Abstract
Detection of antigen-specific CD4+ T cells is facilitated by the use of fluorescently labeled soluble peptide-major histocompatibility complex (MHC) multimers which mirror the antigen specificity of T-cell receptor recognition. We have used soluble peptide-MHC class II tetramers containing peptides from the protective antigen (PA) of Bacillus anthracis to detect circulating T cells in peripheral blood of subjects vaccinated with an anthrax vaccine. PA-specific HLA class II-restricted T lymphocytes were isolated which displayed both TH1- and TH2-like characteristics, indicating heterogeneity of the lymphocyte lineage within the CD4+ response. Presentation of antigen to these T-cell clones by HLA-matched antigen-presenting cells exposed to the intact PA protein confirmed that the identified epitopes are indeed naturally processed by the human immune system. Specific tetramer-derived T-cell profiling may be useful for monitoring helper CD4+ T-cell responses to anthrax vaccination.
Bacillus anthracis is a gram-positive nonmotile rod-shaped spore-forming bacterium found in soil throughout the world. Cutaneous, gastrointestinal, or inhalational infection of Bacillus anthracis causes three different forms of the disease anthrax. Occurring most commonly in animals, anthrax is rare in humans and was contracted primarily by the handling of infected animals or animal products, until its development as a biological weapon. The anthrax vaccine (anthrax vaccine absorbed [AVA]) is a cell-free filtrate of Bacillus anthracis containing protective antigen (PA) as the principal immunogen, and numerous efforts are under way to modify or replace this vaccine with improved or PA-specific alternatives. We describe a general approach for identifying CD4+ T-cell epitopes associated with immune responses to the PA of Bacillus anthracis, using soluble peptide-major histocompatibility complex (pMHC) tetramers and T-cell cloning. Current immune assays for monitoring anthrax exposure or anthrax vaccine efficacy utilize antibody responses, with no reliable or standard method for the T-lymphocyte profile. Nevertheless, for improved vaccine efficacy, it is important to optimize T-cell help, and assays to directly measure the helper T compartment are needed (1).
MHC proteins bind peptide antigens for presentation to T lymphocytes, enabling the development of technologies using soluble pMHC for analysis of T-cell specificity. Soluble pMHC class I tetramers have been used extensively for studies of CD8+ T cells and are particularly informative for evaluating the class I-restricted response to viral infection and viral vaccines (5, 9, 10, 21). This utility reflects the importance of lysis of virus-infected host cells by CD8+ cytolytic T cells for protective cellular immune responses to most viruses. In contrast, bacterial infections are cleared predominantly by other immune mechanisms, including a role for the CD4+ T-cell compartment generating cytokines and providing T-cell help for the antibody response. In order to efficiently monitor this latter pathway, various lymphocyte activation assays have been used, often involving the activation or proliferation of CD4+ T cells in vitro as an indirect measure of antigen-specific responses. With the development of pMHC class II tetramers, it is now possible to directly assay the CD4+ T-cell population for antigen-specific phenotypes (2, 3, 7, 12-14, 22).
Routine use of pMHC class II tetramers is challenged by two important biological and technical issues: first, antigen-specific CD4+ T cells are present in the peripheral blood at a very low frequency, generally between 1:3,000 and 1:30,000 cells; second, the avidity of T-cell receptor (TCR)-pMHC recognition tends to be heterogeneous, with functionally relevant cells often showing low-avidity recognition properties. In our previous studies of class II-restricted CD4+ T-cell responses to antigens associated with autoimmune diseases, we developed a protocol to compensate for these challenges, in which peripheral blood lymphocytes are exposed to class II-restricted peptides, followed by selection of cells using flow cytometry to isolate the rare cells which bind specific pMHC tetramers (16, 17). Cloning of the tetramer-positive lymphocytes sorted by flow cytometry provides a population of expanded antigen-specific T cells suitable for further analysis. We have now used this antigen-MHC-directed system to identify and characterize CD4+ T cells responding to PA.
Epitopes on PA that are recognized by neutralizing antibodies have been defined (reviewed in reference 11), but factors affecting the development of antigen-specific T-cell-dependent help for the production of anti-PA antibodies and memory B cells, and the T-cell epitopes that are recognized by these helper T cells, are uncharacterized. Humans immunized with AVA develop antibodies and a detectable proliferative response to PA, but the magnitude of the proliferative response is relatively low (4). These findings, and the need for annual boosters to maintain antibodies, suggest that the antigen-specific memory B-cell and T-cell responses to Bacillus anthracis protective antigen are relatively weak. Defining these responses and determining if enhancement of T-cell immunity can also improve efficacy against Bacillus anthracis infections could lead to improved vaccines.
MATERIALS AND METHODS
Peptide binding assays.
Competitive binding assays were used to identify class II-binding epitopes from PA. All peptides used in this work were synthesized on an Applied Biosystems 432A peptide synthesizer (Foster City, CA). As previously described for studies of other antigens (6, 18, 20), purified soluble HLA class II (50 nM) was incubated with 0.001 to 10 μM nonbiotinylated PA peptides of interest, as well as a known positive control peptide, in binding buffer (1 mM PefaBloc, 0.75% n-octyl-β-d-glucopyranoside [OG], 150 mM citrate-phosphate, pH 5.4) for 1 h at 37°C. A biotinylated competitor peptide was then added and incubated overnight at 4°C. For each different HLA class II molecule, the competitor was a standard reference peptide unrelated to PA, and the optimal concentration was determined in a previous binding assay. The biotinylated competitor peptides used were as follows. For HLA DRB1*0101, DRB1*0405, DRB1*0701, and DRB1*1101, hemagglutinin peptide sequence 306 to 318 (HA 306-318) (0.1 μM) was used. The rubella virus strain M33 E1 protein sequence 254-266 (5 μM) was used for DRB1*0301, and glutamic acid decarboxylase 65 (Gad65) 555-567 (557I) (0.01 μM) was used for DRB1*0401. For DRB1*0404, DRB1*1501, and DRB1*1301, the competitor peptides were outer-surface protein A (OspA) 163-175 of Borrelia burgdorferi (1 μM), myelin basic protein (MBP) 84-102 (0.1 μM), and HA 306-318 (1 μM), respectively. The next day, the binding reaction was neutralized by an equal volume of 50 mM Tris (pH 8) containing 0.75% OG. The class II molecules were captured on a high-binding polypropylene flat-bottom plate (Corning, Corning, NY) using anti-class II antibodies (L243; ATCC, Manassas, VA) for 4 h at room temperature or overnight at 4°C. After plates were washed, europium-labeled streptavidin was added, and the plates were developed with europium activation buffer, using a Wallac Victor fluorometer (Perkin-Elmer, Downers Grove, IL). From the binding curves, the inhibitory concentration was calculated as the amount of nonbiotinylated peptide that reduced binding of the biotinylated standard by 50%.
Vaccination and sample collection.
Peripheral blood was obtained with informed consent from a normal volunteer laboratory worker (HLA DRB1*1302 DRB1*0407) who received conventional AVA (BioPort Corp., Lancing, MI) as prophylaxis while working in a high-risk laboratory facility. The individual received the full schedule of five subcutaneous immunizations and was given a booster within 2 years prior to sample collection.
In vitro expansion culture.
For studies of fresh blood, peripheral blood mononuclear cells (PBMC) were separated by gradient centrifugation (Lymphoprep; Nycomed, Oslo, Norway); for experiments with frozen PBMC, cells were thawed in 10% fetal bovine serum (FBS) with 20 U/ml DNase (Worthington Biochemical Corp., Lakewood, NJ). PBMCs (3.5 million) were cultured per well in a 24-well plate with pooled PA peptides (10 μg/ml each) and medium (10% pooled human serum) in RPMI medium containing l-glutamine and HEPES with 1 mM pyruvate, 0.01 U/ml penicillin, and 0.01 μg/ml streptomycin. Interleukin 2 (IL-2; 1-to-20 final dilution; Hemagen, Columbia, MD) was added on day 7, and medium was replenished between days 9 and 11. At day 13, the cultured PBMC were harvested, and tetramer analysis was performed.
Tetramer preparation.
The production of MHC class II tetramers is described elsewhere (14). Briefly, DRB1*0404 or DRB1*1302 monomers containing a biotinylation sequence at the 3′ end were generated in a Cu-inducible Drosophila melanogaster expression vector. The monomers were purified and biotinylated prior to peptide loading for 48 to 72 h at 37°C, after which the tetramers were assembled by the addition of phycoerythrin (PE)-labeled streptavidin.
Tetramer analysis.
Cells were washed in Dulbecco's phosphate-buffered saline (D-PBS) and resuspended in fresh medium at 2 to 6 million cells per ml for staining with PE-labeled DRB1*1302 or DRB1*0404 tetramers. PE-labeled tetramers (10 μg/ml) were added, and the samples were incubated for 2.5 h at 37°C. Fluorescein isothiocyanate (FITC)- or peridinin chlorophyll protein (PerCP)-labeled anti-CD4 was added for 30 min on ice. After samples were washed with D-PBS containing 1% FBS (HyClone, Logan, VT), the cells were analyzed using a Becton Dickinson FACSCalibur flow cytometer, and CellQuest (Becton Dickinson, Franklin Lakes, NJ) and FlowJo (Ashland) were used for data analysis. The numbers reported in the dot plots shown in Fig. 2 to Fig. 4 are percentages based on cells in the live lymphocyte gate, and the quadrants are based on single-color control samples.
FIG. 2.
Flow cytometry of PBMC from an AVA-vaccinated donor, using PE-labeled PA tetramers (vertical axis) and PerCP- or allophycocyanin-labeled anti-CD4 staining (horizontal axis). (A) Frozen PBMC were thawed and incubated in the presence of PA peptides. After 12 to 14 days of expansion, the sample was stained with DRB1*1302 PA tetramers and a negative control tetramer (DRB1*1302 mortalin10A; peptide sequence, AIKGAVVGIALG). (B) Fresh PBMC were cultured with PA peptides for 12 to 14 days and stained with the specific DRB1*0404 PA 112-127 tetramer or the negative control tetramer DRB1*0404 mortlin10A. The DRB1*0404 tetramers were added in an attempt to detect a response mediated through the DRB1*0407 haplotype of the patient, since DRB1*0407 tetramers were not available. The numbers reported are percentages based on cells in the live lymphocyte gate.
FIG. 4.
Characterization of the BRI4PA clones by tetramer staining and proliferation to PA 112-127. (A) BRI4PA clones were generated by single-cell sorting the tetramer-positive PA 112-127 population, followed by 12 to 14 days of mitogen stimulation and 12 to 14 days of antigen-specific stimulation. The clones were tetramer stained with DRB1*0404 mortalin10A, as a negative control, and the DRB1*0404 PA112-127 specific tetramer; subsequently, samples were stained with anti-CD4 monoclonal antibody and analyzed by flow cytometry. (B) BRI4PA clones were exposed to PA 112-127 in the presence of human DRB1*1302, DRB1*0404, and DRB1*0407 antigen-presenting cells (APC); [3H]thymidine incorporation after 72 h is shown. (C) BRI4PA.18 cells were labeled with CFSE prior to specific stimulation with 10 μg/ml PA 112-127 and irradiated DRB1*0407 PBMC. Six days after stimulation, the sample was tetramer stained with the same tetramers as described for panel A. The percentages reported for flow cytometry are based on all cells in the live lymphocyte gate. On the histogram plot, the black line is the CFSE profile of the culture containing PA 112-127, and the gray line is the profile of a culture without peptide.
T-cell sorting and cloning.
Tetramer-positive samples were then sorted using a FACSVantage cell sorter (Becton Dickinson) to enrich for tetramer-positive cells. Following sorting, T cells were expanded with two rounds of stimulation for 11 to 14 days, using 5 μg/ml phytohemagglutinin (Sigma, St. Louis, MO) and 10 U/ml IL-2 for the first round and, in some cases, specific antigen for the second round. If a specific stimulation was carried out, HLA DRB1-matched irradiated PBMC were used with 10 μg/ml peptide. Wells with positive growth were then evaluated for T-cell function.
Proliferation assay.
To confirm peptide specificity and evaluate cytokine profiles produced in response to antigen, T-cell clones were evaluated using proliferation assays. Clones were harvested from days 11 to 14 poststimulation, washed with D-PBS, and resuspended in medium. Fifty thousand cells were added per well in a round-bottom 96-well plate. PBMC matched with the tetramer for HLA type were used as antigen-presenting cells, pulsed with antigen (at least 1 h for peptide and at least 4 hours for whole protein) at 37°C prior to irradiation with 5,000 rads. Recombinant PA was provided by NIAID's BEI Resources program (Manassas, VA). Antigen-pulsed presenting cells (100,000 per well) were added, and supernatants were harvested at 48 h for a cytokine assay (Cytometric Bead Array; BD Biosciences, San Jose, CA). [3H]thymidine was added to the wells, and proliferation was measured 20 to 24 h later by scintillation counting.
Magnetic bead separation for CD14+ cells.
PBMC were washed and suspended in separation buffer (0.5% bovine serum albumin [Sigma, St. Louis, MO], 2 mM EDTA [Sigma, St. Louis, MO] in D-PBS [pH 7.2]) at 125 million cells per ml. Cells were incubated with CD14 Microbeads (Miltenyi Biotech, Auburn, CA) at 20 μl per 10 million cells for 15 min at 4 to 8°C. The labeled cells were separated over a magnetic LS column (Miltenyi Biotech, Auburn, CA). All fractions were collected and analyzed for purity by flow cytometry using FITC-labeled CD19, PE-labeled CD14, allophycocyanin-labeled CD3, and PerCP-labeled CD4 antibodies.
Carboxyfluorescein diacetate succinimidyl ester staining.
For analysis of proliferation by flow cytometry, T-cell clones were washed two times with PBS, followed by incubation with 200 nM carboxyfluorescein diacetate succinimidyl ester (CFSE) (Invitrogen, Carlsbad, CA) for 10 min at 37 oC. FBS was added for 2 min, and the cells were washed once with D-PBS and two more times with medium. CFSE-labeled cells (250,000) were then cultured in a 48-well plate in the presence of irradiated PBMC with 10 μg/ml PA peptide. Seven days later, the clones were tetramer and anti-CD4 stained as described above.
RESULTS
Selection of PA peptides.
Two strategies were used to select PA antigenic epitopes. First, nine HLA class II molecules were studied in peptide binding assays, in order to identify candidate PA peptide epitopes suitable as potential antigens for T-cell recognition within a genetically diverse population. The sequence of the PA protein was interrogated using MHC class II-binding predictive algorithms (19), followed by synthesis of the putative epitopes and evaluation by a competitive peptide-binding assay. Five peptides plus a positive control were tested for binding to nine different HLA DR molecules (Table 1). The strongest binding interactions occurred between PA peptides 373-393 and the HLA DR1, DR7, and DR13 molecules and between PA peptides 595-605 and the HLA DR4 molecules. At least one PA peptide sequence was identified which bound each HLA DRB1 type tested, except for HLA DRB1*0405. The PA 373-393 sequence encompasses two potential HLA binding epitopes, which were each tested separately using PA 374-385 and PA 381-392. In most cases, the PA 373-393 displayed better binding than the two shorter segments. An example of these competitive binding determinations is shown in Fig. 1 for peptide interactions with HLA DR13 molecules. As a second strategy, a panel of 16-mer overlapping peptides from the PA protein was tested for stimulation of PBMC in vitro; one peptide sequence, PA 112-127, was identified with cells from an HLA DRB1*0407-positive donor.
TABLE 1.
IC50 of PA peptide binding to HLA class II moleculesa
| Peptideb | Result for HLA DRB1 molecule tested
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| DR*0101 | DR*0301 | DR*0401 | DR*0404 | DR*0405 | DR*0701 | DR*1101 | DR*1302 | DR*1501 | |
| PA 112-127 | N | N | N | — | — | — | — | N | — |
| PA 373-393 | 0.25 | 8 | N | — | — | 0.1 | >10 | 0.2 | — |
| PA 374-385 | 5 | N | N | 6.6 | N | 0.08 | N | N | N |
| PA 382-392 | N | N | N | 6.4 | N | N | >10 | 2 | 1.8 |
| PA 579-591 | — | 5 | N | — | — | — | 5 | >10 | — |
| PA 595-605 | N | N | 0.76 | 5 | N | N | N | N | N |
| Positive control | 0.25 | >10 | 0.5 | 0.01 | 1 | 10 | 1 | 10 | 0.225 |
| Positive control peptide | HA | Rubella | HA | Gad5571 | HA | HA | HA | HA | MBP |
Values for 50% inhibitory concentration (IC50) are given in μM peptide. N, no binding; —, no suitable motif/not tested.
Peptide sequences: PA 112-127, RLYQIKIQYQRENPTE; PA 373-393, PIYNVLPTTSLVLGKNQTLAT; PA 374-385, IYNVLPTTSLVL; PA 381-392, SLVLGKNQTLAT; PA 579-591, DKIKLNAKMNILI; PA 595-605, RFKYDRNNIAV; HA 306- 318, PKYVKQNTLKLAT; Rubella 254-266, RLRLVDADDPLLR; Gad65 555-567 (557I), NFIRMVISNPAAT; MBP 87-96, VHFFKNIVTP.
FIG. 1.
Competition binding of PA peptides to HLA DRB1*1302. Six PA peptides and a positive control (HA 306-317) were each incubated with purified recombinant HLA DRB1*1302 at the concentration shown and then challenged with 0.1 μM biotinylated HA 306-318. The DRB1 molecules were captured with anti-MHC monoclonal antibodies on an enzyme-linked immunosorbent assay plate, followed by the addition of europium-labeled streptavidin, and developed using europium activation buffer.
Class II pMHC tetramers identify antigen-specific CD4+ T cells following vaccination with AVA.
PBMC from an HLA DRB1*0407 DRB1*1302 heterozygous individual recently vaccinated with AVA were tested for response to the panel of putative PA peptide epitopes. After stimulation in vitro with the PA peptides, lymphocytes were stained with either DR4 or DR13 tetramers loaded with each of the candidate epitopes. Flow cytometry profiles of the PE-labeled tetramer binding are shown in Fig. 2 for the DR13 PA 381-392 and PA 373-393 tetramers (Fig. 2A) and for the DR4 PA 112-127 tetramer (Fig. 2B). In each case, control staining of the same cell sample using an HLA-matched but peptide-mismatched tetramer is also shown.
Very-low-level staining with the DR13 tetramer was observed, with the PA 381-392 tetramer binding only slightly higher than the background binding seen with the control tetramer. Reactivity with the DR4 tetramer was seen by using PA 112-127, with more tetramer-positive cells, particularly in the CD4 population. These profiles were suggestive of low-level (i.e., low-frequency) T-cell responses to the specific PA peptides, so each putative tetramer-stained sample was subjected to cell sorting using a high-speed flow cytometer, followed by expansion and analysis of the tetramer-binding population.
Figure 3 summarizes the findings after cells were sorted within the 1.26% positive-stained population using the DRB1*1302 PA 381-392 tetramer. First, the positive tetramer-sorted population was expanded using mitogen stimulation and then reanalyzed (Fig. 3A). Readily apparent tetramer staining with the DRB1*1302 PA 381-392 tetramer as well as with the longer, overlapping peptide PA 373-393 was seen, verifying the enrichment of the previously rare antigen-specific population. From the tetramer-positive population, a number of clones were then generated by several rounds of antigen-specific stimulation of single cells, in which clones BRI13PA.66 and BRI13PA.155 were representative examples. The specificity of these clones was confirmed by antigen-specific proliferation using human DRB1*1302 PBMC as antigen-presenting cells (Fig. 3B) and by specific PA tetramer binding (Fig. 3C). Interestingly, both clones recognized both PA 381-392 and PA 373-393, although there were subtle differences between the proliferative and the tetramer binding results: in a proliferation assay, the BRI13PA clones responded more to PA 373-393 than to PA 381-392, while the tetramer staining intensities with these long and short peptides were fairly equal.
FIG. 3.
Characterization of the DRB1*1302 PA-positive tetramer population, BRI13PA cells, by tetramer staining and proliferation assay. (A) Tetramer staining of the bulk-sorted DRB1*1302 PA 381-392 sample after expansion with mitogen stimulation. The DRB1*1302 tetramers individually loaded with mortalin10A (Negative Control peptide) or PA peptides were incubated with cells for 2.5 h at 37°C, followed by CD4 staining for 30 min on ice. (B) BRI13PA clones proliferated to specific PA peptides in the presence of irradiated DRB1*1302 PBMC. [3H]thymidine was added 48 h after antigen and detected by scintillation counting 20 to 24 h later. Greater proliferation to the longer PA 373-393 peptide sequence correlates with the relative strength of peptide binding. (C) BRI13PA clones were stained with a negative control tetramer (DRB1*1302 mortalin10A; peptide sequence, AIKGAVVGIALG), DRB1*1302 PA 373-393, and DRB1*1302 PA 381-392 and analyzed by flow cytometry. The flow cytometry plots show cells in the live lymphocyte gate.
CD4+ T-cell clones were also generated using the DRB1*0404 PA 112-127 tetramer by single-cell sorting the 2.59% tetramer-positive population from the initial PBMC sample. After two rounds of expansion in culture, the clones were evaluated for specificity. Figure 4A shows the positive DRB1*0404 PA 112-127 tetramer staining of four representative examples, referred to as the BRI4PA clones. Proliferation assays were performed using both DRB1*0407 and DRB1*0404 human antigen-presenting cells, since the patient HLA genotype is DRB1*0407. The BRI4PA clones responded to PA 112-127 in the context of both DRB1*0404 and DRB1*0407 but not with DRB1*1302, demonstrating promiscuity in DRB1*04 subtype recognition but not extending to DRB1*1302 (Fig. 4B). Direct confirmation that these clones both bind tetramers and proliferate to PA 112-127 is shown in Fig. 4C, in which the BRI4PA.18 clone was CFSE labeled and then stimulated with PA 112-127 loaded on DRB1*0407 PBMC for 6 days. Tetramer staining shows that all BRI4PA.18 cells are tetramer positive and respond to PA 112-127 through CFSE dilution.
Characterization of the PA-specific CD4+ T-cell response.
Supernatants from the proliferation assays whose results are shown in Fig. 3B and Fig. 4B were analyzed for the production of the cytokines gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), IL-10, IL-5, IL-4, and IL-2. All BRI13PA clones made a large amount of IFN-γ and a small amount of TNF-α, requiring a fairly high peptide concentration for stimulation. An example is shown in Fig. 5 for one of these clones, reflecting a characteristic TH1 phenotype. In contrast, the BRI4PA clones had a TH2 cytokine profile. Large amounts of IL-5 were detected at the high peptide concentrations for all the BRI4PA clones, along with low levels of IL-4 (Fig. 5). The same cytokine profiles were seen when antigen was presented with the DRB1*0404 and DRB1*0407 antigen-presenting cells.
FIG. 5.
Cytokines produced by the BRI13PA and BRI4PA clones. Supernatants from every condition and clone shown in Fig. 3B and Fig. 4B were tested for a panel of T-cell-derived cytokines using the BD Biosciences Cytometric Bead Array. Each set of clones produced similar cytokines, and the two graphs shown are representative examples. APC, antigen-presenting cell.
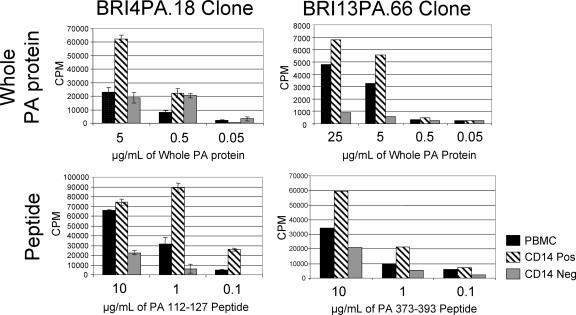
The PA epitope specificities of these clones were defined by synthetic peptides corresponding to MHC-binding regions of the PA protein. To investigate the likelihood that these epitopes actually represent naturally processed fragments of PA, we incubated full-length recombinant PA protein with HLA DRB1*0407 or DRB1*1302 antigen-presenting cells and subsequently tested the BRI4PA and BRI13PA T-cell clones for activation, with no exposure to individual peptides. As shown in Fig. 6, efficient presentation was observed for all cases for antigen presentation by unfractionated PBMC, by CD14+ cells (predominantly monocytic), and by CD14− cells (enriched for B cells). Using the CD19 antibody as a B-cell marker, the B-cell enrichment of the CD14− fraction was verified by flow cytometry (data not shown).
FIG. 6.
Proliferative response of PA-specific clones presented with whole-PA protein by HLA DRB1*1302 or DRB1*0407 antigen-presenting cells. PBMC, CD14+ PBMC, and CD14− PBMC, separated with magnetic beads, were pulsed with PA protein antigen for at least 4 hours prior to irradiation. The HLA DRB1-matched antigen-presenting cells were added at a 1:1 volume with 50,000 cloned T cells of each specificity. At 48 h, [3H]thymidine was added, and incorporation was measured by scintillation counting 20 to 24 h later. Top panels, recombinant PA; bottom panels, synthetic peptides.
DISCUSSION
A major component of the antibody-mediated immune response to B. anthracis and to the anthrax vaccine is directed to PA, making this protein a useful target for diagnostic monitoring of infection and vaccine efficacy. T-cell help for antibody production comes largely from the CD4+ subset of T lymphocytes, which derive their specificity from recognizing peptides from the PA protein bound and presented by the MHC class II molecules of the host. It is therefore useful to consider the specificity of this T-cell helper compartment and the type of T-cell response which is elicited on exposure to B. anthracis or vaccine. We utilized a direct immunodiagnostic approach, utilizing fluorescently labeled soluble peptide-MHC tetrameric ligands, which were successful in identifying antigen-specific CD4+ T cells from peripheral blood following anthrax immunization. Epitopes derived from PA were recognized by HLA and peptide-specific T lymphocytes, which displayed both TH1- and TH2-like characteristics, indicating heterogeneity of the lymphocyte lineage within the CD4+ response. These differences in CD4+ response might be important in determining the potency of antibody responses to epitope-based vaccines. Cloned T cells maintained their antigen specificity, thus defining several peptides which represent potential helper epitopes for the immune response to B. anthracis PA. Presentation of antigen to these clones by HLA-matched antigen-presenting cells exposed to the intact PA protein confirmed that the epitopes which were identified are indeed naturally processed by the human immune system.
The strategy illustrated in this study represents a general approach to epitope identification for vaccine and infectious disease immune profiling. The most direct way to evaluate a T-cell response which occurs at low frequency in blood is to directly interrogate individual T cells within the large pool of multispecific circulating lymphocytes. The tetramer-based method described here has several differences compared to other assays, such as enzyme-linked immunospot assay and limiting-dilution cloning, which have been successfully used in the past. First, the use of computer-assisted predictive algorithms, followed by analysis of MHC binding, facilitates the analysis of large, complex proteins or proteomes, by generating a panel of potential epitopes prior to the more laborious functional cellular studies. Second, the primary outcome measure is the binding properties of the T cell itself, thereby providing a direct measurement, rather than an indirect readout potentially sensitive to bystander effects. Third, the method directly generates antigen-specific human T-cell clones, which can then be used for further studies of lineage and function.
A commitment has been made in the United States to vaccinate large numbers of persons against anthrax. However, the current AVA regimen is administered as six subcutaneous injections and requires annual boosters. Passive protection studies of animals and some epidemiological data from workers in at-risk industries indicate that antibodies to PA correlate with immunity to anthrax. However, very little is known about the T-cell response, particularly in humans. An Institute of Medicine report in 2002 recommended that efforts be made to develop immune correlates of vaccine response, which could be used to guide next-generation vaccine development (8). Anthrax is well suited to the use of a tetramer-based monitoring strategy, since PA is known to be the major immunogenic component of AVA, and it is highly conserved in sequence among all reported anthrax strains (15). Sequence conservation is important, since antigenic variation due to selective pressure might defeat peptide-based strategies for immune profiling or protection. Our initial panel of potential epitopes corresponds to HLA genotypes carried by the majority of the population, and the examples of successful tetramer-derived T-cell profiling presented in this study demonstrate the feasibility of this approach.
Acknowledgments
This work was supported by grant AI059798 and a SERCEB grant from the National Institutes of Health.
Recombinant Bacillus anthracis protective antigen protein was provided by the Biodefense and Emerging Infections Research Resources Repository.
We thank K. Arumuganatha for expert assistance in cell sorting; Vivian Gersuk and the BRI Clinical Core Laboratory for HLA typing; Jason Berger for peptide synthesis; Tuan Nguyen, Kelly Geubtner, and Sharon Kochik for technical assistance; and Janice Abbas and Ellen Corke for preparation of the manuscript.
Editor: D. L. Burns
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Allen, J. S., A. Skowera, G. J. Rubin, S. Wessely, and M. Peakman. 2006. Long-lasting T cell responses to biological warfare vaccines in human vaccinees. Clin. Infect. Dis. 43:1-7. [DOI] [PubMed] [Google Scholar]
- 2.Buckner, J. H., U. Holzer, E. J. Novak, H. Reijonen, W. W. Kwok, and G. T. Nepom. 2002. Defining antigen-specific responses with human MHC class II tetramers. J. Allergy Clin. Immunol. 110:199-208. [DOI] [PubMed] [Google Scholar]
- 3.Day, C. L., N. P. Seth, M. Lucas, H. Appel, L. Gauthier, G. M. Lauer, G. K. Robbins, Z. M. Szczepiorkowski, D. R. Casson, R. T. Chung, S. Bell, G. Harcourt, B. D. Walker, P. Klenerman, and K. W. Wucherpfennig. 2003. Ex vivo analysis of human memory CD4 T cells specific for hepatitis C virus using MHC class II tetramers. J. Clin. Investig. 112:831-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Everson, M. P., K. Shi, P. Aldridge, A. A. Bartolucci, and W. D. Blackburn. 2002. Immunological responses are not abnormal in symptomatic Gulf War veterans. Ann. N. Y. Acad. Sci. 966:327-342. [DOI] [PubMed] [Google Scholar]
- 5.Gupta, M., P. Greer, S. Mahanty, W. J. Shieh, S. R. Zaki, R. Ahmed, and P. E. Rollin. 2005. CD8-mediated protection against Ebola virus infection is perforin dependent. J. Immunol. 174:4198-4202. [DOI] [PubMed] [Google Scholar]
- 6.Hill, C. M., A. Liu, K. W. Marshall, J. Mayer, B. Jorgensen, B. Yuan, R. M. Cubbon, E. A. Nichols, L. S. Wicker, and J. B. Rothbard. 1994. Exploration of requirements for peptide binding to HLA DRB1*0101 and DRB1*0401. J. Immunol. 152:2890-2898. [PubMed] [Google Scholar]
- 7.Holzer, U., W. W. Kwok, G. T. Nepom, and J. H. Buckner. 2003. Differential antigen sensitivity and costimulatory requirements in human Th1 and Th2 antigen-specific CD4+ cells with similar TCR avidity. J. Immunol. 170:1218-1223. [DOI] [PubMed] [Google Scholar]
- 8.Institute of Medicine. 2002. The anthrax vaccine. Is it safe? Does it work? Summary of the Committee to Assess the Safety and Efficacy of the Anthrax Vaccine. National Academies Press, Washington, DC.
- 9.Krebs, P., E. Scandella, B. Odermatt, and B. Ludewig. 2005. Rapid functional exhaustion and deletion of CTL following immunization with recombinant adenovirus. J. Immunol. 174:4559-4566. [DOI] [PubMed] [Google Scholar]
- 10. Kuzushima, K., N. Hayashi, A. Kudoh, Y. Akatsuka, K. Tsujimura, Y. Morishima, and T. Tsurumi. 2003. Tetramer-assisted identification and characterization of epitopes recognized by HLA A*2402-restricted Epstein-Barr virus-specific CD8+ T cells. Blood 101:1460-1468. [DOI] [PubMed] [Google Scholar]
- 11.Leppla, S. H., J. B. Robbins, R. Schneerson, and J. Shiloach. 2002. Development of an improved vaccine for anthrax. J. Clin. Investig. 110:141-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macaubas, C., J. Wahlstrom, A. P. Galvao da Silva, T. G. Forsthuber, G. Sonderstrup, W. W. Kwok, R. H. DeKruyff, and D. T. Umetsu. 2006. Allergen-specific MHC class II tetramer+ cells are detectable in allergic, but not in nonallergic, individuals. J. Immunol. 176:5069-5077. [DOI] [PubMed] [Google Scholar]
- 13.Mallone, R., and G. T. Nepom. 2004. MHC Class II tetramers and the pursuit of antigen-specific T cells: define, deviate, delete. Clin. Immunol. 110:232-242. [DOI] [PubMed] [Google Scholar]
- 14.Novak, E. J., A. W. Liu, G. T. Nepom, and W. W. Kwok. 1999. MHC class II tetramers identify peptide-specific human CD4(+) T cells proliferating in response to influenza A antigen. J. Clin. Investig. 104:R63-R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Price, L. B., M. Hugh-Jones, P. J. Jackson, and P. Keim. 1999. Genetic diversity in the protective antigen gene of Bacillus anthracis. J. Bacteriol. 181:2358-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reijonen, H., and W. W. Kwok. 2003. Use of HLA class II tetramers in tracking antigen-specific T cells and mapping T-cell epitopes. Methods 29:282-288. [DOI] [PubMed] [Google Scholar]
- 17.Reijonen, H., R. Mallone, A. K. Heninger, E. M. Laughlin, S. A. Kochik, B. Falk, W. W. Kwok, C. Greenbaum, and G. T. Nepom. 2004. GAD65-specific CD4+ T-cells with high antigen avidity are prevalent in peripheral blood of patients with type 1 diabetes. Diabetes 53:1987-1994. [DOI] [PubMed] [Google Scholar]
- 18. Steere, A. C., B. Falk, E. E. Drouin, L. A. Baxter-Lowe, J. Hammer, and G. T. Nepom. 2003. Binding of outer surface protein A and human lymphocyte function-associated antigen 1 peptides to HLA-DR molecules associated with antibiotic treatment-resistant Lyme arthritis. Arthritis Rheum. 48:534-540. [DOI] [PubMed] [Google Scholar]
- 19.Sturniolo, T., E. Bono, J. Ding, L. Raddrizzani, O. Tuereci, U. Sahin, M. Braxenthaler, F. Gallazzi, M. P. Protti, F. Sinigaglia, and J. Hammer. 1999. Generation of tissue-specific and promiscuous HLA ligand databases using DNA microarrays and virtual HLA class II matrices. Nat. Biotechnol. 17:555-561. [DOI] [PubMed] [Google Scholar]
- 20.Wicker, L. S., S. L. Chen, G. T. Nepom, J. F. Elliott, D. C. Freed, A. Bansal, S. Zheng, A. Herman, A. Lernmark, D. M. Zaller, L. B. Peterson, J. B. Rothbard, R. Cummings, and P. J. Whiteley. 1996. Naturally processed T cell epitopes from human glutamic acid decarboxylase identified using mice transgenic for the type 1 diabetes-associated human MHC class II allele, DRB1*0401. J. Clin. Investig. 98:2597-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu, Y., J. Zhang, S. Chen, A. Chen, L. Wang, J. Li, T. Zhao, L. Zou, Y. Tang, L. Tingrong, and F. Wang. 2004. Frequencies of epitope-specific cytotoxic T lymphocytes in active chronic viral hepatitis B infection by using MHC class I peptide tetramers. Immunol. Lett. 92:253-258. [DOI] [PubMed] [Google Scholar]
- 22.Yang, J., E. A. James, L. Huston, N. A. Danke, A. W. Liu, and W. W. Kwok. 2006. Multiplex mapping of CD4 T cell epitopes using class II tetramers. Clin. Immunol. 120:21-32. [DOI] [PubMed] [Google Scholar]