-Rad) maintained at 14°C, using 1% PFGE-certified agarose (Bio-Rad) and 0.5× Tris-buffered EDTA. The conditions used for PFGE, transfer of electrophoresed DNA to nylon membranes (Roche), and probe hybridization were as previously described (13). After probing, PFGE Southern blots were developed using reagents from the DIG DNA labeling and detection kit (Roche). When a blot was reprobed for the presence of additional genes, the blot was first stripped with 0.2 N NaOH and 0.1% sodium dodecyl sulfate and then incubated with prehybridization solution. Prior to hybridization with the second probe, the stripped blot was exposed to film to confirm that all signal had been removed during stripping. Probe hybridization was visualized with CSPD, a ready-to-use substrate from Roche.
Restriction endonuclease digestion of pulsed-field gel plugs.
DNA plugs, prepared as described above, were washed carefully at room temperature with 0.1× Tris-EDTA buffer at least three times (15 min for each wash). A plug slice (corresponding to approximately one-sixth of the plug) was incubated overnight at proper temperature (as instructed by the manufacturer) in 30 μl of 10× commercial restriction endonuclease buffer, 4 μl of enzyme (NcoI, ClaI, StuI, BamHI, KpnI, PstI, SmaI, ApaI, BstXI, SalI, and XbaI; all from New England Biolabs), and 266 ml of distilled water. These digested plugs were then electrophoresed in a CHEF-DR II PFGE system using the following running conditions: 14°C, 6 V, and a 1- to 12-s pulse for 15 h.
Overlapping PCR analyses to determine whether type E isolates carry the conserved or variable region of pCPF5603 or pCPF4969.
For these short-range PCRs, template DNA was obtained from C. perfringens colony lysates, as described previously (30). Each PCR mixture contained 2 μl of template DNA, 17 μl of TAQ Complete 1.1 X Master Mix (Gene Choice, Frederick, MD), and 1 μl of each primer pair (1 μM final concentration). Primers used for investigating whether type E isolates carry the variable region of pCPF5603 or pCPF4969 were listed previously (21). PCR conditions for these reactions were also described previously (21). Primers used for investigating whether type E isolates carry the conserved region of pCPF5603/pCPF4969 are listed in reference 21 and Table S2 in the supplemental material. Each PCR mixture contained 2 μl of template DNA, 17 μl of TAQ Complete 1.1 X Master Mix (Gene Choice, Frederick, MD), and 1 μl of each primer pair (1 μM final concentration). The reaction mixtures, with a total volume of 20 μl, were placed in a thermal cycler (Techne) and subjected to the following amplification conditions: 1 cycle of 95°C for 2 min; 35 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min 40 s; and a single extension of 72°C for 10 min. PCR products were then electrophoresed on a 1% agarose gel, which was stained with ethidium bromide.
Sequencing of the iap/ibp locus in type E isolates.
C. perfringens DNA was isolated from type E strain NCIB10748 using the MasterPure gram-positive DNA purification kit (Epicentre). The primers dcm-F (5′-TAATATTCCTTTCAGGCTTTC-3′) and iota-R (R5′-TAAATTAAAACCCTATAAACGC-3′) were designed based upon previous results (3, 21). Those primers were added (at a 5 μM final concentration) to a PCR mixture containing 5 μl of purified NCIB10748 DNA template and Roche long-range PCR kit ingredients. The reaction mixtures, with a total volume of 50 μl, were placed in a thermal cycler (Techne) and subjected to the following amplification conditions: 1 cycle of 95°C for 3 min; 40 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 5 min; and a single extension of 68°C for 15 min. PCR products were then electrophoresed on a 1% agarose gel, which was stained with ethidium bromide. This PCR product was cloned into a TOPO vector (Invitrogen), which was sent for sequencing to the University of Pittsburgh Core Sequencing Facility.
Overlap PCR analyses of the iap/ibp locus in type E isolates.
Primers overlapping the iap/ibp plasmid region spanning from dcm to the IS1151-like sequence downstream of the silent cpe sequences were designed based upon sequencing results obtained for this region (this study and reference 4; see also Table S3 in the supplemental material). Template DNA for these short-range PCRs was obtained from colony lysates, prepared as described previously (30). Each PCR mixture contained 2 μl of template DNA, 17 μl of TAQ Complete 1.1 X Master Mix (Gene Choice, Frederick, MD), and 1 μl of each primer pair (1 μM final concentration). PCR amplification was then performed in a Techne (Burkhardtsdorf, Germany) thermocycler using the following conditions: 94°C for 2 min; 35 cycles of 94°C for 30 s, 54°C for 1 min, and 72°C for 1 min 30 s; and a single extension of 72°C for 10 min. PCR products were separated on 1% agarose gels and visualized with ethidium bromide staining.
PCR evaluation of iap/ibp-carrying circular intermediate formation.
Each PCR mixture contained 5 μl of template DNA (freshly prepared lysate from an overnight brain heart infusion culture of isolate 853), 44 μl of TAQ Complete 1.1 X Master Mix (Gene Choice, Frederick, MD), and 1 μl of each primer pair (1 μM final primer concentration). Primers used in these studies included P1-R and P8-F from Table S3 in the supplemental material and L1-F (5′-TGTATCCAGTGATATTATTAC-3′) and L1-R (5′-TTAGAAGAATATAAGAAAGAAC-3′). The PCRs that were run included reaction R1 (using primers L1-R and P8-F), R2 (using primers L1-R and L1-F), R3 (using primers P1-R and L1-F), and R4 (using primers P1-R and P8-F). PCR amplification was then performed in a Techne thermocycler using the following conditions: 94°C for 2 min; 35 cycles of 94°C for 30 s, 54°C for 30 s, and 72°C for 2 min 30 s; and a single extension of 72°C for 15 min. PCR products were separated on 1.5% agarose gels and visualized with ethidium bromide staining. PCR products were also excised from the gel, ligated into a Topo vector, and sequenced.
Nucleotide sequence accession number.
The sequencing results were deposited in GenBank under accession number EF179380.
RESULTS
Pulsed-field gel Southern blot examination of iap-carrying plasmids in type E isolates.
Previous studies by several laboratories (3, 8, 10, 12, 13, 18, 21) have demonstrated that Southern blot assays of pulsed-field gels provide useful insights into the size, gene carriage, and diversity of C. perfringens virulence plasmids. Therefore, this technique was applied to a collection of type E isolates in order to (i) evaluate whether selected genes are commonly present among type E isolates; (ii) determine whether those genes, when present, are located on the chromosome or on plasmids; and (iii) evaluate the size and diversity of type E virulence plasmids.
Our initial pulsed-field Southern blot analyses focused on the iap/ibp plasmid, whose presence is required for the iota-toxin production that defines type E isolates. For this experiment, undigested DNA from eight type E isolates was subjected to PFGE under conditions allowing undigested plasmid DNA (but not undigested chromosomal DNA) to enter the pulsed-field gels (3, 8, 10, 12, 13, 18, 21). The result of Southern blot assays demonstrated (Fig. 1A), for all eight type E isolates, that an iap-specific probe hybridized with undigested DNA that had entered the pulsed-field gels, confirming previous studies (3) showing that the iap gene is plasmid borne in all these type E isolates. As a negative control, the iap probe did not hybridize with DNA from type A isolate F5603, which is iap negative by PCR (data not shown).
FIG. 1.
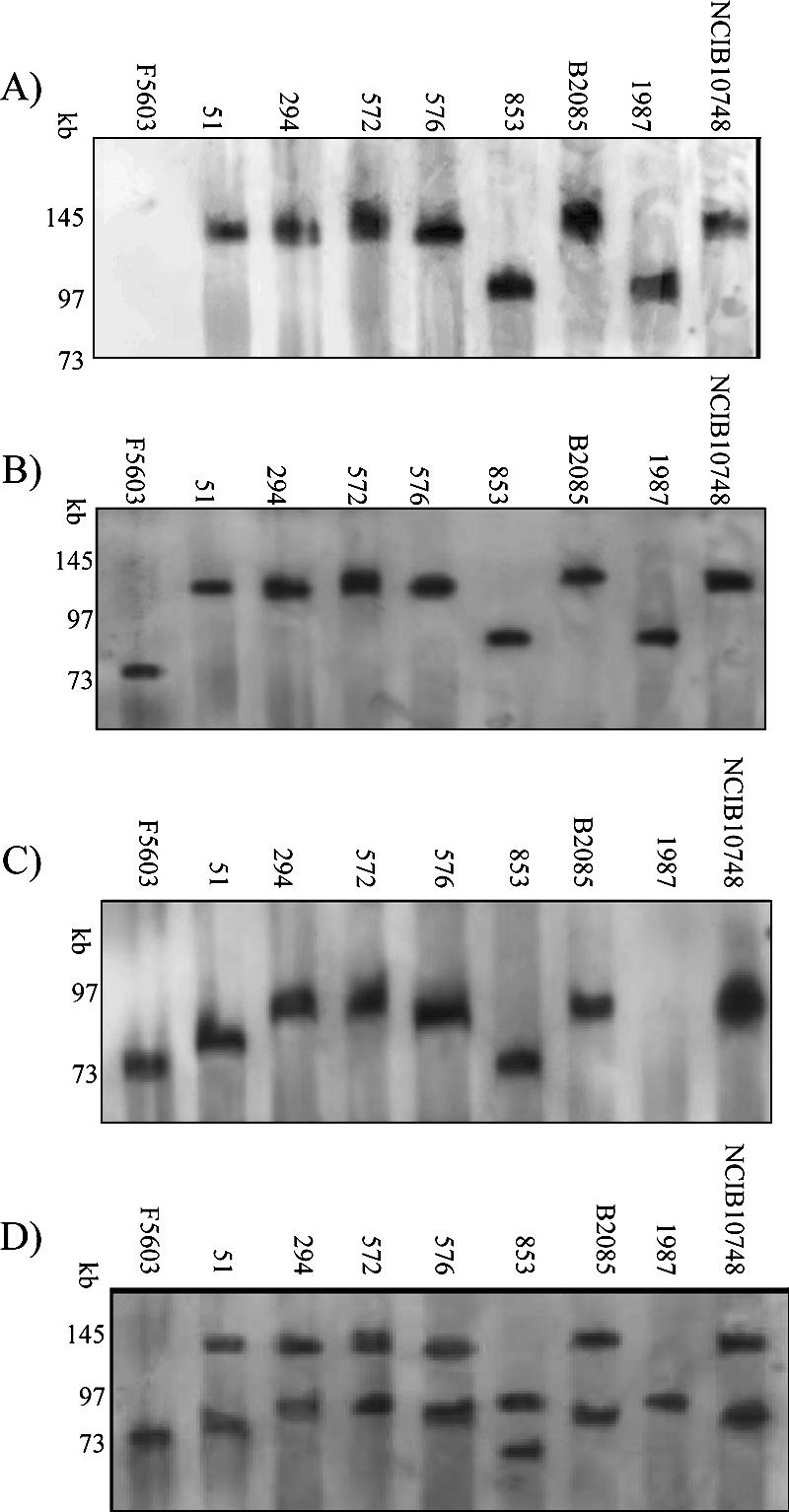
Southern blot of a pulsed-field gel electrophoresed with DNA from specified type E isolates and then hybridized with an iap probe (A). After stripping, the blot was rehybridized with a cpe probe (B). After a final stripping, the blot was rehybridized with a cpb2 probe (C). An overlay of the panel B and panel C blots demonstrates that cpb2 is located on a different plasmid than the iota-toxin locus (D). Numbers at left of each blot indicate migration of size markers in kb.
It has also been established previously that, on these pulsed-field Southern blots, probes hybridize predominately to the linearized plasmid form, thus allowing an accurate determination of plasmid size (10, 12, 13, 18, 21). This approach had previously shown that the iap/ibp plasmid in NCIB10748 is ∼130 kb in size (18), but the size of the iap/ibp plasmid in other type E isolates had not been evaluated prior to our current study. As shown in Fig. 1A, this Southern blot revealed the existence of size variations among iap/ibp plasmids of different type E isolates. Specifically, in six of the eight type E isolates surveyed, the iap/ibp virulence plasmid was ∼135 kb, while the other two type E plasmids were found to carry an iap/ibp plasmid of only ∼97 kb. It is noteworthy that the ∼135-kb size determined in the current study for the iap/ibp plasmid of NCIB10748 closely matched the previous ∼130-kb size estimate for this plasmid (18).
Previous Western blotting and DNA sequencing results (3) demonstrated that the eight type E isolates surveyed each carry silent cpe sequences near their iap/ibp genes. When the Fig. 1A Southern blot was stripped and rehybridized with a cpe probe, the results obtained were consistent with those previous findings, i.e., for each of these eight type E isolates, a hybridization signal was observed at the same blot location containing the iap/ibp plasmid (Fig. 1B). The specificity of this cpe probe hybridization with DNA from the type E isolates (and cpe-positive type A isolate F5603) was supported by the failure of the probe to react with DNA from cpe-negative type A isolate ATCC 3624 or strain 13 (data not shown).
Pulsed-field gel Southern blot analyses of type E isolates to assess their plasmid carriage of cpb2, lam, ure, IS1151-like sequences, and tcp conjugative transfer genes.
Other laboratories have reported (17) that type E isolates often carry the cpb2 gene encoding beta2-toxin, a potential accessory toxin for virulence. However, to our knowledge, it had not yet been determined whether the cpb2 gene in type E isolates is plasmid borne and, if so, whether this toxin gene is present on the iap plasmid. To address those questions, the Fig. 1B Southern blot was stripped and reprobed with a cpb2-specific probe. The results obtained (Fig. 1C and Table 1) demonstrate that, in seven of the eight type E isolates surveyed, the cpb2 gene is present on plasmids that range in size from ∼70 to ∼97 kb. The single type E isolate (1987) whose DNA failed to hybridize with the cpb2 probe on this Southern blot also tested cpb2 negative by PCR (data not shown). In contrast to the limited diversity of iap/ibp plasmid sizes detected among the surveyed type E isolates in Fig. 1A, five differently sized cpb2-carrying plasmids were demonstrated among the seven cpb2-positive type E isolates. Furthermore, each of the seven cpb2-positive type E isolates clearly carried its cpb2 gene on a plasmid distinct from the iap/ibp plasmid. As specificity controls, the cpb2 probe was shown to hybridize with F5603 DNA (Fig. 1C), which carries cpb2 on pCPF5603 (21), but failed to hybridize with DNA from cpb2-negative isolate ATCC 3624 (data not shown).
TABLE 1.
Sizes of virulence plasmids among type E isolates
| Strain | Type | Size(s) (kb) of plasmid(s) carrying gene or element:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| iap/ibp | cpe | ure | lam | cpb2 | tcpH | tcpF | IS1151 | ||
| F5603 | A | 75 | 75 | 75 | 75 | 75 | |||
| 51 | E | 135 | 135 | 135 | 135 | 85 | 135, 65 | 135, 65 | 135, 85 |
| 294 | E | 135 | 135 | 135 | 135 | 97 | 135 | 135 | 135, 97 |
| 572 | E | 135 | 135 | 135 | 135 | 97 | 135, 65 | 135, 65 | 135, 97 |
| 576 | E | 135 | 135 | 135, 90 | 135 | 90 | 135, 90 | 135, 90 | 135, 90 |
| 853 | E | 97 | 97 | 97 | 97 | 70 | 97, 70 | 97, 70 | 97, 70 |
| 1987 | E | 97 | 97 | 97 | 97 | 97 | 97 | ||
| B2085 | E | 135 | 135 | 135 | 135 | 90 | 135, 65 | 135, 65 | 135, 90 |
| 10748a | E | 135 | 135 | 135 | 135 | 90 | 135, 65 | 135, 65 | 135, 90 |
NCIB10748.
Two other potential virulence genes, i.e., ure and lam, have been detected in one or a few type E isolates (12, 14, 16, 18); in addition, ure genes were shown to be plasmid borne in two type E isolates, including NCIB10748 (12). However, it had not yet been systematically investigated whether ure and lam are commonly present in type E isolates and, if so, whether they are present on the iap/ibp plasmid (although one study [12] suggested that ure might be on the iap/ibp plasmid of NCIB10748). Therefore, pulsed-field gel Southern blot assays were performed using lam or ure probes, which demonstrated (Table 1) the presence of lam in all eight type E isolates surveyed. Furthermore, for each type E isolate surveyed, the lam probe hybridized to the same blot location containing the iap/ibp plasmid. In contrast, DNA from one type E isolate (1987) failed to hybridize to the ure probe (Table 1); isolate 1987 was also PCR negative for ure, while the other seven isolates were PCR positive (data not shown). For the seven ure-positive type E isolates, the ure probe consistently hybridized to the same blot location containing the iap/ibp plasmid. In addition, type E isolate 576 was also found to carry a second, smaller ure plasmid (Table 1). As a negative control for probe specificity, the ure and lam probes each failed to hybridize with DNA from F5603, which is ure negative and lam negative by PCR (data not shown).
Our previous sequencing studies (3) had demonstrated the presence of an IS1151-like sequence ∼1 kb downstream of the silent cpe sequences in the iap/ibp locus of these eight surveyed type E isolates. Consistent with that previous finding, all eight type E isolates hybridized to an IS1151-like specific probe to the same blot location containing the iap/ibp plasmid. For seven of these type E isolates (the exception being 1987, the only cpb2-negative type E isolate surveyed), the IS1151-like probe also hybridized to a second plasmid. The additional IS1151-like carrying plasmids, which ranged in size from 70 to 97 kb, consistently matched the size of the cpb2 plasmid present in each of those seven isolates (Table 1).
The conjugative mechanism for C. perfringens plasmid pCW3 (and probably pCPF4969 and pCPF5603 [21]) was recently shown (1) to involve a Tn916-related tcp locus, and similar tcp genes had been previously detected in one type E isolate (21). Since we previously hypothesized that iap/ibp plasmids are conjugative (3), Southern blot analyses of pulsed-field gels were performed to evaluate whether tcp conjugative transfer genes are commonly present in type E isolates and, if so, whether they are associated with the iap/ibp plasmid in our eight surveyed type E isolates. Using probes for two different tcp genes, each known to be essential for pCW3 plasmid conjugation (1), hybridization was observed with DNA from all eight type E isolates (Table 1). Furthermore, the tcpH and tcpF probes hybridized to the same blot location containing the iap plasmid for each of the type E isolates surveyed. In addition, both the tcpH and tcpF probes also hybridized to a second plasmid in six of the eight type E isolates. For five of those six type E isolates, their second tcpH- and tcpF-carrying plasmid was clearly distinct from the cpb2 plasmid. However, for isolate 576, the tcpH, tcpF, and cpb2 probes all hybridized to a plasmid(s) of ∼90 kb.
As controls for tcpH and tcpF probe specificity, similar Southern blot assays of pulsed-field gels demonstrated that (i) both tcp probes hybridized to pCPF5603, which has been shown by sequencing to carry tcpH and tcpF (21), but (ii) both tcp gene probes failed to hybridize with DNA from strain 13, whose single plasmid is devoid of the tcp locus (28).
Pulsed-field gel Southern blot analysis of restriction endonuclease-digested DNA to distinguish whether comigrating genes in type E isolates are present on the same plasmid or two similarly sized plasmids.
The pulsed-field gel Southern blotting results presented in Fig. 1 and Table 1 could indicate either that (i) ure, lam, tcpF, tcpH, and IS1151-like are often found on the iap/ibp-carrying plasmid or (ii) in type E isolates, those genes are often present on plasmids that share a similar size but are distinct from iap/ibp plasmids. To help discriminate between those two possibilities, DNA from representative type E isolates 853 and NCIB10748 was subjected to digestion with a restriction enzyme battery. In this experiment (10), the migration of two genes present on the same plasmid should consistently exhibit the same response (sensitivity or insensitivity) to digestion with each restriction endonuclease. However, if one gene showed no change in migration after digestion with a particular restriction endonuclease while migration of the second gene was affected by the same enzyme, this would indicate that the two genes are present on similarly sized, but distinct, plasmids.
When this analysis was performed with the representative type E isolates 853 and NCIB10748, the results obtained supported the presence of lam, ure, tcpH, and tcpJ on the iap/ibp-carrying plasmid of both isolates (Fig. 2 and data not shown). Specifically, the migration of plasmid DNA carrying iap, lam, ure, tcpH, and tcpJ was affected by NcoI, ClaI, and StuI but not by BamHI, KpnI, PstI, SmaI, ApaI, BstxI, SalI, or XbaI.
FIG. 2.
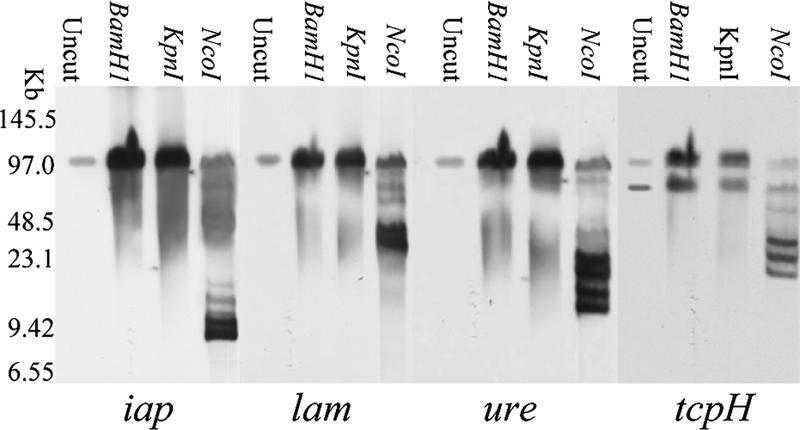
Southern blot of a pulsed-field gel electrophoresed with DNA from type E isolate 853 and then digested with specified restriction enzymes for 16 h. The Southern blot was first hybridized with an iap probe and then sequentially stripped and rehybridized with lam, ure, and tcpH probes.
PCR analyses to determine whether the pCPF4969/pCPF5603 conserved region is present among type E isolates.
An internal open reading frame (ORF)-specific PCR analysis showed (Table 2) that either all (isolate 853) or most (the remaining seven type E isolates) of 14 surveyed ORFs in the conserved region of pCPF4969/pCPF5603 are also present in the eight surveyed type E isolates. Notably, these analyses confirmed the pulsed-field Southern blotting results indicating that all eight type E isolates carry the tcp ORFs known to mediate conjugative transfer of C. perfringens plasmid pCW3 and (probably) pCPF4969 and pCPF5603.
TABLE 2.
Results for individual ORF PCRsa
| Gene | F5603 | 853 | 1987 | 51 | 572 | B2085 | NCIB10748 | 294 | 576 |
|---|---|---|---|---|---|---|---|---|---|
| Conserved region | |||||||||
| dcm | + | + | + | + | + | + | + | + | + |
| cpe | + | + | + | + | + | + | + | + | + |
| IS1151 | + | + | + | + | + | + | + | + | + |
| ftsK | + | + | + | + | + | + | + | + | + |
| tcpC | + | + | + | + | + | + | + | + | + |
| tcpH | + | + | + | + | + | + | + | + | + |
| tcpF | + | + | + | + | + | + | + | + | + |
| dam | + | + | + | + | + | + | + | + | + |
| cna | + | + | + | + | + | + | + | + | + |
| srt | + | + | + | + | + | + | + | + | + |
| lexA-1 | + | + | + | + | + | + | + | + | + |
| pspC | + | + | + | + | + | + | + | + | − |
| hel | + | + | + | + | + | + | + | + | − |
| flgJ | + | + | − | − | + | + | + | − | − |
| Variable region | + | + | + | + | + | + | + | + | − |
| capK | |||||||||
| pri | + | + | − | + | − | − | − | − | − |
| comEC | + | + | + | + | − | − | − | − | − |
| his | + | + | − | − | − | − | − | − | − |
| thiF | + | + | − | − | − | − | − | − | − |
| cpb2 | + | + | − | + | + | + | + | + | + |
| ure | + | + | − | + | + | + | + | + | + |
+, present; −, absent.
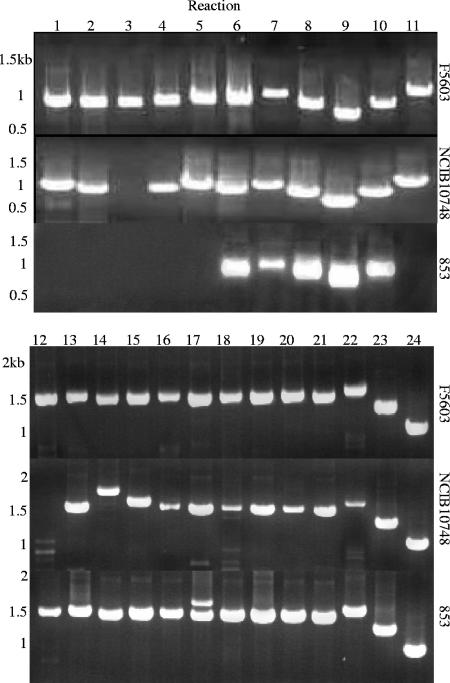
Overlapping PCR analyses were then performed to assess the arrangement of those conserved-region ORFs in the surveyed type E isolates; this 24-reaction analysis, which spans from an IS1151 located near cpe to the start of the pCPF5603 or pCPF4969 variable region, revealed (Fig. 3; see also Table S4 in the supplemental material) that all eight isolates carry a tcp region arranged similarly to that present on pCPF5603, rather than the slightly different tcp locus arrangement of pCPF4969. This assay also clearly demonstrated (Fig. 3 and data not shown) that one type E isolate (NCIB10748) carries virtually the entire pCPF4969/pCPF5603 conserved region, with the remaining seven isolates carrying most of the pCPF4969/pCPF5603 conserved region.
FIG. 3.
Overlapping PCR amplification of the conserved region of pCPF5603/pCPF4969 using a previously described primer battery (21) and one described in this study (see Table S2 in the supplemental material). DNA template included F5603 DNA (top), NCIB10748 DNA (middle), and 853 DNA (bottom). Marker sizes are shown at left.
PCR analyses to determine whether the pCPF4969 or pCPF5603 variable region is present among type E isolates.
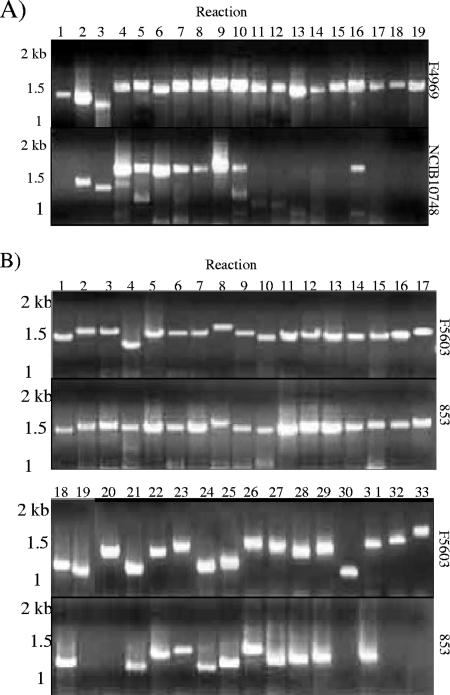
The presence of the pCPF4969 variable region in the eight type E isolates was evaluated using a previously described 24-reaction, overlapping PCR assay that spans the entire pCPF4969 variable region (21). This PCR battery amplified only two of the expected 24 PCR products from seven type E isolates (see Table S5 in the supplemental material). However, this assay indicated that NCIB10748 apparently carries a more substantial portion (from reaction 2 to reaction 10) of the pCPF4969 variable region (Fig. 4A). A Southern blot assay of a pulsed-field gel with a probe to reaction 6 of the pCF4969 variable-region overlapping PCR assay confirmed that pCFP4969 variable-region sequences are present on the iap/ibp plasmid of NCIB10748.
FIG. 4.
Overlapping PCR amplification of the pCPF4969 (A) or pCPF5603 (B) variable region using DNA from type E isolates NCIB10748 and 853. No substantial amplification was observed using DNA from the other six surveyed type E isolates. Marker sizes are shown at left.
A second, 33-reaction overlapping PCR battery spanning the entire pCPF5603 variable region (21) was then performed to survey the presence of the pCPF5603 variable region in the surveyed type E isolates. With this PCR battery, type E isolate 853 showed significant amplification over a substantial portion of the pCPF5603 variable region (Fig. 4B; see also Table S6 in the supplemental material), while isolate 1987 supported amplification of reactions 21 to 29 of this assay. The other six isolates amplified only a few products (never representing more than three adjacent sequence reactions) in this assay. To assess whether the amplified pCPF5603 reactions of isolates 853 and 1987 were present on the iap/ibp plasmid, a pulsed-field gel was run with DNA from those two isolates and hybridized with a probe to the putative comEC ORF of the pCPF5603 variable region. That probe failed to react with NCIB10748 DNA, consistent with Table 2 results, but hybridized to a plasmid of about 97 kb in isolates 1987 and 853, matching the size of the iap/ibp plasmid in both those isolates.
Further characterization of the iap/ibp locus in type E isolates.
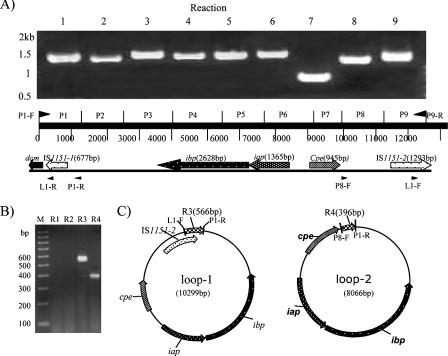
The iap/ibp locus, previously shown to be highly conserved among these eight type E isolates (3), consists of an IS1151-like sequence (now referred to as IS1151-2) upstream of silent cpe sequences, along with the iota-toxin genes and silent cpe sequences (Fig. 5A). Since the plasmid-borne cpe gene in both pCPF4969 and pCPF5603 is present near dcm sequences (13, 21), long-range PCR analyses were performed to evaluate whether the iap/iab genes of type E isolates might also be located near a dcm gene. Those long-range PCR assays successfully linked dcm to the iap gene for all eight surveyed type E isolates (data not shown). This reaction produced a 4.3-kb PCR product that, when sequenced from NCIB10748, revealed that the iap/ibp locus also contains a second, partial copy of IS1151 (now termed IS1151-1) located between dcm and iap, as shown in Fig. 5A.
FIG. 5.
Further characterization of the iap/ibp locus arrangement and demonstration that this locus can exist as circular intermediates that could be transposition intermediates. Based upon sequencing of a long-range PCR product connecting dcm to the iap/ibp locus in NCIB10748 (not shown), an overlapping PCR assay was designed and applied to the eight surveyed type E isolates. Results for NCIB10748 are shown in panel A; similar results were also observed with the other seven isolates. Panel B shows results from a PCR assay that used primer sets that amplify products from the iap/ibp locus of NCIB10748 when this locus is present in a small circular form but not in a linear form. The primer sets used are R1 (primers L1-R and P8-F), R2 (L1-R and L1-F), R3 (P1-R and L1-F), and R4 (P1-R and P8-F); locations of primers are shown in panel A. Panel C shows the expected products from small circular forms containing IS1151-2-ibp-iap-silent cpe sequences or ibp-iap-silent cpe sequences. The identity of these products was confirmed by DNA sequencing.
This new information demonstrating that the iap/ibp locus is flanked by IS1151-like sequences could indicate that the iap/ibp genes are present on an integrated transposon. To start addressing whether the iap/ibp locus of type E isolates can be mobilized, we employed PCR methods similar to those used previously (4) to demonstrate that the chromosomal cpe locus of type A isolates can form circular species that could represent transposition intermediates. Specifically, these PCRs employed primers that are unable to amplify a product from a linear iap/ibp locus but yield PCR products if this region can form circular intermediates. Those analyses successfully amplified several PCR products from all eight type E isolates (Fig. 5B). When two of those products (from isolate 853) were sequenced, they corresponded to circular products containing either iap-ibp-silent cpe sequences or iap-ibp-silent cpe-IS1151-2 sequences (Fig. 5C).
DISCUSSION
Despite the significance of type E isolates as veterinary pathogens, their virulence plasmids had received only limited research attention. In the current study, Southern blot assays of pulsed-field gels revealed previously unappreciated size, sequence, or arrangement variations among the iota-toxin-encoding plasmids of type E isolates. Most surveyed type E isolates were found to carry their iap genes (and, by extension, ibp genes) on ∼135-kb plasmids, which represent the largest virulence plasmids identified, to date, in C. perfringens. However, precedent exists for even larger virulence plasmids in other gram-positive bacteria: e.g., pXO1 of Bacillus anthracis is ∼182 kb (24). Our results also identified variations in gene sequence or arrangement, even among iap/ibp plasmids of similar size. For example, an overlapping PCR assay amplified some pCPF4969 variable-region sequences from NCIB10748 but not from the other type E isolates carrying an ∼135-kb iap/ibp plasmid.
Results from this study have also clearly established that iap/ibp plasmids often, although not universally (this study and reference 15), carry lam and ure as additional potential virulence genes. The common presence of lam on iap/ibp plasmids is interesting since the ιa and ιb components of iota-toxin must be proteolytically activated to gain toxicity (2) and lambda-toxin is a metalloprotease known to activate iota-toxin (15). Therefore, the presence of lam on many iap plasmids may ensure that those type E strains can activate their iota-toxin. Urease also could contribute to C. perfringens virulence, since this enzyme is a proven virulence factor for other gastrointestinal pathogens (9). Conversely, urease conceivably might inhibit type E isolate virulence by raising the microenvironment pH, thus interfering with internalization of ιa into the host cell cytoplasm, since that process involves endosomal acidification (2). These alternatives should be resolved by future studies.
Our results confirmed previous surveys (17) indicating that many type E isolates carry the cpb2 gene, although we also identified a cpb2-negative type E isolate. Interestingly, Southern blots clearly showed that, when present, the cpb2 gene in type E isolates is present on a different plasmid than that carrying the iap/ibp, ure, and lam genes, i.e., many type E isolates maintain at least two distinct potential virulence plasmids. In that respect, type E isolates resemble those type A isolates carrying pCPF4969-like cpe plasmids and a second, distinct cpb2 plasmid (13). A novel aspect of the type E cpb2 plasmids is their large size, which extends up to ∼97 kb; in contrast, the largest identified cpb2-carrying plasmids of type A isolates are only 75 kb (13). Further studies should examine the relationship between the cpb2 plasmids of different type E isolates, their relationship to the cpb2 plasmids of type A isolates, and how commonly the large type E cpb2 plasmids encode additional virulence factors. However, our results already suggest that the cpb2 plasmid of isolate 576 carries a second potential virulence gene, i.e., ure. Interestingly, the presence of this ure+ cpb2+ plasmid in isolate 576 represents only one of several differences detected in our study between that isolate and NCIB10748, which should be the same isolate supplied from different lab sources. PFGE gels comparing MluI- or ApaI-digested DNA from NCIB10748 and 576 (not shown) suggested that these isolates are closely related but also revealed some minor banding differences. Collectively, these results suggest that 576 and NCIB10748 share a common origin but now exhibit some differences among their plasmids.
Our current results demonstrate that many iap/ibp plasmids carry a tcp locus similar to that present on known C. perfringens conjugative plasmids (21), predicting that these iap/ibp plasmids should also be conjugative. The current study also found that many type E isolates possess more than one tcp-carrying plasmid, suggesting that a single C. perfringens cell can carry multiple conjugative plasmids. For example, the cpb2 plasmid of isolate 853 also may be conjugative, marking the first identification of a potential conjugative plasmid carrying cpb2 in the absence of cpe or silent cpe sequences.
Results from the current and previous (3) studies indicate that the iap/ibp locus in many type E isolates is flanked by upstream and downstream IS1151-related sequences. This arrangement could indicate either that (i) the iap/ibp genes (and silent cpe sequences) are part of an integrated composite transposon that had flanking IS1151-like sequences or that (ii) an element containing IS1151-iap-ibp inserted near the cpe promoter on a pCPF5603-like plasmid (21), where IS1151-like sequences were already present downstream of the cpe gene. Interestingly, another laboratory reported that type E isolate CN5056 has an IS1469, rather than IS1151, sequence downstream of its silent cpe sequences in the iap/ibp locus (26). In our current study, PCR indicated (not shown) that at least six of the eight surveyed type E isolates do not carry IS1469. Further surveys should determine whether CN5056 is a unique type E isolate or represents a second, common lineage of type E isolates. Our current results further suggest that IS1151-related sequences are also present on the cpb2 plasmid in most type E isolates. Future studies should determine if those IS1151-like sequences are associated with cpb2, which could help explain the broad distribution of the cpb2 gene among C. perfringens isolates.
Our initial model for type E isolate evolution proposed that a mobile genetic element carrying iap/ibp had inserted near the promoter of a plasmid-borne cpe gene in a type A isolate, inactivating that cpe gene and permitting the accumulation of additional mutations. To account for the presence of highly conserved mutations in the silent cpe sequences of our eight chromosomally distinct type E isolates, we further proposed that this progenitor iap/ibp plasmid had then conjugatively transferred to a number of other type A isolates, converting those isolates to type E. The current results support certain aspects of our previous model. For example, our PCR results showed that all of the surveyed type E iap/ibp plasmids, but particularly the ∼97-kb iap/ibp plasmids of 853 and 1987, share some sequence similarity with the cpe plasmids of type A isolates. The current study also found that these iap/ibp plasmids typically carry a tcp locus, which predicts that these plasmids should be conjugative (1). Finally, supporting the model's suggestion that a mobile genetic element carrying the iap/ibp genes had inserted into a cpe plasmid of a type A isolate, our current results revealed that (i) iap/ibp genes can be found in circular forms that could be transposition intermediates and (ii) a downstream, partial IS1151-like sequence is present that could once have helped mediate iap/ibp insertion into a cpe plasmid. Interestingly, we found that those IS1151-1-iap-ibp sequences are present near dcm sequences in type E isolates, further supporting the dcm region as an apparent hot spot for toxin gene insertion onto C. perfringens plasmids (21).
However, our current results also revealed some inconsistencies with our previous model for type E isolate evolution. Most notably, that model had predicted that iap/ibp plasmids should all be similar, yet our current studies demonstrated substantial size and sequence or arrangement variations among type E iap/ibp plasmids. One possible explanation for this discrepancy vis-à-vis our original model would be for the ∼97-kb iap/ibp plasmids to represent a progenitor iap plasmid, arising from the 75-kb pCPF5603 cpe plasmid acquiring an iap/ibp-carrying mobile genetic element. Whether that element also carried lam and ure or whether those (and other) genes were acquired later in independent insertional events remains to be determined. In this scenario, the ∼135-kb iap/ibp plasmids present in the other six surveyed type E isolates might simply represent an ∼97-kb iap/ibp plasmid that was further enlarged by acquiring additional DNA sequences. If so, some of the original pCPF5603 DNA was lost or rearranged during this enlargement process, since the ∼135-kb iap/ibp plasmids lack nearly all of the pCPF5603 variable-region genes found in the ∼97-kb iap/ibp plasmids. An alternative model is that the iap/ibp locus later remobilized from the ∼97-kb plasmid and then reinserted onto another C. perfringens plasmid or plasmids to give rise to the ∼135-kb iap plasmids. This second possibility is supported by the presence of multiple large tcp-carrying plasmids in some type E isolates that could be targets for iap/ibp element insertion. However, this model would not readily explain why both the ∼97-kb and ∼135-kb iap plasmids share some cpe plasmid sequences or carry ure and lam, unless those two genes had comobilized with the iap/ibp genes. In any event, it is notable that all large C. perfringens plasmids studied to date (this study; 1, 3, 21, 28), whether toxin or antibiotic resistance encoding, carry a similarly arranged region containing cna, srt, and dam ORFs, suggesting a common evolutionary core for many large C. perfringens plasmids. Sequencing of several type E plasmids would help to resolve the remaining questions regarding the evolution of iap plasmids among different type E strains and resolve whether the very large iap plasmids carry additional, still-unrecognized, virulence genes.
As mentioned in the introduction, iap/ibp-like genes are also found in C. spiroforme and C. difficile (2), including the newly emergent C. difficile epidemic strains (29). In fact, our type E evolution model had proposed that type E isolates originally acquired their iap/ibp genes from another clostridial species. This possibility is supported by our results suggesting that the iap/ibp genes are associated with mobilizable genetic elements. This apparent association of iap/ibp genes with mobilizable elements may also help to explain the distribution of iap/ibp-like genes among several clostridial species. For example, two clostridial species in the gastrointestinal tract or soil may have exchanged a conjugative plasmid carrying an iap/ibp-like genetic element, which could then have mobilized and stably inserted onto the chromosome or another plasmid in the recipient clostridial species. Future studies should compare the iap/ibp-like genes found in other clostridial species with those found in C. perfringens type E isolates to gain further insights into clostridial virulence evolution.
Supplementary Material
Acknowledgments
This work was generously supported by grants R37AI19844 and RO1AI056177 from the National Institute of Allergy and Infectious Diseases and by grant 2005-53201-15387 from the Ensuring Food Safety Program of the United States Department of Agriculture.
We thank J. Glenn Songer for originally supplying most of the type E isolates and Derek Fisher for helpful experimental suggestions.
Editor: D. L. Burns
Footnotes
Published ahead of print on 29 January 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Bannam, T. L., W. L. Teng, D. Bulach, D. Lyras, and J. I. Rood. 2006. Functional identification of conjugation and replication regions of the tetracycline resistance plasmid pCW3 from Clostridium perfringens. J. Bacteriol. 188:4942-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, H., K. Aktories, M. R. Popoff, and B. G. Stiles. 2004. Binary bacterial toxins: biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol. Mol. Biol. Rev. 68:373-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billington, S. J., E. U. Wieckowski, M. R. Sarker, D. Bueschel, J. G. Songer, and B. A. McClane. 1998. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin sequences. Infect. Immun. 66:4531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4.Brynestad, S., and P. E. Granum. 1999. Evidence that Tn5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. FEMS Microbiol. Lett. 170:281-286. [DOI] [PubMed] [Google Scholar]
- 5.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. The enterotoxin (CPE) plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brynestad, S., B. Synstad, and P. E. Granum. 1997. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 143:2109-2115. [DOI] [PubMed] [Google Scholar]
- 7.Bueschel, D. M., B. H. Jost, S. J. Billington, H. T. Trinh, and J. G. Songer. 2003. Prevalence of cpb2, encoding beta2 toxin, in Clostridium perfringens field isolates: correlation of genotype with phenotype. Vet. Microbiol. 94:121-129. [DOI] [PubMed] [Google Scholar]
- 8.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with nonfoodborne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, C. M., and S. E. D'Orazio. 1993. Bacterial ureases: structure, regulation of expression and role in pathogenesis. Mol. Microbiol. 9:907-913. [DOI] [PubMed] [Google Scholar]
- 10.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Carnard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 11.Daube, G., P. Simon, and A. Kaeckenbeeck. 1993. IS1151, an IS-like element of Clostridium perfringens. Nucleic Acids Res. 21:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupuy, B., G. Daube, M. R. Popoff, and S. T. Cole. 1997. Clostridium perfringens urease genes are plasmid-borne. Infect. Immun. 65:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, D. J., K. Miyamoto, B. Harrision, S. Akimoto, M. R. Sarker, and B. A. McClane. 2005. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56:747-762. [DOI] [PubMed] [Google Scholar]
- 14.Gibert, M., C. Jolivet-Reynaud, and M. R. Popoff. 1997. Beta2 toxin, a novel toxin produced by Clostridium perfringens. Gene 203:65-73. [DOI] [PubMed] [Google Scholar]
- 15.Gibert, M., S. Perelie, G. Daube, and M. R. Popoff. 1997. Clostridium spiroforme toxin genes are related to C. perfringens iota toxin genes but have a different genomic localization. Syst. Appl. Microbiol. 20:337-347. [Google Scholar]
- 16.Jin, F., O. Matsushita, S. Katayama, S. Jin, C. Matsushita, J. Minami, and A. Okabe. 1996. Purification, characterization, and primary structure of Clostridium perfringens lambda-toxin, a thermolysin-like metalloprotease. Infect. Immun. 64:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jost, B. H., S. J. Billington, H. T. Trinh, D. M. Bueschel, and J. G. Songer. 2005. Atypical cpb2 genes, encoding beta2-toxin in Clostridium perfringens isolates of nonporcine origin. Infect. Immun. 73:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katayama, S. I., B. Dupuy, G. Daube, B. China, and S. T. Cole. 1996. Genome mapping of Clostridium perfringens strains with I-Ceu I shows many virulence genes to be plasmid-borne. Mol. Gen. Genet. 251:720-726. [DOI] [PubMed] [Google Scholar]
- 19.McClane, B. A., F. A. Uzal, M. F. Miyakawa, D. Lyerly, and T. Wilkins. 2006. The enterotoxic clostridia, p. 698-752. In M. Dworkin, S. Falkow, E. Rosenburg, K. H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, vol. 4. Springer-Verlag, New York, NY. [Google Scholar]
- 20.Miyamoto, K., G. Chakrabarti, Y. Morino, and B. A. McClane. 2002. Organization of the plasmid cpe locus of Clostridium perfringens type A isolates. Infect. Immun. 70:4261-4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyamoto, K., D. J. Fisher, J. Li, S. Sayeed, S. Akimoto, and B. A. McClane. 2006. Complete sequencing and diversity analysis of the enterotoxin-encoding plasmids in Clostridium perfringens type A non-food-borne human gastrointestinal disease isolates. J. Bacteriol. 188:1585-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naylor, C. E., J. T. Eaton, A. Howells, N. Justin, D. S. Moss, R. W. Titball, and A. K. Basak. 1998. Structure of the key toxin in gas gangrene. Nat. Struct. Biol. 5:738-746. [DOI] [PubMed] [Google Scholar]
- 24.Okinaka, R. T., K. Cloud, O. Hampton, A. R. Hoffmaster, K. K. Hill, P. Keim, T. M. Koehler, G. Lamke, S. Kumano, J. Mahillon, D. Manter, Y. Martinez, D. Riche, R. Svensson, and P. J. Jackson. 1999. Sequence and organization of pXO1, the large Bacillus anthracis plasmid harboring the anthrax toxin genes. J. Bacteriol. 181:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petit, L., M. Gilbert, and M. R. Popoff. 1999. Clostridium perfringens: toxinotype and genotype. Trends Microbiol. 7:104-110. [DOI] [PubMed] [Google Scholar]
- 26.Popoff, M. R., and B. G. Stiles. 2005. Clostridial toxins vs. other bacterial toxins, p. 323-385. In P. Dürre (ed.), Handbook on clostridia. CRC Press, Taylor & Francis Group, Boca Raton, FL.
- 27.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ. Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thorpe, C. M., and S. L. Gorbach. 2006. Update on Clostridium difficile. Curr. Treatment Options Gastroenterol. 9:265-271. [DOI] [PubMed] [Google Scholar]
- 30.Wen, Q., K. Miyamoto, and B. A. McClane. 2003. Development of a duplex PCR genotyping assay for distinguishing Clostridium perfringens type A isolates carrying chromosomal enterotoxin (cpe) genes from those carrying plasmid-borne enterotoxin (cpe) genes. J. Clin. Microbiol. 41:1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.