Abstract
Mycoplasma arthritidis is a naturally occurring murine pathogen, and the disease model has been used extensively to understand inflammatory mechanisms. Recently, Triton X-114 extracts of a virulent strain of M. arthritidis were found to be more potent in activating macrophages than were those from an avirulent strain, suggesting a role in disease. Here, octyl glucoside extraction of cells was used to identify four distinct bioactive moieties, with molecular masses of ∼41, 37, 34, and 17 kDa. Their bioactivities were resistant to proteinase K but were destroyed by alkaline hydrolysis and oxidation. As for MALP-2, all were dependent upon Toll-like receptor 2, but unlike MALP-2, they were also dependent upon CD14. The M. arthritidis lipoproteins exhibited infrared absorbances at 2,900 cm−1 and 1,662 cm−1, similar to those seen in Pam3-Cys-Ser-(Lys)4. Edman degradation failed to reveal N-terminal sequences, suggesting that they were blocked and therefore might be triacylated. However, mass spectrometry of fragments revealed that the 41-kDa moiety, which binds to serum apolipoprotein A-1, had similarity with the recently described MlpD lipoprotein of M. arthritidis.
Mycoplasmas, or mollicutes, are the smallest self-replicating prokaryotes and are common parasites of humans, animals, and plants. Mycoplasma arthritidis infections in rodents serve as animal models for a chronic arthritis that closely resembles human rheumatoid arthritis, necrotizing fasciitis-like syndrome, and acute toxic shock syndrome (6). Virulence factors include a superantigen (SAg), the M. arthritidis mitogen MAM (2), which utilizes Toll-like receptor 2 (TLR2) and TLR4 (14); two membrane lipoproteins, Maa1 (32) and Maa2 (34); and bacteriophage MAV1 (28). MAM is a potent activator of T cells, B cells, and macrophages (2). Although MAM can induce arthritis when injected directly into rat joints, systemic administration into rodents is without significant clinical effects. However, major changes in immune function do occur after intravenous injection of MAM (15), and MAM can trigger and exacerbate autoimmune collagen-induced arthritis (3). The inability of MAM to cause overt clinical disease by itself supports the notion that other virulence factors participate in the inflammatory effects of whole organisms.
We recently reported that Triton X-114 extracts of whole cells of a virulent strain of M. arthritidis contained components that activated macrophages through TLR2 and caused dendritic cell maturation, as evidenced by upregulation of surface expression of major histocompatibility complex class II, CD40, B7-1, and B7-2 (4). These activities were not due to contamination with MAM or lipopolysaccharide (LPS) and were significantly less potent in extracts from an avirulent strain of M. arthritidis, suggesting an association with disease. Other mycoplasmas have also been shown to possess macrophage-activating components, including MALP-2 (16), a lipopeptide derivative of MALP-404 from M. fermentans; fibroblast- and macrophage-stimulating lipopeptide FSL-1 (23) from M. salivarium; and a partially purified 29-kDa lipoprotein from M. hominis (19). The role of these potential inflammatory molecules in clinical disease remains to be defined in part because of the absence of a suitable animal model. However, such a model is available for M. arthritidis (2, 5); prior to initiating these studies, the first goal would be to purify and characterize the bioactive components of this organism. In the present study, we purified four macrophage-activating components from M. arthritidis, the activities of which are all TLR2 and CD14 dependent. We also show that one, OGex A (see below), which has binding avidity for serum apolipoprotein A-1 (9), is related to the MlpD, MlpE, and MlpF lipoproteins of unknown function, recently described by Washburn et al. (31, 35).
MATERIALS AND METHODS
Organism and culture conditions.
Mycoplasma arthritidis 158p10p9 (8) was grown in modified Edward medium (7) consisting of PPLO broth supplemented with 15% (vol/vol) heat-inactivated horse serum, 1.5% yeast extract (vol/vol) (Invitrogen Corp.), 0.25% (wt/vol) l-arginine HCl, 0.001% (wt/vol) NAD, and 500 U of penicillin G per ml. To adapt the organisms to a serum-free medium, the serum content was gradually reduced to 0% and was replaced with 3% (wt/vol) bovine serum albumin and 0.4% (vol/vol) of a 250× cholesterol lipid concentrate (Invitrogen) based on that previously described (30). Cultures of M. arthritidis grew up to 2 × 108 CFU/ml after 48 to 72 h of incubation in serum-free broth, i.e., ∼10-fold less than in Edward medium. The organisms were harvested by centrifugation at 27,000 × g for 30 min, washed three times with normal saline (NS), concentrated 100-fold, and frozen at −70°C until use.
Chemicals, enzymes, and antibodies.
Pam3-Cys-Ser-(Lys)4 (Pam3CSK4), a synthetic bacterial lipopeptide analogue from Escherichia coli (1); MALP-2, a 2-kDa synthetic lipopeptide from M. fermentans; and LPS from E. coli R515 were purchased from Alexis Biochemicals (San Diego, CA). Endotoxin-free NS was from Baxter Healthcare (Dearfield, IL). Polymyxin B, n-octyl-β-glucopyranoside (OG), and proteinase K PK-5056 (PK) were from Sigma (St. Louis, MO). Monoclonal antibodies (MAb) against the Maa1 or Maa2 lipoproteins from M. arthritidis were prepared as described previously (29). Horseradish peroxidase-conjugated anti-mouse immunoglobulin G was from eBioscience (San Diego, CA). Fluorescein isothiocyanate-conjugated anti-human CD25 MAb (clone M-A251) and purified rat anti-mouse CD14 MAb (clone 4C1/CD14) were from BD Biosciences (San Jose, CA).
Mouse antisera were prepared against five recombinant M. arthritidis lipoproteins, MlpA, MlpC, MlpD, MlpE, and MlpF (31, 35). Mlp genes, minus their signal peptide-encoding regions, were amplified by PCR with primers placing restriction sites at 5′ and 3′ ends. PCR primer sequences for Mlp genes are listed in Table 1. Amplification conditions were maintained as described previously (34). Amplicons were inserted into the appropriate pRSET vectors (Invitrogen); recombinant proteins were expressed in E. coli BL21(DE3)/pLysS and purified as described by the manufacturer (Invitrogen). Protein emulsions in incomplete Freund's adjuvant were injected subcutaneously at ∼50 μg into each of five male BALB/c mice. Mice were boosted 2 weeks later by intraperitoneal injection of 50 μg in phosphate-buffered saline. Antibody was monitored by enzyme-linked immunosorbent assay (ELISA). Pooled sera were collected from each group.
TABLE 1.
PCR primers for mlp gene amplification
| Primera | Sequence (5′-3′)b | Reference or GenBank accession no. |
|---|---|---|
| mlpA-f | CTCggatccGCGATAATACCGAGAAAAAGCC | Unpublished |
| mlpA-r | CAGgaattcTTTCTTGCTAAAGAACCTTG | AF154924 (OrfA) |
| mlpC-f | CAGggatccGATAATACTGATAAAAAACTAG | 31 |
| mlpC-r | CTGgaattcCATTGCTATTTTTGCTTGTTTCTCC | 31 |
| mlpD-f | CGCcagctgATAATACTAATAAAAAACTAGAAGAACC | 31 |
| mlpD-r | CTATTTGGaagcttCTATAGC | 31 |
| mlpEF-fb | CGGctgcagATAATACATATAAAAAACC | AY544183 |
| mlpE-r | GGCggtaccCTATTTTGAAGCTTCTTCGGC | AY544183 |
| mlpF-r | GGCgaattcTTATTGTTTAAAATTCTTTAAC | DQ451673 |
f, forward; r, reverse. The same forward primer was used for both mlpE and mlpF.
Restriction sites added by PCR (except for mlpD-r) for cloning into expression vectors are italicized and in lowercase. The HindIII site in mlpD-r occurs naturally at the 3′ end of mlpD.
Mice.
Female C57BL/6 and C57BL/6 CD14−/− knockout (KO) mice were purchased from Jackson Laboratories (Bar Harbor, ME); C57BL/6 TLR2−/− KO mice were from Thomas Hawn (University of Washington School of Medicine, Seattle, WA), courtesy of Shizuo Akira (Osaka University, Japan). All KO mice were bred in the animal care facility of the University of Utah under specific-pathogen-free conditions in compliance with the Animal Welfare Act and were used at 8 to 12 weeks of age. BALB/c mice for production of anti-Mlp antisera were purchased from Harlan Sprague-Dawley (Indianapolis, IN) and housed in the University of South Dakota animal care facility.
Cell culturing for cytokine induction and ELISA.
Mouse peritoneal adherent cells and mouse macrophage RAW 264.7 lines were prepared as described previously (15). Levels of tumor necrosis factor alpha (TNF-α), interleukin-6, and/or interleukin-12 p40 in culture supernatants were determined by using ELISA kits purchased from eBioscience (San Diego, CA) or BD Biosciences. For each experiment, triplicate cell suspensions were treated with each agonist or control prior to ELISA, and results are expressed as means ± standard deviations. CHO cells stably transfected with murine CD14 and ELAM-CD25 and with TLR2 were provided by D. Golenbock (University of Massachusetts Medical Center, Worcester, MA), courtesy of Janis Weiss (University of Utah Health Sciences Center), and were used as described previously (14).
Isolation and purification of the bioactive components.
The lipoprotein fraction was extracted from M. arthritidis by OG as previously described (23, 24). Briefly, OG extracts containing (lipo)proteins (OGex) were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and were transferred to a 0.45-μm-pore-size nitrocellulose membrane (Bio-Rad, Hercules, CA), which was cut into 2-mm strips that were each dissolved in 1 ml dimethyl sulfoxide. The (lipo)protein-coated particles, formed by dropwise addition of 3 ml 0.05 M sodium carbonate buffer (pH 9.6) to the dissolved membrane, were washed three times with NS and tested for the ability to induce RAW 264.7 cells to secrete TNF-α. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of extracted strips was repeated, and the gel was stained with zinc (Bio-Rad); the bioactive (lipo)protein bands were excised, and the gel slices were eluted by Electro-Eluter (Bio-Rad). The resulting lipoproteins were precipitated and washed with ice-cold acetone; purity was confirmed by SDS-PAGE and silver staining.
Characterization of lipoproteins.
The effect of proteolytic digestion on bioactivity was determined by incubating lipoproteins at 1 μg/ml with 5 μg/ml PK (Sigma) for 1 h at 37°C followed by 100°C for 15 min to inactivate the enzyme. Determination of the susceptibility of lipoproteins to alkaline hydrolysis, indicating acyl groups, and to oxidation, indicating presence of thioesters, was performed as described previously (4). Determination of infrared (IR) absorbance spectra to identify bioactive groups was performed on dried fractions of each lipoprotein in KBr pellets by use of a Bruker model IFS 88 spectrometer. Flow cytometry to determine TLR-2 expression on transfected CHO cells exposed to agonists was carried out as previously described (4).
Partial amino acid sequences were examined by Edman degradation using lipoproteins blotted onto Immobilon polyvinylidene difluoride (PVDF) membranes that were analyzed by use of an ABI Procise sequencer (Applied Biosystems, Foster City, CA) or by mass spectrometry of tryptic digests using an ion trap mass spectrometer (LCQ-Decca; Thermo-Finnigan Corp., Mountain View, CA). DNA sequencing of genes encoding Mlp proteins was performed by the Iowa State University DNA Synthesis and Sequencing Facility.
Western and dot blot analyses were performed with PVDF membranes (Millipore, Bedford, MA), horseradish peroxidase-conjugated anti-mouse immunoglobulin G (eBioscience), and chemiluminescent substrate (Pierce).
RESULTS AND DISCUSSION
Isolation and purification of bioactive lipoproteins from M. arthritidis.
OGex from washed cells of the virulent 158p10p9 strain of M. arthritidis induced TNF-α production by murine RAW 264.7 macrophages in a dose-dependent manner, with a concentration of 0.2 to 1.0 μg/ml being optimal (Fig. 1A). The activity in the extracts was not due to contamination with endotoxin or the MAM SAg, as determined by methods previously described (4).
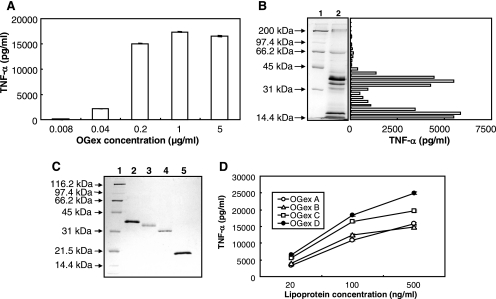
FIG. 1.
Purification of bioactive components from M. arthritidis. The following purification data are representative of repeated OG extractions on two separate batches of organisms. (A) Dose dependency of OGex induction of TNF-α in murine RAW 264.7 cells. (B) SDS-PAGE of OGex. Lane 1, protein standards; lane 2, OGex. The gel was blotted to nitrocellulose; 2-mm strips were excised, dissolved in dimethyl sulfoxide, and precipitated; and the (lipo)protein-coated particles were tested for TNF-α production in culture supernatants of RAW cells. (C) SDS-PAGE of purified lipoproteins stained with silver. Lane 1, protein standards; lane 2, OGex A; lane 3, OGex B; lane 4, OGex C; lane 5, OGex D. (D) Dose response of TNF-α production by purified OGex A, B, C, and D in RAW 264.7 cells.
SDS-PAGE of OGex revealed the presence of seven major bands (Fig. 1B). When the gel was blotted to a nitrocellulose membrane and strips therefrom extracted, the TNF-α-inducing bioactivity was found in two major areas, corresponding to approximately 42 to 32 kDa and 20 to 14 kDa, the latter comprising the dye front (Fig. 1B). Bands occurring in the regions of activity were identified by negative zinc staining and were extracted by electroelution. Repeat SDS gels and extraction revealed the presence of four homogeneous components that corresponded to molecular masses of ∼ 41, 37, 34, and 17 kDa (Fig. 1C), which we designated OGex A, B, C, and D, respectively. All four purified lipoproteins activated RAW 264.7 cells in a dose-dependent manner, with activity levels being similar (Fig. 1D).
Characterization of M. arthritidis lipoproteins.
Before characterizing these moieties in more detail, we first tested whether they might be related to the Maa1 and Maa2 M. arthritidis lipoprotein adhesins, virulence factors for disease caused by M. arthritidis (33). We concluded that there was no relationship since the molecular masses of Maa1 and Maa2 were markedly greater (86 and 56 kDa, respectively), and Western blotting of OGex A, B, C, and D individually with specific monoclonal antisera to Maa1 and Maa2 provided no evidence of cross-reactivity (data not shown). Also, peptide fragments of OGex did not match adhesin sequences in the MASCOT database (see below).
Since a number of studies have shown that the bioactivities of lipoproteins derived from mycoplasmas or other bacteria (10, 16, 23) are borne on small lipopeptides, we treated the M. arthritidis lipoproteins with PK or NS as a control. The protein bands of all moieties were totally digested by PK (Fig. 2A), and all of the bioactivity now migrated to the dye front on blotted and extracted gels (Fig. 2A). Importantly, PK treatment did not decrease the bioactivity of the lipoproteins compared with that seen for NS-treated lipoproteins (Fig. 2B).
FIG. 2.
Properties of OGex. (A) SDS-PAGE of OGex. Lane 1, protein standards; lane 2, OGex treated with PK. PK-treated OGex were blotted to cellulose nitrate strips, extracted, and tested for activity, all of which migrated to the dye front. (B) OGex were incubated with NS or with PK and, after being heated to 100°C for 15 min, were assayed for the ability to induce TNF-α in RAW cell cultures. (C) Effect of alkaline hydrolysis on the activity of 50 ng/ml of OGex A, B, C, and D, as indicated by the ability to stimulate TNF-α production by RAW 264.7 cells. (D) Effect of H2O2 treatment on bioactivity of OGex A, B, C, and D. (C and D) Tests for lipid groups were carried out on three separate cell suspensions stimulated with each of the treated OGex. Mean results ± standard deviations are shown.
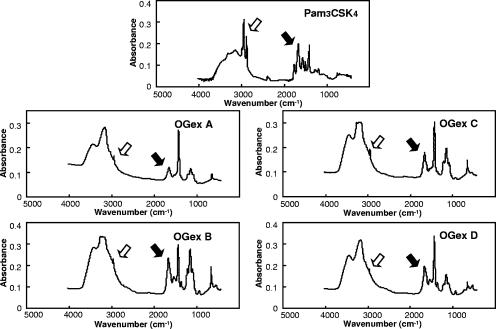
To confirm that the active moieties were associated with lipids, we demonstrated that all four purified components were susceptible to alkaline hydrolysis, indicating a role for fatty acid acyl groups (Fig. 2C), and were also susceptible to oxidation with H2O2, indicating involvement of thioesters (Fig. 2D). The presence of these bioactive lipid groups was confirmed by IR spectrometry by showing that, as for the bioactive synthetic Pam3CSK4 lipopeptide, the spectra of OGex A, B, C, and D exhibited absorbances at ∼1,700 cm−1 and ∼2,900 cm−1 (Fig. 3), again suggesting the presence of acyl chains and thioester bonds, respectively, that are characteristic of microbial lipoproteins (10). Interestingly, the overall profiles of the four lipoproteins were quite similar although not identical.
FIG. 3.
IR spectra of Pam3CSK4 and OGex A, B, C, and D. White arrows show the signals at about 2,900 cm−1, suggesting the presence of fatty acid acyl chains. Black arrows show the signals at about 1,700 cm−1, suggesting the presence of ester bonds.
Macrophage activation by M. arthritidis lipoproteins is dependent on TLR2 and CD14 coreceptors.
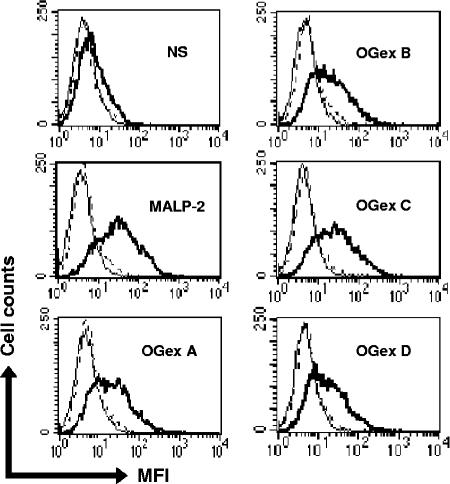
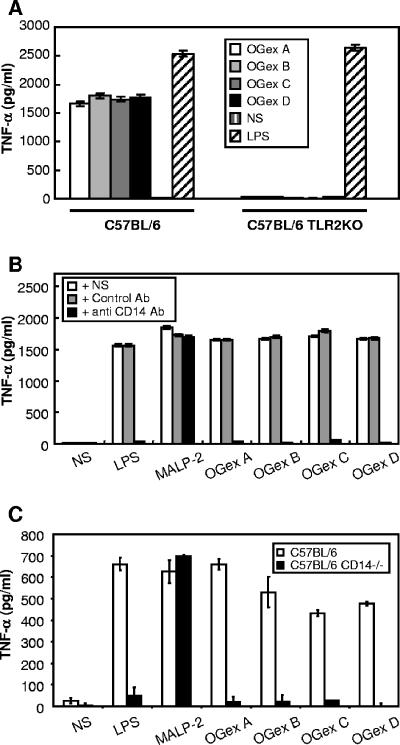
A wide range of bioactive bacterial lipoproteins are known to be dependent upon TLRs (11, 13, 25), and preliminary findings suggested that the bioactivity of Triton X-114 extracts of M. arthritidis might also be dependent upon TLR2 (4). To determine TLR usage by the purified lipoproteins of M. arthritidis, we tested their ability to up-regulate CD25 on CHO cells transfected with TLR2 and CD14, which acts as an indicator of cell activation of NF-κB through TLR2. CD25 expression on these CHO cells was up-regulated by MALP-2 as well as by all OGex moieties, indicating activation through TLR2 (Fig. 4). We also demonstrated that whereas peritoneal macrophages from wild-type C57BL/6 mice produced high levels of TNF-α, those from C57BL/6 TLR2−/− KO mice failed to produce significant amounts (P < 0.0005). In contrast, the levels of TNF-α induced by LPS, which utilizes TLR4, were not significantly different (P > 0.9) in macrophages from either TLR2+/+ or TLR2−/− mice (Fig. 5A).
FIG. 4.
Flow cytometric analysis of CHO/TLR2 cells. The cells were stained with fluorescein isothiocyanate-labeled anti-human CD25 MAb and analyzed by flow cytometry for the expression of the CD25 transgene, an indicator of TLR activation. The activation of cells was expressed by mean fluorescence intensity (MFI). Thin lines, no antibody; broken lines, isotype control antibody; thick lines, anti-human CD25 MAb.
FIG. 5.
(A) TNF-α production by peritoneal macrophages of C57BL/6 mice or C57BL/6 TLR2 KO mice stimulated with LPS, NS, or OGex A, B, C, or D. The concentration of LPS and OGex A, B, C, and D was 100 ng/ml. Peritoneal adherent cells were stimulated for 18 h with inducers, and the amounts of TNF-α in cell culture supernatants were tested by ELISA. The data are representative of two experiments using four to five mice each. The experiment was repeated twice. (B) RAW 264.7 cells were preincubated with NS, isotype control antibody (Ab), or anti-mouse CD14 MAb (10 μg/ml) for 1 h. Then, they were stimulated with LPS (10 ng/ml), MALP-2, or OGex A, B, C, or D (20 ng/ml) for 18 h. (C) Peritoneal macrophages from C57BL/6 or C57BL/6 CD14 KO mice were stimulated with 50 ng/ml LPS, MALP-2, or OGex A, B, C, or D, and culture supernatants were tested for the presence of TNF-α. Representative data from two similar experiments using four to five mice each are shown.
Toll-like receptors are now known to be key molecules in innate immunity, recognizing pathogen-associated molecular patterns on microbial as well as endogenous agonists (13, 26). Microbial lipoproteins, such as those from Treponema pallidum, OspA from Borrelia burgdorferi (22), possibly a lipoprotein of M. penetrans (24), and Pam3CSK4 (1), are triacylated. The triacylated lipoproteins possess two acyl fatty acids attached to the cysteine residue at the N terminus of the molecule through a sulfhydryl group and the third added to the free N terminus by acyl transferase during cleavage of the mature molecule from the signal peptide (20). In the case of MALP-2 from M. fermentans (16) and probably also the FSL lipopeptide from M. salivarium (23), the third acyl group is missing, due to an absence of acyl transferase. Characteristically, triacylated lipoproteins use TLR1 with TLR2 and diacylated lipoproteins use TLR6 with TLR2, although recent studies indicate that the N-terminal cysteine peptide region can also influence TLR usage (18, 27). In addition, at least one other coreceptor may be required for agonist recognition, since triacylated lipoproteins and LPS also require CD14 (12, 17, 22). The requirement for CD14 by diacylated lipoproteins has been more controversial (17, 21; P. F. Muhlradt, personal communication).
In view of the above-mentioned details, several approaches were used to determine the comparative usages of CD14 by OGex and MALP-2, using LPS as a positive control for CD14 usage. First, we examined the effect of anti-CD14 antibody on TNF-α induction by each of the agonists. In a preliminary dose-response experiment, we established that 10 μg/ml of anti-CD14 antibody was the lowest concentration that reduced TNF-α levels induced by OGex to background levels. Anti-CD14 antibody alone failed to influence spontaneous TNF-α release by cells in the absence of stimulants. However, antibody to CD14, which, as expected, completely inhibited TNF-α production by murine RAW 264.7 cells in response to LPS (P < 0.0005), also significantly inhibited (P < 0.0005) TNF-α production by the OGex A, B, C, and D lipoproteins. In contrast, the response to the MALP-2 lipopeptide was not significantly inhibited (P > 0.05) by antibody to CD14 (Fig. 5B), even when doses as low as 0.25 ng/ml of MALP-2 were used (data not shown). To confirm this observation, we compared TNF-α production from peritoneal macrophages isolated from C57BL/6 wild-type mice with those from C57BL/6 CD14 KO mice (Fig. 5C). The absence of CD14 virtually abolished the ability of the M. arthritidis lipoproteins (P < 0.02) and LPS (P < 0.05) to induce TNF-α in resident peritoneal macrophages, whereas the response to MALP-2 was not decreased by absence of CD14 (P > 0.05). We conclude that cytokine induction by OGex is CD14 dependent and thus quite unlike that of the MALP-2 diacylated lipopeptide, which, in our hands, did not require CD14.
Relationship of OGex A, B, C, and D with other lipoproteins.
Edman degradation to derive N-terminal sequences of OGex A, B, C, and D failed, suggesting that the N-terminal cysteines were blocked and, thus, likely triacylated. This finding supports our results described above indicating that all OGex require CD14. Trypsin digests of OGex A, B, C, and D were also analyzed by mass spectrometry for internal fragment sequences that matched proteins in the MASCOT database (Fig. 6). Two fragments found in OGex A (LLELNILK and LLELNDLK) were identical to two sequences in M. arthritidis MlpD and, except for a single amino acid, in MlpE (formerly L-Rep [31]). Similar sequences were subsequently found in the newly identified lipoprotein MlpF (Fig. 6). These three Mlp proteins share extensive DNA and amino acid sequence homology and contain two domains of tandem, nonidentical repeats. The OGex A-like sequences are located near the beginning of repeat units in the second domain. Although we obtained some information on peptide fragment sequences from OGex B, C, and D, their amino acid sequences did not match any in the MASCOT databases.
FIG. 6.
Amino acid sequence comparisons between OGex A fragments and MlpD, MlpE, and MlpF. Amino acid residues identical to one or both of the OGex A peptides are shaded black.
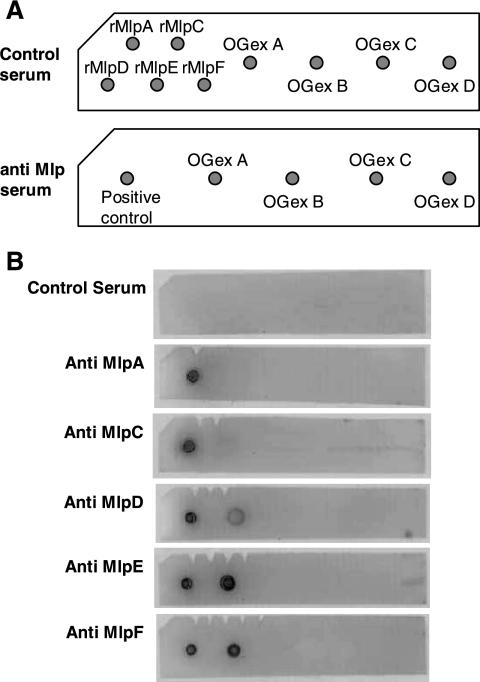
To confirm the relatedness between OGex A and the Mlp lipoproteins, OGex A, B, C, and D were blotted onto PVDF membranes and tested for their reactivity to MlpA, MlpC, MlpD, MlpE, and MlpF antibodies and to preimmune sera (Fig. 7) . A sixth Mlp, MlpB, has also been identified. We were unable to express it in E. coli; however, its deduced amino acid sequenced has been determined (GenBank accession no. AF154924, OrfA′). OGex A reacted with anti-MlpD, -MlpE, and -MlpF, but OGex B, C, and D did not. None reacted with the preimmune serum. The results indicate that OGex A is closely related to the MlpD, MlpE, and MlpF lipoproteins of M. arthritidis but that MlpA and MlpC might be distinct.
FIG. 7.
Dot blot analysis of OGex A, B, C, and D. Recombinant MlpA, MlpC, MlpD, MlpE, and MlpF lipoproteins and OGex A, B, C, and D were placed on a PVDF membrane and exposed to anti-Mlp antiserum. (A) Schema of protein spotting patterns. (B) PVDF membranes of dot blot analysis. Antigens were detected by use of anti-Mlp serum. The amount of protein used was 2 μg/dot. The experiment was conducted three times using different antigen concentrations but with similar results. rMlp, recombinant Mlp.
The bioactive lipid moieties of microbial lipoproteins are characteristically borne on the N-terminal cysteine residue of the molecule. An analysis of the sequences of the mature MlpD, MlpE, and MlpF lipoproteins revealed that all possessed only one cysteine residue and that the adjacent region, which in MlpD was CDNTNKKLEEPKKE, was highly conserved with MlpE and MlpF. We therefore propose that this region likely represents the bioactive lipopeptide contained in OGex A. Interestingly, although MlpA, MlpB, and MlpC share less sequence similarity with the other Mlp proteins, they all bear N-terminal cysteine regions which are very similar to those in MlpD, MlpE, and MlpF. This raises the possibility that all members of the Mlp family of lipoproteins may have bioactivity mediated by a common lipopeptide. Inasmuch as MlpA, MlpB, and MlpC exhibit molecular masses similar to those of OGex D, B, and C, we cannot exclude the possibility that they may in fact be related, despite our preliminary failure to detect sequence similarity by mass spectrometry.
Concluding remarks.
These studies have established that M. arthritidis possesses multiple macrophage-activating lipoproteins that appear to be related to the Mlp family of lipoproteins (31). Furthermore, the OGex moieties appear to be quite distinct from the MALP-404 lipoprotein of M. fermentans and its synthetic lipopeptide derivative, MALP-2, in that they are likely triacylated and have a requirement for CD14, whereas the MALP molecules are diacylated and are independent of CD14 for cytokine induction. Although the function of the M. arthritidis Mlp lipoproteins was recently considered to be unknown (31), the present study suggests that they have inflammatory potential and thus might play a role in disease mediated by this organism. Preliminary studies have indeed shown an association with the potency of the lipoproteins and organism virulence (4). Of importance is their dependency on TLR2, a molecule that is instrumental in M. arthritidis-mediated inflammatory disease (14, 15). In this regard, it should be reemphasized that the M. arthritidis SAg MAM can regulate the immune system not only through TLR2 but also through TLR4 (14). The additional finding that MAM engagement of TLR4 can down-regulate inflammatory disease mediated by M. arthritidis (B. C. Cole and H.-H. Mu, unpublished observations) suggests that both MAM and OGex play a significant role in disease pathogenesis. Furthermore, concurrent work has demonstrated that OGex A lipopeptides can bind to apolipoprotein A-1, a component of high-density-lipoprotein cholesterol, which also suggests a mechanism whereby microbial agonists might render host lipoproteins inflammatory, a process which in this case may contribute to atherosclerosis (9).
Acknowledgments
This work was supported by the Nora Eccles Treadwell Foundation, by NIH grant AR02255, by a grant from the North Central Chapter of the Arthritis Foundation, and by a grant from the South Dakota Health Research Foundation. B.C.C. is the recipient of the Nora Eccles Harrison Chair in Rheumatology.
Amino acid sequence analysis and IR spectrometry were done by University of Utah HSC Core Research Facilities. We also thank Chris Li and John Huntinghouse for excellent technical assistance.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Braun, V., and K. Rehn. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 10:426-438. [DOI] [PubMed] [Google Scholar]
- 2.Cole, B., A. Sawitzke, and H.-H. Mu. 2000. Mycoplasma arthritidis and its superantigen M. arthritidis mitogen as a model for inflammatory and autoimmune disease, p. 93-107. In M. W. Cunningham and R. S. Fujinami (ed.), Effects of microbes on the immune system. Lippincott, Williams and Wilkins, Philadelphia, PA.
- 3.Cole, B. C., and M. M. Griffiths. 1993. Triggering and exacerbation of autoimmune arthritis by the Mycoplasma arthritidis superantigen MAM. Arthritis Rheum. 36:994-1002. [DOI] [PubMed] [Google Scholar]
- 4.Cole, B. C., H.-H. Mu, N. D. Pennock, A. Hasebe, F. V. Chan, L. R. Washburn, and M. R. Peltier. 2005. Isolation and partial purification of macrophage- and dendritic cell-activating components from Mycoplasma arthritidis: association with organism virulence and involvement with Toll-like receptor 2. Infect. Immun. 73:6039-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, B. C., H.-H. Mu, and A. D. Sawitzke. 2000. The mycoplasma superantigen MAM: role in arthritis and immune-mediated disease. Int. J. Med. Microbiol. 290:489-490. [DOI] [PubMed] [Google Scholar]
- 6.Cole, B. C., L. R. Washburn, and D. Taylor-Robinson. 1985. Mycoplasma-induced arthritis, p. 108-160. In S. Razin and M. F. Barile (ed.), Mycoplasma pathogenicity, vol. IV. Academic Press, Inc., New York, NY. [Google Scholar]
- 7.Edward, D. G. 1947. A selective medium for pleuropneumonia like organisms. J. Gen. Microbiol. 1:238-243. [DOI] [PubMed] [Google Scholar]
- 8.Golightly-Rowland, L., B. C. Cole, J. R. Ward, and B. B. Wiley. 1970. Effect of animal passage on arthritogenic and biological properties of Mycoplasma arthritidis. Infect. Immun. 1:538-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hasebe, A., N. D. Pennock, H. H. Mu, F. V. Chan, M. L. Taylor, and B. C. Cole. 2006. A microbial TLR2 agonist imparts macrophage-activating ability to apolipoprotein A-1. J. Immunol. 177:4826-4832. [DOI] [PubMed] [Google Scholar]
- 10.Kiura, K., H. Kataoka, T. Nakata, T. Into, M. Yasuda, S. Akira, N. Inoue, and K. Shibata. 2006. The synthetic analogue of mycoplasmal lipoprotein FSL-1 induces dendritic cell maturation through Toll-like receptor 2. FEMS Immunol. Med. Microbiol. 46:78-84. [DOI] [PubMed] [Google Scholar]
- 11.Lien, E., T. J. Sellati, A. Yoshimura, T. H. Flo, G. Rawadi, R. W. Finberg, J. D. Carroll, T. Espevik, R. R. Ingalls, J. D. Radolf, and D. T. Golenbock. 1999. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 274:33419-33425. [DOI] [PubMed] [Google Scholar]
- 12.Manukyan, M., K. Triantafilou, M. Triantafilou, A. Mackie, N. Nilsen, T. Espevik, K. H. Wiesmuller, A. J. Ulmer, and H. Heine. 2005. Binding of lipopeptide to CD14 induces physical proximity of CD14, TLR2 and TLR1. Eur. J. Immunol. 35:911-921. [DOI] [PubMed] [Google Scholar]
- 13.Medzhitov, R., and C. Janeway, Jr. 2000. The Toll receptor family and microbial recognition. Trends Microbiol. 8:452-456. [DOI] [PubMed] [Google Scholar]
- 14.Mu, H.-H., N. D. Pennock, J. Humphreys, C. J. Kirschning, and B. C. Cole. 2005. Engagement of Toll-like receptors by mycoplasmal superantigen: downregulation of TLR2 by MAM/TLR4 interaction. Cell. Microbiol. 7:789-797. [DOI] [PubMed] [Google Scholar]
- 15.Mu, H.-H., A. D. Sawitzke, and B. C. Cole. 2001. Presence of Lpsd mutation influences cytokine regulation in vivo by the Mycoplasma arthritidis mitogen superantigen and lethal toxicity in mice infected with M. arthritidis. Infect. Immun. 69:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muhlradt, P. F., M. Kiess, H. Meyer, R. Sussmuth, and G. Jung. 1997. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J. Exp. Med. 185:1951-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakata, T., M. Yasuda, M. Fujita, H. Kataoka, K. Kiura, H. Sano, and K. Shibata. 2006. CD14 directly binds to triacylated lipopeptides and facilitates recognition of the lipopeptides by the receptor complex of Toll-like receptors 2 and 1 without binding to the complex. Cell. Microbiol. 8:1899-1909. [DOI] [PubMed] [Google Scholar]
- 18.Omueti, K. O., J. M. Beyer, C. M. Johnson, E. A. Lyle, and R. I. Tapping. 2005. Domain exchange between human Toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J. Biol. Chem. 280:36616-36625. [DOI] [PubMed] [Google Scholar]
- 19.Peltier, M. R., A. J. Freeman, H.-H. Mu, and B. C. Cole. 2005. Characterization and partial purification of a macrophage-stimulating factor from Mycoplasma hominis. Am. J. Reprod. Immunol. 54:342-351. [DOI] [PubMed] [Google Scholar]
- 20.Sankaran, K., and H. C. Wu. 1994. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269:19701-19706. [PubMed] [Google Scholar]
- 21.Schroder, N. W., H. Heine, C. Alexander, M. Manukyan, J. Eckert, L. Hamann, U. B. Gobel, and R. R. Schumann. 2004. Lipopolysaccharide binding protein binds to triacylated and diacylated lipopeptides and mediates innate immune responses. J. Immunol. 173:2683-2691. [DOI] [PubMed] [Google Scholar]
- 22.Sellati, T. J., D. A. Bouis, R. L. Kitchens, R. P. Darveau, J. Pugin, R. J. Ulevitch, S. C. Gangloff, S. M. Goyert, M. V. Norgard, and J. D. Radolf. 1998. Treponema pallidum and Borrelia burgdorferi lipoproteins and synthetic lipopeptides activate monocytic cells via a CD14-dependent pathway distinct from that used by lipopolysaccharide. J. Immunol. 160:5455-5464. [PubMed] [Google Scholar]
- 23.Shibata, K., A. Hasebe, T. Into, M. Yamada, and T. Watanabe. 2000. The N-terminal lipopeptide of a 44-kDa membrane-bound lipoprotein of Mycoplasma salivarium is responsible for the expression of intercellular adhesion molecule-1 on the cell surface of normal human gingival fibroblasts. J. Immunol. 165:6538-6544. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu, T., Y. Kida, and K. Kuwano. 2004. Lipid-associated membrane proteins of Mycoplasma fermentans and M. penetrans activate human immunodeficiency virus long-terminal repeats through Toll-like receptors. Immunology 113:121-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeda, K., and S. Akira. 2001. Regulation of innate immune responses by Toll-like receptors. Jpn. J. Infect. Dis. 54:209-219. [PubMed] [Google Scholar]
- 26.Takeda, K., and S. Akira. 2001. Roles of Toll-like receptors in innate immune responses. Genes Cells 6:733-742. [DOI] [PubMed] [Google Scholar]
- 27.Takeuchi, O., T. Kawai, P. F. Muhlradt, M. Morr, J. D. Radolf, A. Zychlinsky, K. Takeda, and S. Akira. 2001. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13:933-940. [DOI] [PubMed] [Google Scholar]
- 28.Voelker, L. L., K. E. Weaver, L. J. Ehle, and L. R. Washburn. 1995. Association of lysogenic bacteriophage MAV1 with virulence of Mycoplasma arthritidis. Infect. Immun. 63:4016-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Washburn, L. R., S. Hirsch, and L. L. Voelker. 1993. Mechanisms of attachment of Mycoplasma arthritidis to host cells in vitro. Infect. Immun. 61:2670-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Washburn, L. R., J. H. Hughes, and N. L. Somerson. 1978. Mycoplasma growth factors in bovine serum fraction. J. Bacteriol. 135:818-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Washburn, L. R., E. J. Miller, S. Mukherjee, and D. Dannenbring. 2004. Mycoplasma arthritidis bacteriophage MAV1 prophage integration, deletions, and strain-related polymorphisms. Plasmid 52:31-47. [DOI] [PubMed] [Google Scholar]
- 32.Washburn, L. R., E. J. Miller, and K. E. Weaver. 2000. Molecular characterization of Mycoplasma arthritidis membrane lipoprotein MAA1. Infect. Immun. 68:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Washburn, L. R., and E. J. Weaver. 1997. Protection of rats against Mycoplasma arthritidis-induced arthritis by active and passive immunizations with two surface antigens. Clin. Diagn. Lab. Immunol. 4:321-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Washburn, L. R., K. E. Weaver, E. J. Weaver, W. Donelan, and S. Al-Sheboul. 1998. Molecular characterization of Mycoplasma arthritidis variable surface protein MAA2. Infect. Immun. 66:2576-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Washburn, L. R. 2006. Abstr. 16th Int. Congr. Int. Org. Mycoplasmol., abstr. 166.