Abstract
Campylobacter jejuni is a major worldwide cause of enteric illnesses. Adult immunocompetent mice are not susceptible to C. jejuni infection. However, we show here that mice deficient in the adaptor protein myeloid differentiation factor 88 (MyD88), which is required for signaling through most Toll-like receptors, can be stably colonized by C. jejuni but not by isogenic derivatives carrying mutations in known virulence genes. We also found that Nramp1 deficiency increases the mouse susceptibility to C. jejuni infection when administered systemically. These results indicate that MyD88-deficient mice could be a useful model to study C. jejuni colonization and reveal a potential role for Nramp1 in the control of this bacterial pathogen.
Campylobacter jejuni is a leading cause of food-borne illnesses in the United States and Europe (3). Its ability to stably and asymptomatically colonize a great variety of warm-blooded animals, including some important food-producing species, has contributed significantly to the high incidence of infections in human populations (3). Despite the availability of the nucleotide sequences of the genomes of several C. jejuni strains (11, 20, 38), relatively little is known about the mechanisms utilized by these bacteria to colonize the host and cause disease. A number of studies have demonstrated the importance in C. jejuni pathogenesis of various surface determinants, protein glycosylation, motility, and the ability to enter intestinal epithelial cells (5, 14, 16-18, 23-25, 34, 45, 48). However, much less is known about host determinants that contribute to the control of C. jejuni infections. Toll-like receptors (TLRs) are essential components of the innate immune system (2, 33). These receptors recognize conserved bacterial molecules orchestrating a variety of responses that are important to control microbial infections. TLR5, for example, recognizes conserved residues in bacterial flagellin (21). However, C. jejuni flagellin is not recognized by TLR5 due to differences in amino acid sequences that are crucial for receptor recognition (4, 46). Since TLR5 is expressed in intestinal epithelial cells, C. jejuni's lack of agonistic capacity for this TLR may constitute a specific adaptation that could benefit the ability of this pathogen to colonize the intestine and evade recognition by the innate immune system. However, C. jejuni encodes other potential agonists of TLRs as well as specific adaptations that result in the stimulation of innate-immunity outputs in a manner that is apparently independent of TLR signaling (46). Mice are refractory to C. jejuni infection and therefore can only be very transiently colonized by these bacteria, with no underlying detectable pathology (49). Here we report that myd88−/− mice, which are deficient for TLR signaling, can be efficiently and persistently colonized by C. jejuni. We also found that a loss-of-function mutant in nramp1, which encodes a divalent cation transporter essential for the control of infection by several intracellular pathogens (10), significantly increased the susceptibility of myeloid differentiation factor 88 (MyD88)-deficient mice to C. jejuni systemic infections. Furthermore, strains carrying mutations in genes previously shown to be important for C. jejuni colonization or virulence were defective in their ability to colonize myd88−/− mice, indicating that these animals can serve as a useful model to study this important enteric pathogen.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Wild-type C. jejuni 81-176 has been described previously (28). The isogenic ΔflaA mutant has been previously described (46), and isogenic mutants carrying insertions in the pglF or Cj1418c gene were generated by inserting the transposon Tn552kan-Campy within the coding sequence of each gene as described elsewhere (7). The exact positions of the transposon insertions were determined by nucleotide sequence analysis, which indicated that the insertions resulted in the inactivation of the respective genes. C. jejuni strains were routinely grown on tryptic soy broth agar supplemented with 5% sheep blood or in brain heart infusion broth at 37°C under 10% CO2 or low-oxygen conditions (GasPak Plus; BD-Diagnostic Systems, New Jersey). When appropriate, kanamycin (50 μg ml−1) was added.
Mutant complementation.
A new complementation vector was constructed to allow the constitutive expression of genes by chromosomal integration. The Cj1553c open reading frame, which encodes a homologue of hsdM, a putative type I restriction enzyme, was chosen as a site of integration since this gene does not affect C. jejuni virulence (data not shown). The Cj1553c locus was PCR amplified with the primers ROW-FWD (ACGCGTCGACTTAGGATATGCCTGATTTT) and ROW-REV (GCTCAGACAGTTTTGATTGGATTTTA) and cloned into the suicide vector pGK2003 (15). A chloramphenicol acetyltransferase gene (without a transcription terminator) was amplified from pRY109 (47) with the oligonucleotides DHO142 (GCTCTAGACCGTCGTCGGTATCGTATGGAG) and DHO143 (GCTCTAGACTAGTCTCGAGCGGCCGCCTAGGCCATGGTTATTTATTCAGCAAGTCTTGTAA) by PCR and inserted into the single AvrII site of Cj1553c, resulting in plasmid pSB3021. The primer DHO143 contains multiple restriction sites (NcoI, AvrII, NotI, XhoI, and SpeI) that are unique for pSB3021, allowing the insertion of genes downstream of the chloramphenicol acetyltransferase cassette. Therefore, these genes are coexpressed with the chloramphenicol resistance marker. To complement the pglF mutant strain, the pglF gene of C. jejuni 81-176 was amplified with its own Shine-Dalgarno sequence by using the primers DHO250 (CATGCCATGGGTTTGTGAAATTTCAAAACTGATCTTA) and DHO251 (CCGCTCGAGTTATACACCTTCTTTATTGTGTTTAAATT) and cloned into the NcoI/XhoI sites of pSB3021. The resulting plasmid was verified by sequencing and transformed into the pglF mutant of C. jejuni 81-176. Transformants were selected on tryptic soy agar-blood plates supplemented with kanamycin (50 μg/ml) and chloramphenicol (7.5 μg/ml).
Preparation of bone marrow-derived macrophages, bacterial infections, Erk activation assay, and cytokine measurements.
myd88+/+ and myd88−/− mice were sacrificed, and femurs and tibias were excised and flushed with Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), penicillin (100 U ml−1), and streptomycin (50 μg ml−1). Cells were spun down, resuspended in bone marrow-derived macrophage (BMDM) differentiation medium (DMEM containing 20% FBS, 30% L-cell supernatant, penicillin [100 U ml−1], and streptomycin [50 μg ml−1]), and plated on non-tissue culture-treated 10-cm plastic dishes. The cells were fed fresh BMDM differentiation medium on day 3 to 4 to allow further differentiation until day 6 to 7. BMDMs were then seeded at 106 cells per well in a six-well tissue culture dish. For Erk activation assays, BMDMs were washed three times with Hanks balanced salt solution (HBSS), and infected with the different strains at multiplicity of infections (MOIs) of 50 and 20 for C. jejuni and Salmonella enterica serovar Typhimurium, respectively. At the indicated times, macrophages were lysed in sample buffer, equal amounts of cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the activation of Erk was analyzed by Western immunoblotting using a monoclonal antibody specific for the phosphorylated (activated) form of this kinase. To determine the total levels of Erk, blots were stripped and reprobed with an antibody directed against Erk. For cytokine measurements, BMDMs were infected with an MOI of 100 for 1 h. The wells were washed with HBSS, and the medium was replaced with 1.5 ml of DMEM with 10% FBS and containing gentamicin (100 μg ml−1). Supernatants were collected following an additional 7 h of incubation at 37°C with 5% CO2 and centrifuged for 10 min at 12,000 × g to pellet residual bacteria and cell debris. The levels of tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) were determined by enzyme-linked immunosorbent assay using a commercial kit (BD Biosciences, Pharmingen) according to the instructions of the manufacturer.
C. jejuni mouse infections.
The myd88−/− nramp1−/− mouse strain was obtained from S. Akira (1). This mouse has been backcrossed into a C57Black/6 background, which is nramp1−/−. The nramp1+/+ derivatives of these mice were obtained by backcrossing a wild-type nramp1 allele into the same background. Animals were backcrossed five times. The nramp1 and myd88 genotypes were confirmed by nucleotide sequencing and PCR, respectively, as previously described (29). Sex- and age (6 to 8 weeks)-matched mice were infected orally by stomach gavage with 109 CFU or intraperitoneally with 106 CFU of the different C. jejuni strains. At the indicated time points, fresh feces were collected, weighed, dissolved in brain heart infusion broth, and plated to determine the number of CFU per gram of feces. When appropriate, animals were sacrificed, organs removed and homogenized, and the bacterial loads in different tissues determined by serial plating on blood agar containing the appropriate antibiotics. For mixed infections, equal numbers (109 and 106 bacterial CFU orally and intraperitoneally, respectively) of C. jejuni 81-176 and its isogenic mutants were simultaneously administered to sex-matched 6-week-old myd88−/− mice. Colonization by the different strains was monitored by enumerating the number of CFU in the feces of the inoculated animals. To differentiate between mutant and wild-type bacteria, equal amounts of homogenized feces were plated on blood agar plates containing Campylobacter-selective supplements (Oxoid SR0167E) with or without kanamycin (50 μg ml−1). To differentiate between the mutant and complemented strains, total bacteria were selected for on blood agar plates containing Campylobacter-selective supplements (Oxoid SR0167E), and the numbers of CFU were compared to those recovered on blood agar plates containing chloramphenicol (7.5 μg/ml), which select for only the complemented stain. At the end of the experiment, mice were sacrificed and the bacterial loads in the intestines were determined by plating on selective plates as described above. Statistical analysis of the results was carried out with the Wilcoxon (Mann-Whitney) rank test.
Enumeration of C. jejuni loads in BMDMs by flow cytometry.
BMDMs from wild-type and nramp1−/− mice were seeded at density of 105 cells per well on a 24-well dish and infected at an MOI of 20. Following a 1-h incubation at 37°C and 5% CO2, the BMDMs were washed with HBSS, and DMEM containing 10% FBS and 100 μg ml−1 gentamicin was added to each well. Cells were washed again and lysed at the designated time points in 500 μl of 0.05% sodium deoxycholate in phosphate-buffered saline (PBS). The cell lysates were collected and subjected to a low-speed spin (1,000 rpm) for 2 min to remove large cell debris. Supernatants were collected, and intracellular bacteria were isolated by a 2-min high-speed spin (10,000 rpm). The isolated bacterial pellet was resuspended in 500 μl filter-sterilized staining buffer (PBS containing 1 mM EDTA and 0.01% Tween). The bacteria were then stained with the reagents of a cell viability kit (BD Biosciences, San Jose, CA), which distinguishes live and dead cells by using a thiazole orange (TO) solution, which stains all bacteria, and propidium iodide (PI), which stains only dead bacteria. TO and PI were added to final concentrations of 53 nM and 11 μM, respectively, in accordance to the manufacturer's instructions. After 5 min of staining, bacteria were pelleted, washed once in PBS, resuspended in 1 ml of PBS, and analyzed by flow cytometry. The absolute count of live and dead bacteria was carried out by addition of 50 μl of a liquid suspension of a known number of fluorescent beads (supplied in the kit from BD Biosciences, San Jose, CA), following the manufacturer's instructions. Samples were analyzed on a FACScalibur flow cytometer. TO fluoresces primarily in FL1 and FL2; PI fluoresces primarily in FL3. An side-scatter threshold was used, and cells and beads were gated using scatter and FL2, which detects the TO fluorescence and therefore the total bacterial population. In order to best discriminate between live and dead populations, a plot of FL1 versus FL3 was used and live and dead populations were gated within this plot (dead cells, FL3+; live cells, FL1+). To determine the concentrations of the cell populations, the following equation was used: number of events in cell region/number of events in bead region × number of beads per test/test volume × dilution factor = concentration of cell population. A plot was generated after using this equation to calculate the number of viable bacteria (in triplicate wells) in both nramp1+/+ and nramp1−/− BMDMs at each time point.
Macrophage transduction and fluorescence microscopy.
BMDMs were isolated from wild-type mouse femurs as described above. Isolated cells were spun down and resuspended in cell supernatants containing pseudotyped recombinant murine leukemia virus (MLV) expressing Nramp1-green fluorescent protein (GFP) fusion protein. The Nramp1-GFP virus stocks were generated by transfecting a 10-cm dish of 293 cells (50% confluence) with 4 μg pMLV GagPol, 4 μg of pVSV-G along with 4 μg pLZRS encoding Nramp1-GFP, and 12 μl of FuGENE6 (Roche Diagnostics, Indianapolis, IN). At 24 hours after transfection, cultures were split into two 10-cm dishes, and after an additional 48 h, the virus-containing supernatants were harvested, filtered, and frozen at −80°C in 3- to 4-ml aliquots. Transduction of BMDMs was carried out as follows. Freshly isolated bone marrow cells were infected with the recombinant MLV for 2 h at 4°C in a rotating wheel and subsequently plated on petri dishes in BMDM-differentiating medium for 6 to 7 days. Once differentiated, BMDMs were plated on coverslips that were placed on 24-well culture dishes at a cell density of 105 cells per well. Prior to bacterial infection, BMDMs were washed three times with HBSS, infected with C. jejuni at an MOI of 10 for 30 min, washed again with HBSS, and incubated at 37°C with 5% CO2 for 1 h. After fixation in 4% paraformaldehyde, coverslips were washed three times in PBS and incubated in rabbit anti-C. jejuni serum for 30 min. The coverslips were then washed three times in PBS and incubated in secondary AlexaFluor 594 goat anti-rabbit immunoglobulin G (Molecular Probes, Eugene, OR). After three subsequent washes, the coverslips were mounted on glass slides. Images were acquired on a Nikon TGE2000-U Eclipse inverted microscope fitted with a Micromax Princeton digital camera controlled by the Metamorph software package, version 6.1 (Universal Imaging Corp., Downingtown, PA).
RESULTS
Impaired Erk activation and proinflammatory cytokine production in MyD88-deficient mouse BMDMs after C. jejuni infection.
We previously reported that in cultured intestinal epithelial cells, C. jejuni activates both the Erk and p38 mitogen-activated protein kinase pathways leading to the production of IL-8 (46). These responses were independent of TLR signaling, since these cells express only TLR5, which is not activated by C. jejuni flagellin, the agonist for this receptor (4, 46). To investigate the potential involvement of TLRs in C. jejuni infections, we compared Erk activation and proinflammatory cytokine production after infection of BMDMs from wild-type and MyD88-deficient macrophages. Erk activation and proinflammatory cytokine production are well-established indicators of TLR stimulation, and MyD88 is an adaptor protein that is essential for signaling through most TLRs (2). BMDMs were infected with wild-type C. jejuni or an isogenic ΔflaA strain, which is defective for stimulation of mitogen-activated protein kinase activation and proinflammatory cytokine production in intestinal epithelial cells (46). Erk stimulation was assessed at different times after infection by Western immunoblotting with an antibody specific to the phosphorylated (activated) form of this kinase. As shown in Fig. 1, infection with both wild-type and ΔflaA C. jejuni efficiently stimulated Erk activation in myd88+/+ BMDMs. Erk activation was detected as early as 20 min after infection, and maximum stimulation was seen 40 min after C. jejuni infection. In contrast, Erk activation was severely impaired in myd88−/− BMDMs at all time points after infection. These observations indicate that C. jejuni stimulates innate immune responses in macrophages through TLR signaling.
FIG. 1.
MyD88 is required for C. jejuni-induced Erk activation. BMDMs obtained from myd88+/+ and myd88−/− mice were infected with C. jejuni (wild type or ΔflaA as indicated) or S. enterica serovar Typhimurium (as a positive control [35]) for the indicated times, and Erk activation was determined by Western immunoblot analysis of cell extracts using an antibody directed to the phosphorylated (activated) form of Erk (phospho Erk). To ascertain equal loading, blots were reprobed with an antibody directed to Erk (total Erk).
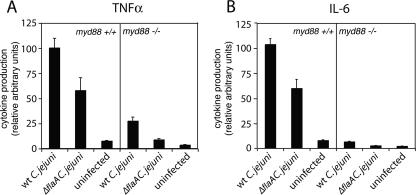
We also investigated the role of MyD88 in proinflammatory cytokine production during C. jejuni infection. To this end, we examined TNF-α and IL-6 secretion in myd88+/+ and myd88−/− BMDMs at 8 h after infection with either wild-type or ΔflaA C. jejuni strains. Proinflammatory cytokine production was significantly reduced (P = 0.003 for TNF-α and P = 0.001 for IL-6 [Student t test]) in myd88−/− macrophages infected with either C. jejuni strain, demonstrating the importance of TLR signaling in the production of proinflammatory cytokines during C. jejuni infection (Fig. 2). Taken together, these data indicate that TLRs can recognize and mediate responses to C. jejuni during infection.
FIG. 2.
MyD88 is required for C. jejuni-induced cytokine production. BMDMs obtained from myd88+/+ and myd88−/− mice were infected with wild-type (wt) C. jejuni or an isogenic ΔflaA mutant, and at 7 h after infection, the levels of TNF-α (A) and IL-6 (B) were determined as indicated in Materials and Methods. Values represent the means ± standard deviations of three independent measurements and were standardized by considering the stimulation of myd88+/+ mice by wild-type C. jejuni to be 100%.
MyD88-deficient mice are efficiently colonized by C. jejuni.
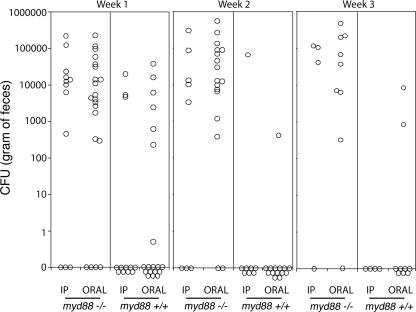
The observation that MyD88 is essential for the stimulation of proinflammatory cytokine production of infected BMDMs in vitro prompted us to investigate the role of this adaptor protein, and by extension the role of TLR signaling, in C. jejuni infections in vivo. myd88+/+ and myd88−/− mice (C57Black/6 nramp1−/− background) were inoculated orally or intraperitoneally with 109 and 106 wild-type C. jejuni organisms, respectively, and bacterial fecal shedding and colonization of different tissues were assessed at different times after infection. C. jejuni were detected in the feces of only a small fraction (∼30%) of the myd88+/+ mice at 1 week after inoculation by either route (Fig. 3). This is consistent with previous observations indicating that C. jejuni is inefficient at colonizing mice (49). In contrast, C. jejuni was detected in the feces of most (∼80%) of the myd88−/− mice after oral or intraperitoneal inoculations (Fig. 3). At 2 and 3 weeks after infection, the detection of C. jejuni in the feces of myd88+/+ animals decreased sharply, and bacteria could be detected in only 1 out of 8 and 1 out of 15 intraperitoneally or orally inoculated animals, respectively. In contrast, the number of myd88−/− mice shedding C. jejuni remained steady, and the actual number of bacteria per gram of feces was much higher than that in the very few wild-type-colonized animals (Fig. 3). Taken together, these data suggest that MyD88 and, by extension, TLR signaling play a critical role in controlling intestinal colonization by C. jejuni.
FIG. 3.
MyD88-deficient mice are efficiently colonized by C. jejuni. myd88+/+ and myd88−/− mice were inoculated orally or intraperitoneally (IP) with the C. jejuni 81-176 wild-type strain. Colonization was evaluated by determining the number of CFU in the feces at different times after infection. Each circle denotes the CFU of an individual animal.
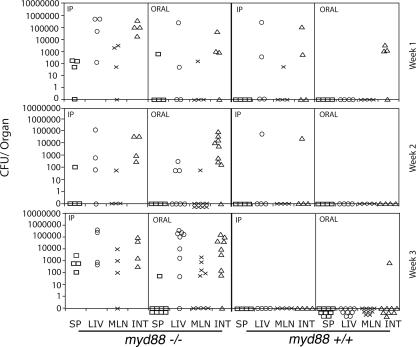
We also investigated the presence of C. jejuni in different organs of myd88+/+ and myd88−/− animals following oral or intraperitoneal inoculation. Significant numbers of bacteria were recovered from spleens, livers, and mesenteric lymph nodes from all myd88−/− mice after both oral and intraperitoneal inoculation (Fig. 4). Systemic tissues were more readily colonized after systemic inoculation, and bacterial loads were always higher in the liver than in any other systemic tissue. Three weeks after bacterial inoculation, large numbers of bacteria were still present in all tissues of most orally or intraperitoneally inoculated myd88−/− mice. In contrast, much lower numbers of bacteria were recovered from only some of the myd88+/+ animals 1 week after oral or intraperitoneal inoculation (Fig. 4). The number of bacteria isolated from myd88+/+ animals steadily decreased over time, and by the third week of infection, all but one animal had apparently cleared the infection and no bacteria were detected in any of the tissues examined (Fig. 4).
FIG. 4.
C. jejuni colonizes the systemic tissues of MyD88-deficient mice. myd88+/+ and myd88−/− mice were inoculated orally or intraperitoneally (IP) with the C. jejuni 81-176 wild-type strain. At different times after infection, the numbers of C. jejuni CFU in the spleen (SP) (squares), liver (LIV) (circles), mesenteric lymph nodes (MLN) (×), and intestine (INT) (triangles) were determined by plating different dilutions of the tissue lysates. Each symbol represents the CFU of an individual animal.
To examine whether intestinal colonization leads to pathology, we examined the histopathology of intestines and livers obtained from C. jejuni-colonized myd88−/− animals. Despite the presence of large bacterial loads (as detected by plating of tissue lysates), no pathology was detected in any of the animals examined (data not shown). This observation is consistent with the fact that no symptoms were detected in any of the infected animals inoculated by either route. Taken together, these results indicate that in the absence of MyD88, C. jejuni is able to infect and stably colonize the mouse intestine and systemic tissues, most prominently the liver. Furthermore, since MyD88 is critical for TLR signaling, these results indicate that TLRs play an important role in the control of C. jejuni infections.
Nramp1 deficiency increases susceptibility to C. jejuni infection.
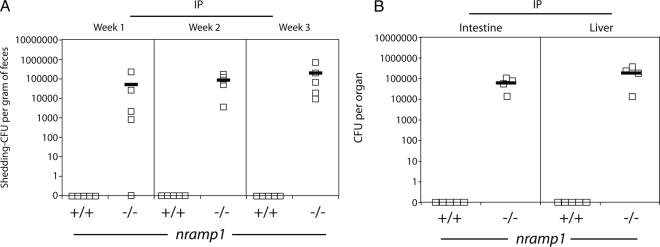
The murine Nramp1 protein has been implicated in resistance to the intracellular pathogens Salmonella spp., Mycobacterium spp., and Leishmania spp. (10). Although it has been shown that Nramp1 is a divalent cation transporter (22), how this function relates to resistance to infection is still unclear. We sought to investigate whether Nramp1 influences the ability of C. jejuni to colonize myd88−/− mice. Intraperitoneally inoculated myd88−/− nramp1−/− mice shed large amounts of C. jejuni in their feces for at least 3 weeks after inoculation. In contrast, no CFU were detected in the feces of myd88−/− nramp1+/+ mice after intraperitoneal inoculation (Fig. 5). Consistent with these results, at 3 weeks after intraperitoneal inoculation, no C. jejuni CFU were detected in the livers and intestines of myd88−/− nramp1+/+ mice, while large numbers of C. jejuni CFU were obtained from the same tissues of myd88−/− nramp1−/− mice (Fig. 5).
FIG. 5.
Nramp1 deficiency increases C. jejuni mouse colonization after systemic administration. myd88−/− nramp1−/− and myd88−/− nramp1+/+ mice were inoculated intraperitoneally (IP) with the C. jejuni 81-176 wild-type strain. Colonization was evaluated by determining the number of CFU in the feces at different times after infection (A). Colonization of systemic tissues was evaluated 4 weeks after infection by determining the number of C. jejuni CFU in the liver and intestine (B). Each square denotes the CFU of an individual animal.
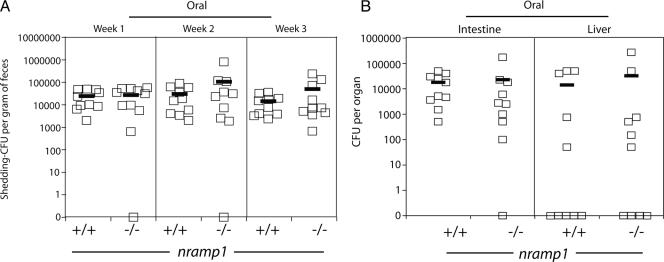
In contrast to the differences observed after intraperitoneal inoculations, no significant differences (P > 0.8) were observed in the levels of C. jejuni colonization of myd88−/− nramp1−/− and myd88−/− nramp1+/+ mice after oral infection. Equivalent numbers of C. jejuni CFU were recovered from feces of orally inoculated myd88−/− nramp1−/− and myd88−/− nramp1+/+ mice (Fig. 6). Furthermore, equivalent numbers of CFU were also recovered from intestines and livers of myd88−/− nramp1−/− and myd88−/− nramp1+/+ mice at 3 weeks after oral inoculation (Fig. 6). These results indicate that Nramp1 plays an important role in the control of C. jejuni after systemic infection but does not seem to influence the course of infection after oral inoculation.
FIG. 6.
Nramp1 deficiency does not alter C. jejuni mouse colonization after oral administration. myd88−/− nramp1−/−, and myd88−/− nramp1+/+ mice were inoculated orally with the C. jejuni 81-176 wild-type strain. Colonization was evaluated by determining the number of CFU in the feces at different times after infection (A). Colonization of systemic tissues was evaluated 4 weeks after infection by determining the number of C. jejuni CFU in the liver and intestine (B). Each square denotes the CFU of an individual animal.
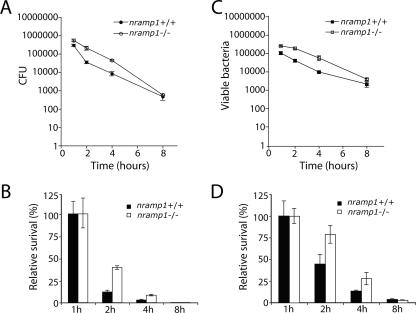
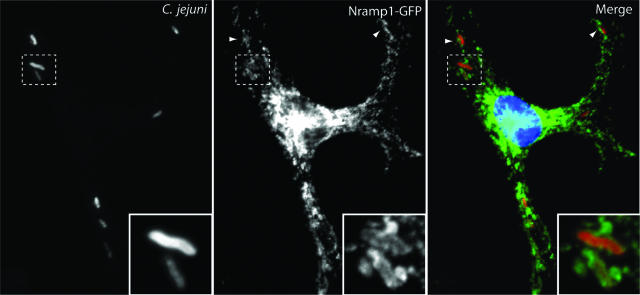
Since Nramp1 is expressed exclusively in cells of the reticuloendothelial system (10), we compared the ability of C. jejuni to infect and survive within BMDMs obtained from nramp1−/− and nramp1+/+ mice. BMDMs obtained from nramp1−/− and nramp1+/+ mice were infected with wild-type C. jejuni, and the viable bacterial loads were measured over time either by determining the CFU on cultured plates or by flow cytometry using live versus dead staining as indicated in Materials and Methods. The number of viable C. jejuni organisms decreased markedly over time in BMDMs obtained from either strain, although significantly higher numbers of CFU were reproducibly recovered from BMDMs derived from nramp1−/− mice at earlier times of infection (Fig. 7) (P = 0.008 at 2 h and P = 0.01 at 4 h of infection [Student t test]). The differences were no longer statistically significant at 8 h of infection. It is possible that the high sensitivity of C. jejuni to macrophage killing may diminish the potential effect of Nramp1 in this assay. We then tested whether Nramp1 could be recruited to the C. jejuni-containing vacuole. We transduced BMDMs with recombinant MLV expressing GFP-tagged Nramp1 and examined its recruitment to the C. jejuni-containing vacuole over time. As shown in Fig. 8, consistent with a possible involvement of Nramp1 in C. jejuni infections, Nramp1-GFP was readily recruited to the C. jejuni vacuole. Taken together, these results implicate Nramp1 in conferring resistance to C. jejuni in a mouse model of infection, adding this pathogen to the limited list of microorganisms whose biology is influenced by this transporter.
FIG. 7.
The absence of Nramp1 results in an increased ability of C. jejuni to survive within macrophages. BMDMs obtained from nramp1+/+ and nramp1−/− mice were infected with C. jejuni, and at different times after infection, the number of intracellular CFU was determined by the gentamicin protection assay (A). The relative survival index (B) represents the ratio between CFU obtained from nramp1+/+ and CFU obtained from nramp1−/− mouse macrophages. Alternatively, viable bacteria were enumerated by flow cytometry (C) as indicated in Materials and Methods. The relative survival index obtained by this method is shown in panel D. Values represent the means ± standard deviations of three independent determinations.
FIG. 8.
Recruitment of Nramp1 to the C. jejuni-containing vacuole. Mouse BMDMs were transduced with a vector encoding Nramp1 fused to GFP. At 24 h after transduction, cells were infected with C. jejuni, and 60 min after infection, cells were fixed, stained with an anti-C. jejuni antibody, and visualized by fluorescence microscopy as indicated in Materials and Methods.
Infection of myd88−/− mice can resolve colonization defects in virulence-attenuated mutants of C. jejuni.
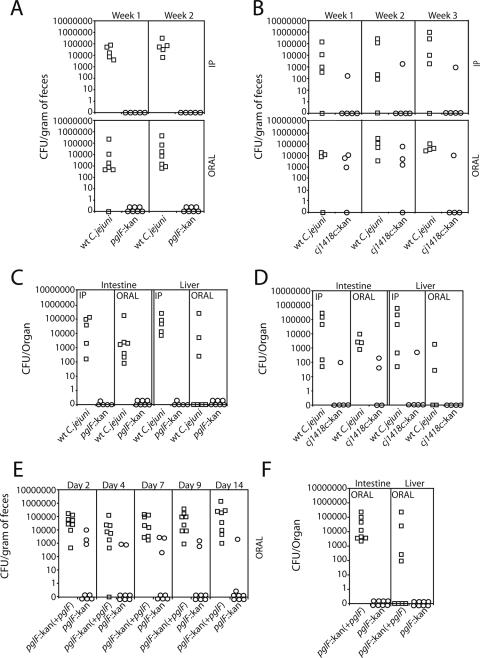
The observation that wild-type C. jejuni can stably colonize MyD88-deficient mice prompted us to examine whether this infection model could be used to detect colonization defects in C. jejuni virulence mutants. We chose to examine the effect of insertion mutations in pglF, a member of a well-characterized locus that encodes a general glycosylation system (31, 44), or in Cj1418c, a member of a gene cluster involved in the synthesis of the extracellular polysaccharide capsule (5, 26, 27). C. jejuni strains carrying mutations in either of these genes (or gene clusters) have been shown to be defective in several in vitro and in vivo virulence assays. We inoculated myd88−/− mice orally or intraperitoneally with equal numbers of wild-type C. jejuni and its isogenic pglF or Cj1418c mutant strain and examined the presence of the different strains in the feces of infected animals over time. Although significant numbers of wild-type C. jejuni CFU were detected in the feces of three out of four of the orally or intraperitoneally infected mice at 1 and 2 weeks after inoculation, no CFU of the pglF mutant strains were detected in any of the inoculated animals (Fig. 9). The mutant phenotype was readily complemented by the reintroduction of the wild-type allele (Fig. 9). Similarly, only one of four mice showed the presence of the Cj1418c mutant in the feces of intraperitoneally or orally inoculated mice at 3 weeks after infection (Fig. 9). Consistent with these observations, at 3 weeks after oral or intraperitoneal inoculation, significant levels of wild-type C. jejuni or the complemented pglF mutant strain were detected in the intestines of all the animals and in the livers of all but three of the inoculated animals. In contrast, no CFU of the pglF mutant strain were recovered from any tissue, and significantly lower levels of the cj1418 mutant were detected only in the intestines of one and two of the intraperitoneally or orally infected animals, respectively (Fig. 9). Taken together, these results indicate that both the pglF and Cj1418 mutants are defective in colonization in this animal model. Furthermore, these results indicate that the MyD88-defective mouse can be used to detect phenotypes of virulence-defective mutants of C. jejuni.
FIG. 9.
C. jejuni virulence mutants are defective for colonization of MyD88-deficient mice. myD88−/− mice were inoculated orally or intraperitoneally (IP) with equal numbers of the C. jejuni 81-176 wild-type (wt) strain and its isogenic derivatives carrying mutations in pglF or Cj1418c. Colonization was evaluated by determining the number of CFU in the feces at different times after infection (A and B). Each circle or square denotes the CFU obtained from an individual animal. Colonization of systemic tissues was evaluated at 3 weeks after infection by determining the number of C. jejuni CFU in the liver and intestine (C and D). To test the complementation of the pglF mutant, myd88−/− mice were inoculated orally with equal numbers of the C. jejuni 81-176 pglF::kan mutant strain and its complemented derivative [pglF::kan(+pglF)] (E and F). Each circle or square denotes the CFU obtained from an individual animal.
DISCUSSION
TLRs have been shown to be very important for the detection of microbial products and the coordination of the innate immune responses to many microbial pathogens (2, 33). Consistent with this observation, mice defective in MyD88, which is essential for signal transduction through most TLRs, are more susceptible to infection by several microbial pathogens (6, 8, 36, 39). We have shown here that MyD88 is also essential for the stimulation of innate immunity outputs by C. jejuni. Consistent with this conclusion, C. jejuni did not stimulate innate immunity responses in BMDMs from myd88−/− mice. Furthermore, in contrast to wild-type mice, myd88−/− mice could be stably colonized by wild-type C. jejuni after oral or intraperitoneal administration. C. jejuni CFU were readily detected in different tissues, such as spleen, liver, intestine, and mesenteric lymph nodes, most often in high numbers. However, despite the high bacterial load, infected mice showed no symptoms. Consistent with this observation, infected tissues showed no discernible histopathology. These results indicate that TLRs are important in the control of C. jejuni colonization, both of the intestine and of systemic tissues.
C. jejuni research has been hampered by the lack of a convenient animal model (49). Although primates or ferrets are good animal models for C. jejuni infection (9, 13, 40, 41), they have some practical disadvantages related to their expense and/or difficulties in their handling or availability. In general, mice constitute the most convenient animal species for the study of microbial pathogens, fundamentally because of the availability of mutant lines, which allow the investigation of very specific aspects of host-pathogen interactions. Adult immunocompetent mice are not generally susceptible to C. jejuni infection, and although they can be transiently colonized, the model is not robust enough to be useful in pathogenesis or colonization studies (49). Certain mutations leading to immunodeficiencies have been shown to increase the susceptibility of mice to C. jejuni infection (12, 19, 32). However, the usefulness of these models is somewhat limited by the fact that these mice are in general difficult to maintain. Our observation that myd88−/− mice can be persistently colonized by C. jejuni, prompted us to test whether this infection model would be robust enough to detect the phenotypes of C. jejuni mutants that have been shown to be defective for colonization in other animal models. We showed that C. jejuni strains carrying mutations in genes required for exopolysaccharide synthesis (Cj1418c) or general glycosylation (pglF), which have been previously shown to be required for colonization or virulence in other models (5, 25, 42), failed to colonize myd88−/− mice. These results indicate that the myd88−/− mouse could be a useful model for the study of C. jejuni colonization.
During the course of these studies, we made the surprising observation that the presence or absence of Nramp1 influences the susceptibility of mice to C. jejuni colonization. Nramp1 is a divalent cation efflux pump localized to the phagosome membrane of neutrophils and macrophages, which has been shown to control the susceptibility of mice to a diverse but limited group of intracellular pathogens, such as Leishmania spp., Mycobacterium spp., and Salmonella enterica (10). Although the mechanism by which this transporter influences the susceptibility of mice is not well understood, studies have suggested that recruitment of Nramp1 to phagosomes modulates its fusogenic properties (10). In addition, since Nramp1 functions as an efflux pump of divalent cations, including Zn2+, Cu2+, Fe2+, and Mn2+, it has been suggested that its bactericidal activities may be associated with its potential ability to remove these rate-limiting nutrients. Nramp1-deficient mice exhibited increased susceptibility to colonization by C. jejuni when administered systemically. This observation suggests that when orally administered, C. jejuni replicates extracellularly or colonizes a compartment which does not express Nramp1. Indeed, C. jejuni has been shown to colonize intestinal mucus both on the outer surface and deep within the intestinal crypts of gnotobiotic or germfree mice (30). Furthermore, although C. jejuni is able to enter intestinal epithelial cells (37, 43), Nramp1 is not expressed in these cells. Nramp1 has been exclusively shown to influence the resistance to pathogens that have an intracellular stage within their life cycle (10). Therefore, the observation that Nramp1 influences the biology of C. jejuni infection suggests that this pathogen must also have an important intracellular stage at some point during its life cycle.
In summary, we have shown that in the absence of MyD88, mice can be persistently colonized by C. jejuni and that this infection model can be used to study the contribution of specific bacterial genes to this phenotype. Furthermore, this infection model has revealed a potential role of Nramp1 in the control of C. jejuni infections.
Acknowledgments
We thank S. Akira for permission to use the myd88−/− mouse strain and R. Medzhitov for providing it. We also thank members of the Galán laboratory for critical review of the manuscript.
D.H. was supported by an EMBO long-term fellowship. This work was supported by a grant from the Ellison Medical Foundation to J.E.G., who is an Ellison Medical Foundation Senior Scholar in Infectious Diseases.
Editor: A. Camilli
Footnotes
Published ahead of print on 28 December 2007.
REFERENCES
- 1.Adachi, O., T. Kawai, K. Takeda, M. Matsumoto, H. Tsutsui, M. Sakagami, K. Nakanishi, and S. Akira. 1998. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity 9:143-150. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 3.Allos, B. M. 2001. Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32:1201-1206. [DOI] [PubMed] [Google Scholar]
- 4.Andersen-Nissen, E., K. Smith, K. Strobe, S. Barrett, B. Cookson, S. Logan, and A. Aderem. 2005. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl. Acad. Sci. USA 102:9247-9252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bacon, D. J., C. M. Szymanski, D. H. Burr, R. P. Silver, R. A. Alm, and P. Guerry. 2001. A phase-variable capsule is involved in virulence of Campylobacter jejuni 81-176. Mol. Microbiol. 40:769-777. [DOI] [PubMed] [Google Scholar]
- 6.Beutler, B., Z. Jiang, P. Georgel, K. Crozat, B. Croker, S. Rutschmann, X. Du, and K. Hoebe. 2006. Genetic analysis of host resistance: Toll-like receptor signaling and immunity at large. Annu. Rev. Immunol. 24:353-389. [DOI] [PubMed] [Google Scholar]
- 7.Colegio, O. R., T. J. ten Griffin, N. D. Grindley, and J. E. Galán. 2001. In vitro transposition system for efficient generation of random mutants of Campylobacter jejuni. J. Bacteriol. 183:2384-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng, C. G., C. A. Scanga, C. M. Collazo-Custodio, A. W. Cheever, S. Hieny, P. Caspar, and A. Sher. 2003. Mice lacking myeloid differentiation factor 88 display profound defects in host resistance and immune responses to Mycobacterium avium infection not exhibited by Toll-like receptor 2 (TLR2)- and TLR4-deficient animals. J. Immunol. 171:4758-4764. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgeorge, R. B., A. Baskerville, and K. P. Lander. 1981. Experimental infection of Rhesus monkeys with a human strain of Campylobacter jejuni. J. Hyg. 86:343-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forbes, J. R., and P. Gros. 2001. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 9:397-403. [DOI] [PubMed] [Google Scholar]
- 11.Fouts, D. E., E. F. Mongodin, R. E. Mandrell, W. G. Miller, D. A. Rasko, J. Ravel, L. M. Brinkac, R. T. DeBoy, C. T. Parker, S. C. Daugherty, R. J. Dodson, A. S. Durkin, R. Madupu, S. A. Sullivan, J. U. Shetty, M. A. Ayodeji, A. Shvartsbeyn, M. C. Schatz, J. H. Badger, C. M. Fraser, and K. E. Nelson. 2005. Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species. PLoS Biol. 3:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, J., A. Rogers, M. Whary, Z. Ge, N. Taylor, S. Xu, B. Horwitz, and S. Erdman. 2004. Gastroenteritis in NF-κB-deficient mice is produced with wild-type Camplyobacter jejuni but not with C. jejuni lacking cytolethal distending toxin despite persistent colonization with both strains. Infect. Immun. 72:1116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, J. G., J. I. Ackerman, N. Taylor, M. Claps, and J. C. Murphy. 1987. Campylobacter jejuni infection in the ferret: an animal model of human campylobacteriosis. Am. J. Vet. Res. 48:85-90. [PubMed] [Google Scholar]
- 14.Fry, B. N., S. Feng, Y. Y. Chen, D. G. Newell, P. J. Coloe, and V. Korolik. 2000. The galE gene of Campylobacter jejuni is involved in lipopolysaccharide synthesis and virulence. Infect. Immun. 68:2594-2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guerry, P., R. A. Alm, M. E. Power, S. M. Logan, and T. J. Trust. 1991. Role of two flagellin genes in Campylobacter motility. J. Bacteriol. 173:4757-4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guerry, P., C. P. Ewing, T. E. Hickey, M. M. Prendergast, and A. P. Moran. 2000. Sialylation of lipooligosaccharide cores affects immunogenicity and serum resistance of Campylobacter jejuni. Infect. Immun. 68:6656-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guerry, P., C. M. Szymanski, M. M. Prendergast, T. E. Hickey, C. P. Ewing, D. L. Pattarini, and A. P. Moran. 2002. Phase variation of Campylobacter jejuni 81-176 lipooligosaccharide affects ganglioside mimicry and invasiveness in vitro. Infect. Immun. 70:787-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hendrixson, D. R., and V. J. DiRita. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol. Microbiol. 52:471-484. [DOI] [PubMed] [Google Scholar]
- 19.Hodgson, A. E., B. W. McBride, M. J. Hudson, G. Hall, and S. A. Leach. 1998. Experimental campylobacter infection and diarrhoea in immunodeficient mice. J. Med. Microbiol. 47:799-809. [DOI] [PubMed] [Google Scholar]
- 20.Hofreuter, D., J. Tsai, R. O. Watson, V. Novik, B. Altman, M. Benitez, C. Clark, C. Perbost, T. Jarvie, L. Du, and J. E. Galán. 2006. Unique features of a highly pathogenic Campylobacter jejuni strain. Infect. Immun. 74:4694-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honko, A., and S. Mizel. 2005. Effects of flagellin on innate and adaptive immunity. Immunol. Res. 33:83-101. [DOI] [PubMed] [Google Scholar]
- 22.Jabado, N., A. Jankowski, S. Dougaparsad, V. Picard, S. Grinstein, and P. Gros. 2000. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192:1237-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, M. A., K. L. Marston, C. A. Woodall, D. J. Maskell, D. Linton, A. V. Karlyshev, N. Dorrell, B. W. Wren, and P. A. Barrow. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect. Immun. 72:3769-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanipes, M. I., L. C. Holder, A. T. Corcoran, A. P. Moran, and P. Guerry. 2004. A deep-rough mutant of Campylobacter jejuni 81-176 is noninvasive for intestinal epithelial cells. Infect. Immun. 72:2452-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karlyshev, A., P. Everest, D. Linton, S. Cawthraw, D. Newell, and B. Wren. 2004. The Campylobacter jejuni general glycosylation system is important for attachment to human epithelial cells and in the colonization of chicks. Microbiology 150:1957-1964. [DOI] [PubMed] [Google Scholar]
- 26.Karlyshev, A. V., O. L. Champion, C. Churcher, J. R. Brisson, H. C. Jarrell, M. Gilbert, D. Brochu, F. St. Michael, J. Li, W. W. Wakarchuk, I. Goodhead, M. Sanders, K. Stevens, B. White, J. Parkhill, B. W. Wren, and C. M. Szymanski. 2005. Analysis of Campylobacter jejuni capsular loci reveals multiple mechanisms for the generation of structural diversity and the ability to form complex heptoses. Mol. Microbiol. 55:90-103. [DOI] [PubMed] [Google Scholar]
- 27.Karlyshev, A. V., D. Linton, N. A. Gregson, A. J. Lastovica, and B. W. Wren. 2000. Genetic and biochemical evidence of a Campylobacter jejuni capsular polysaccharide that accounts for Penner serotype specificity. Mol. Microbiol. 35:529-541. [DOI] [PubMed] [Google Scholar]
- 28.Korlath, J. A., M. T. Osterholm, L. A. Judy, J. C. Forfang, and R. A. Robinson. 1985. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J. Infect. Dis. 152:592-596. [DOI] [PubMed] [Google Scholar]
- 29.Lara-Tejero, M., F. S. Sutterwala, Y. Ogura, E. P. Grant, J. Bertin, A. J. Coyle, R. A. Flavell, and J. E. Galan. 2006. Role of the caspase-1 inflammasome in Salmonella typhimurium pathogenesis. J Exp. Med. 203:1407-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, A., J. L. O'Rourke, P. J. Barrington, and T. J. Trust. 1986. Mucus colonization as a determinant of pathogenicity in intestinal infection by Campylobacter jejuni: a mouse cecal model. Infect. Immun. 51:536-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Linton, D., N. Dorrell, P. G. Hitchen, S. Amber, A. V. Karlyshev, H. R. Morris, A. Dell, M. A. Valvano, M. Aebi, and B. W. Wren. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol. 55:1695-1703. [DOI] [PubMed] [Google Scholar]
- 32.MacKichan, J. K., E. C. Gaynor, C. Chang, S. Cawthraw, D. G. Newell, J. F. Miller, and S. Falkow. 2004. The Campylobacter jejuni dccRS two-component system is required for optimal in vivo colonization but is dispensable for in vitro growth. Mol. Microbiol. 54:1269-1286. [DOI] [PubMed] [Google Scholar]
- 33.Medzhitov, R. 2001. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 1:135-145. [DOI] [PubMed] [Google Scholar]
- 34.Morooka, T., A. Umeda, and K. Amako. 1985. Motility as an intestinal colonization factor for Campylobacter jejuni. J. Gen. Microbiol. 131:1973-1980. [DOI] [PubMed] [Google Scholar]
- 35.Murli, S., R. O. Watson, and J. E. Galán. 2001. Role of tyrosine kinases and the tyrosine phosphatase SptP in the interaction of Salmonella with host cells. Cell Microbiol. 3:795-810. [DOI] [PubMed] [Google Scholar]
- 36.Naiki, Y., K. Michelsen, N. Schroder, R. Alsabeh, A. Slepenkin, W. Zhang, S. Chen, B. Wei, Y. Bulut, M. Wong, E. Peterson, and M. Arditi. 2005. MyD88 is pivotal for the early inflammatory response and subsequent bacterial clearance and survival in a mouse model of Chlamydia pneumoniae pneumonia. J. Biol. Chem. 280:29242-29249. [DOI] [PubMed] [Google Scholar]
- 37.Newell, D. G., and A. Pearson. 1984. The invasion of epithelial cell lines and the intestinal epithelium of infant mice by Campylobacter jejuni/coli. J. Diarrhoeal Dis. Res. 2:19-26. [PubMed] [Google Scholar]
- 38.Parkhill, J., B. W. Wren, K. Mungall, J. M. Ketley, C. Churcher, D. Basham, T. Chillingworth, R. M. Davies, T. Feltwell, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Moule, M. J. Pallen, C. W. Penn, M. A. Quail, M. A. Rajandream, K. M. Rutherford, A. H. van Vliet, S. Whitehead, and B. G. Barrell. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665-668. [DOI] [PubMed] [Google Scholar]
- 39.Philpott, D., and S. Girardin. 2004. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol. Immunol. 41:1099-1108. [DOI] [PubMed] [Google Scholar]
- 40.Russell, R. G., M. J. Blaser, J. I. Sarmiento, and J. Fox. 1989. Experimental Campylobacter jejuni infection in Macaca nemestrina. Infect. Immun. 57:1438-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Russell, R. G., M. O'Donnoghue, D. C. Blake, Jr., J. Zulty, and L. J. DeTolla. 1993. Early colonic damage and invasion of Campylobacter jejuni in experimentally challenged infant Macaca mulatta. J. Infect. Dis. 168:210-215. [DOI] [PubMed] [Google Scholar]
- 42.Szymanski, C. M., D. H. Burr, and P. Guerry. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect. Immun. 70:2242-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szymanski, C. M., M. King, M. Haardt, and G. D. Armstrong. 1995. Campylobacter jejuni motility and invasion of Caco-2 cells. Infect. Immun. 63:4295-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szymanski, C. M., R. Yao, C. P. Ewing, T. J. Trust, and P. Guerry. 1999. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 32:1022-1030. [DOI] [PubMed] [Google Scholar]
- 45.Wassenaar, T. M., N. M. Bleumink-Pluym, and B. A. van der Zeijst. 1991. Inactivation of Campylobacter jejuni flagellin genes by homologous recombination demonstrates that flaA but not flaB is required for invasion. EMBO J. 10:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson, R. O., and J. E. Galan. 2005. Signal transduction in Campylobacter jejuni-induced cytokine production. Cell. Microbiol. 7:655-665. [DOI] [PubMed] [Google Scholar]
- 47.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 48.Yao, R., D. H. Burr, P. Doig, T. J. Trust, H. Niu, and P. Guerry. 1994. Isolation of motile and non-motile insertional mutants of Campylobacter jejuni: the role of motility in adherence and invasion of eukaryotic cells. Mol. Microbiol. 14:883-893. [DOI] [PubMed] [Google Scholar]
- 49.Young, V. B., D. B. Schauer, and A. J. Fox. 2000. Animal models of Campylobacter infection, p. 287-301. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, DC.