Abstract
One of the predominant polymicrobial infections of humans is expressed clinically as periodontal disease. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia have been strongly implicated as members of a pathogenic consortium in the etiology of adult periodontitis. In this study we hypothesized that P. gingivalis, T. denticola, and T. forsythia are synergistic in terms of virulence potential and induce chronic periodontal inflammation that leads to alveolar bone resorption in a polymicrobial infection in rats. Groups of rats were infected with either P. gingivalis, T. denticola, or T. forsythia in monomicrobial infections or with all three species in polymicrobial oral infections with or without Fusobacterium nucleatum. PCR analyses of oral microbial samples demonstrated that rats infected with one bacterium were orally colonized by each of the bacteria during the study interval, and increased serum immunoglobulin G (IgG) antibody levels substantiated the interaction of the host with the infecting bacteria. PCR analyses of the rats with polymicrobial infections demonstrated that most rats were infected with P. gingivalis, T. denticola, and T. forsythia as a consortium. Furthermore, all rats exhibited a significant increase in the level of IgG antibody to the polymicrobial consortium. Radiographic measurement of alveolar bone resorption showed that rats infected with the polymicrobial consortium with or without F. nucleatum exhibited significantly increased alveolar bone resorption compared to the resorption in uninfected control rats, as well as the resorption in rats infected with one of the microbes. These results documented that P. gingivalis, T. denticola, and T. forsythia not only exist as a consortium that is associated with chronic periodontitis but also exhibit synergistic virulence resulting in the immunoinflammatory bone resorption characteristic of periodontitis.
Periodontal diseases, which affect an estimated 116 million Americans (39), produce complex immunoinflammatory lesions, and no single bacterial species is responsible for triggering the destructive host responses. While periodontal diseases are polymicrobial and reflect sequential colonization by a broad array of bacteria in the transition from a healthy subgingival biofilm to a diseased subgingival biofilm (5, 25, 30, 40, 41), the molecular mechanisms and synergism among the genera and species in the disease are not well understood. Three of the periodontal pathogens, Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia, are commonly coisolated or identified in subgingival biofilm samples from adult periodontitis lesions (11, 40, 41). Increased numbers of several other bacterial species, including Campylobacter rectus, Eubacterium nodatum, Prevotella spp., Peptostreptococcus micros, and Streptococcus intermedius, as well as Fusobacterium nucleatum, have also been observed in deep periodontal pockets, and the presence of these organisms is positively correlated with increased probing depth and progressive periodontal ligament attachment loss (22, 30, 40, 41). These bacteria generally represent commensal opportunistic pathogens found at low levels at healthy sites; however, under the appropriate microenvironmental conditions, currently not well defined, they contribute to triggering periodontal disease progression (40, 41).
The human subgingival crevice is a habitat for a complex microbiota and contains more than 600 bacterial species or phylotypes (1, 24). In particular, the number of gram-negative anaerobic species has been shown to increase in subgingival plaque as the severity of periodontal disease increases, reflecting an altered microenvironment and/or contributing to modulation of the milieu with tissue destruction (40). In studies over the last three decades workers have attempted to focus on the microbial ecology of the pathogenic biofilm and to associate specific bacteria or bacterial consortia with progressing disease (15, 30, 40). Socransky et al. (41) described five major complexes observed in subgingival plaque; the “red complex” consisted of P. gingivalis, T. forsythia, and T. denticola, and the level of this complex was commonly elevated in patients with severe periodontitis (38, 40). Each of these species has unique physiological features and an array of potential virulence determinants that could contribute to its ability to colonize disease sites and elicit tissue destruction consistent with periodontitis (15, 16). While not directly linked to this “red complex,” F. nucleatum coaggregates with many oral bacteria, acting as a bridge between early colonizers and late colonizers in the oral cavity (23, 30, 40), contributes to specific interbacterial adhesion and virulence (22), and exhibits biologic activities that elicit alveolar bone resorption (46). However, the mechanisms of interaction of these species as a consortium in the subgingival sulcus and whether they have synergistic pathogenic potential in progressing periodontitis are not known.
Modeling human periodontal disease in animals has been a daunting task due to the host range limitations of the bacterial species, the complex polymicrobial etiology, and challenges in cultivation of fastidious oral anaerobic bacteria (3). Monobacterial periodontal infections primarily using P. gingivalis, Actinobacillus actinomycetemcomitans, and T. forsythia have been studied using rats (12, 35) and mice (4, 36). However, to our knowledge, there has been no study to establish a polymicrobial periodontal disease model in rodents, and this has been due to a lack of knowledge concerning polymicrobial adhesion, metabolism, nutritional interactions, potential synergism, and the requirement for sequential coinfections in vivo (3).
In this study we developed a rat model to examine polymicrobial periodontal disease using P. gingivalis, T. denticola, and T. forsythia as members of a prototype consortium and examined the colonization and infection characteristics of these organisms and their synergistic virulence interactions. We hypothesized that P. gingivalis, T. denticola, and T. forsythia exhibit synergistic virulence for induction of chronic periodontal inflammation leading to alveolar bone resorption, which can be used as a model of polymicrobial disease in rats. Extension of this model should allow future studies that could focus on specific microbial interactions along with detailed investigations of patterns of host responses in gingival tissues that result in the immunoinflammatory lesions of periodontitis.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacteria used in this study were P. gingivalis 381, T. denticola ATCC 35404, T. forsythia ATCC 49307, and F. nucleatum ATCC 49256. P. gingivalis strain 381 was chosen due to its known role in alveolar bone resorption in adult periodontitis and its proven ability to colonize the oral cavities of rodents (12, 19, 21). All bacterial strains were grown under anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37°C in a Coy anaerobic chamber. All media (blood agar and liquid broth) were fully reduced for 24 to 48 h prior to inoculation with bacteria. P. gingivalis was grown for 3 days on CDC anaerobic 5% sheep blood agar plates (Remel, Lenexa, KS), and bacteria were scraped from the agar surface using sterile cotton swabs soaked in mycoplasma broth and suspended in mycoplasma broth. For use as a whole-cell antigen to coat enzyme-linked immunosorbent assay (ELISA) plates, a P. gingivalis culture was grown in mycoplasma broth, enumerated, and resuspended in reduced transport fluid (17). T. forsythia was grown in Trypticase soy agar II basal medium supplemented with yeast extract, Phytone peptone, sheep blood (5%), and N-acetylmuramic acid (NAM) (27, 37) for 3 days, and bacteria were scraped from the agar surface and resuspended in NAM broth. T. denticola was grown in GM-1 broth for 48 to 72 h, log-phase cultures were harvested by centrifugation at 9,000 × g for 15 min at 4°C, and the pellets were resuspended in broth (18, 20). F. nucleatum was grown for 2 days on blood agar plates, and bacteria were scraped from the agar surface and suspended in mycoplasma broth. The amount of each bacterium was determined by counting in a Petroff-Hausser bacterial counting chamber, and the organisms were resuspended at a concentration of 2 × 1010 bacteria per ml. Culture purity and sterility were determined by plating an aliquot of diluted bacteria on blood agar (P. gingivalis), by culturing on NAM plates (T. forsythia), by examination by phase-contrast microscopy (T. denticola), and by Gram staining (19, 20). To obtain whole-cell antigens to coat ELISA plates, a T. forsythia culture was also grown in NAM broth (27, 36).
Monomicrobial and polymicrobial inocula.
For oral monomicrobial infection, each bacterium (2 × 1010 cells per ml) was mixed with an equal amount of sterile 2% carboxymethyl cellulose (CMC) (Sigma, St. Louis, MO) and vortexed at approximately 200 to 300 rpm using the continuous mode for 1 to 2 min inside an anaerobic chamber, and 1 ml was used for infection (1010 cells per ml) by oral gavage. For polymicrobial infection, members of the P. gingivalis-T. denticola-T. forsythia consortium were prepared individually as described above. For oral polymicrobial infection, P. gingivalis was gently mixed with an equal volume of T. denticola, and the organisms were allowed to interact for 5 min; subsequently, T. forsythia was added to the tubes containing P. gingivalis and T. denticola, and the bacteria were mixed gently for 1 to 2 min and allowed to interact for an additional 5 min. An equal volume of sterile 2% CMC was added and mixed thoroughly, and 1 ml (3.3 × 109 cells of P. gingivalis per ml, 3.3 × 109 cells of T. denticola per ml, and 3.3 × 109 cells of T. forsythia per ml) was administered by oral gavage. Similarly, a polymicrobial consortium with F. nucleatum was prepared as described above. After preparation of the P. gingivalis-T. denticola-T. forsythia consortium, F. nucleatum was added and incubated in the anaerobic chamber for approximately 5 min. An equal volume of sterile 2% CMC was added to the consortium and mixed thoroughly, and 1 ml (2.5 × 109 cells of P. gingivalis per ml, 2.5 × 109 cells of T. denticola per ml, 2.5 × 109 cells of T. forsythia per ml, and 2.5 × 109 cells of F. nucleatum per ml) was administered by oral gavage. The bacterial culture growth phase, viability, counts, interaction times, suspension medium, infection dose, and infection procedures were all standardized; i.e., the same preparation and infection protocols were used for all infections throughout the study.
Rat procedures.
Rats are often used in models of experimental periodontitis because it is possible to evaluate clinical disease, strain differences reflecting underlying genetic variation, and the history of the ability to colonize the rat oral cavity with human pathogens (12, 14, 35). The protocol and all rat infection procedures used in this study were approved by the Institutional Animal Care and Use Committee of the University of Kentucky. Female Sprague-Dawley rats (8 to 9 weeks old; Harlan, Indianapolis, IN) were maintained in groups housed under microisolator conditions, were fed standard powdered chow (Teklad Global 18% protein rodent diet 2918; Harlan, Madison, WI), were given H2O ad libitum, and were kept at 25°C with alternating 12-h periods of light and darkness.
Rat oral infections.
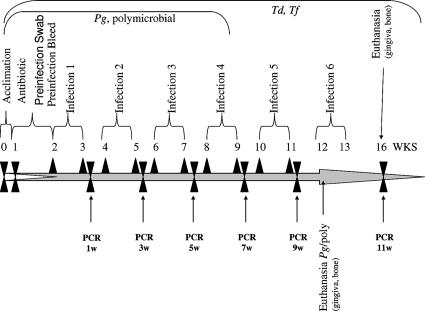
All rats were given kanamycin (20 mg) and ampicillin (20 mg) daily for 4 days in the drinking water (19, 35), and the oral cavity was swabbed with 0.12% chlorhexidine gluconate (PerioGard; Proctor and Gamble, Cincinnati, OH) mouth rinse (19, 35) to suppress the native oral microbiota. Rats were randomly distributed into groups, and this was followed by oral infection. The monomicrobial inocula were administered by oral gavage using approximately 1010 cells per ml of P. gingivalis on five consecutive days in four alternate weeks or using 1010 cells per ml of T. denticola or T. forsythia on five consecutive days in six alternate weeks to maintain stable oral infections in the rats over a 12- to 16-week period (Fig. 1). The polymicrobial inocula were prepared as described above and used for infection by oral gavage on four consecutive days in four alternate weeks.
FIG. 1.
Schematic diagram illustrating the experimental design, including rat acclimation, antibiotic treatment, PerioGard oral swabbing, preinfection oral microbial sample collection, monomicrobial infection (four to six infections) and polymicrobial infection (four infections), oral microbial sample collection (four to six times), PCR analysis, euthanasia, and gingival tissue and alveolar bone collection. For detailed information see Materials and Methods. Pg, P. gingivalis; Td, T. denticola; Tf, T. forsythia.
Oral microbial sampling.
There is an inherent difficulty in actually sampling subgingival biofilms colonizing rodent teeth in model systems of periodontal disease. Moreover, we wanted to monitor the colonization or infection with minimal disruption of the biofilms that developed to enhance the polymicrobial disease outcomes. Thus, the microbial samples collected were obtained by standard methods of oral swabbing. This approach was supported by the fact that in rodents and humans most subgingival bacteria can also be detected in saliva (34), as well as on other surfaces in the oral cavity (e.g., mucosa and tongue) (1). Oral microbial samples from isoflurane-anesthetized rats (anesthetic effect in 1 to 2 min) were collected using sterile cotton swabs gently rubbed over the molar and premolar teeth before and after infection. A total of four postinfection microbial samples were collected the following week from all rats infected with P. gingivalis alone, and six microbial samples were collected from all rats infected with T. denticola alone or T. forsythia alone. Similarly, four postinfection microbial samples were collected the following week from all rats infected with the polymicrobial consortium (Fig. 1). The microbial samples collected on cotton swabs were gently swirled with a circular motion four or five times, the swabs were squeezed, and the contents were suspended in 300 μl of Tris-EDTA buffer and stored at −20°C until DNA isolation. Rats were euthanized with an overdose of carbon dioxide and cervical dislocation as recommended by the American Veterinary Medical Association. Since previous studies clearly showed that naïve Sprague-Dawley rats have no immunoglobulin G (IgG)-specific antibodies to the human oral pathogens P. gingivalis, T. denticola, T. forsythia, and F. nucleatum (19), blood was collected by cardiac puncture after euthanasia and allowed to clot at 4°C, and the serum was separated after centrifugation and stored at −20°C. Rat skulls were removed, autoclaved, and defleshed, and maxillae and mandibles were hemisected and trimmed for evaluation of alveolar bone resorption by radiographic analysis. The periodontal disease outcomes (alveolar bone resorption) for groups of rats infected with the individual species or the polymicrobial consortium were compared to the periodontal disease outcomes for control uninfected rats.
Monitoring bacterial colonization and infection by PCR.
DNA was isolated from rat oral microbial samples using a Wizard genomic DNA purification kit (Promega, Madison, WI). The microbial samples from rats infected with the polymicrobial consortium were centrifuged at 18,000 × g for 2 min in a microcentrifuge, and the supernatants were discarded. Three hundred microliters of a nucleus lysis solution was added to each preparation and mixed gently by pipetting. The mixture was incubated for 5 min at 80°C in a water bath and cooled to room temperature, and 1.5 μl of an RNase solution was added; then the mixture was incubated at 37°C for 30 min and cooled to room temperature. One hundred microliters of a protein precipitation solution was added, the mixture was vortexed, and the sample was kept in an ice bath for 5 min. The mixture was centrifuged at 18,000 × g for 3 min, the supernatant was transferred to a microtube containing 300 μl of 100% isopropanol, mixed by inversion, and centrifuged again for 2 min, and the supernatant was discarded. The pellet was washed in 300 μl of 70% ethanol and centrifuged at 18,000 × g for 2 min, and the supernatant was discarded. The pellet was air dried and resuspended in 50 μl of DNase- and RNase-free water. Finally, the extracted DNA was stored at −20°C. The standard genomic DNA of P. gingivalis, T. denticola, T. forsythia, and F. nucleatum were also extracted by using the same procedure from 24- to 72-h pure cultures. Subsequently, PCR was performed using 10 μl of DNA in a 30-μl (final volume) mixture containing 10 pM primer, each deoxynucleotide triphosphate at a concentration of 1 mM, and 1.5 mM MgCl2. Two units of Taq DNA polymerase (Invitrogen, Life Technologies) in the manufacturer's buffer was used with an Eppendorf Mastercycler PCR system. The following program was used: 94°C for 5 min, followed by 94°C for 30 s, 52°C (P. gingivalis and T. forsythia), 62°C (T. denticola), or 60°C (F. nucleatum) for 1 min, and 72°C for 1 min for 35 to 40 cycles and then a final cycle of 72°C for 7 min. The 16S rRNA gene species-specific PCR oligonucleotide primers (2, 9, 29, 33) used for all the bacteria are shown in Table 1. After amplification PCR products (10 to 20 μl) were detected by 1.5% agarose gel electrophoresis in Tris-borate-EDTA buffer at 100 V for a maximum of 1 h using a 1-kb plus DNA ladder as a molecular weight marker. The results were documented using the BioDoc-It imaging system (UVP, Upland, CA). The genomic DNA extracted from P. gingivalis, T. denticola, T. forsythia, and F. nucleatum served as positive controls, and a PCR performed with no template DNA was used as the negative control. Each PCR assay with the standard DNA was sensitive enough to detect 0.05 pg of DNA (data not shown). Different numbers of PCR cycles (35 to 40 cycles) were standardized to produce detectable amplicons with the smallest amount of template DNA (0.05 pg).
TABLE 1.
Bacterial species-specific primers used in PCR
| Bacterium | Primer sequence (5′ to 3′) | Strand | Size (bp) | Conditions
|
|
|---|---|---|---|---|---|
| Melting temp (°C) | No. of cycles | ||||
| Porphyromonas gingivalis | 5′ GGT AAG TCA GCG GTG AAA CC 3′ | + | 600 | 55 | 40 |
| 5′ ACG TCA TCC ACA CCT TCC TC 3′ | − | ||||
| Treponema denticola | 5′ TAA TAC CGA ATG TGC TCA TTT ACA T 3′ | + | 860 | 62 | 40 |
| 5′ CTG CCA TAT CTC TAT GTC ATT GCT CTT 3′ | − | ||||
| Tannerella forsythia | 5′ AAA ACA GGG GTT CCG CAT GG 3′ | + | 426 | 55 | 40 |
| 5′ TTC ACC GCG GAC TTA ACA GC 3′ | − | ||||
| Fusobacterium nucleatum | 5′AGA GTT TGA TCC TGG CTC AG 3′ | + | 360 | 60 | 35 |
| 5′ GTC ATC GTG CAC ACA GAA TTG CTG 3′ | − | ||||
Antibody analysis.
Serum from rats infected with one microbe (n = 4 to 9) or with multiple microbes (n = 11) was used to determine IgG antibody titers to P. gingivalis, T. forsythia, T. denticola, and F. nucleatum by a standard ELISA protocol (18, 19), and this provided an additional marker of infection. Briefly, P. gingivalis, T. forsythia, T. denticola, and F. nucleatum were grown in liquid cultures as described above, cells were harvested by centrifugation, and the pellets were washed with sterile phosphate-buffered saline (17). The cells were treated overnight with 0.5% buffered formal saline (FK cells), washed, and diluted to obtain an optical density at 600 nm of 0.3 for use as a coating antigen. Purity and sterility (blood agar plating) were confirmed, and the antigens were stored at −20°C. Rat serum diluted 1:100 was incubated in wells of antigen-coated microtiter plates for 2 h on a rotator at room temperature along with a purified rat IgG standard (ICN/Cappel, Durham, NC) to create a gravimetric curve for antibody quantification. The serum was washed off, and affinity-purified goat anti-rat IgG conjugated to biotin was added to the plates and incubated for 2 h. Streptavidin-alkaline phosphatase (Sigma) was added to the wells, incubated overnight, and washed, and the assay preparation was developed with p-nitrophenolphosphate (Sigma). The assay reactions were stopped by addition of 1 N NaOH, and the plates were evaluated using a Dynex MRX4000 reader and Revelation software for analysis. The titer of rat serum IgG antibody was extrapolated by performing a four-parameter logistic fit curve analysis. Antigen-coated wells in the absence of serum were used as a negative background control. Mean serum IgG antibody titers were derived from triplicate determinations for each rat serum.
Radiographic assessment of alveolar bone resorption.
The hemisected maxillae and mandibles were trimmed to reduce the buccolingual dimensions so that the teeth were close to the radiographic film. This trimming process did not alter the relationship of the teeth to the alveolar bone, as only buccolingual bone not directly in contact with the teeth was trimmed. Each jaw was secured to Kodak Ultra Speed size 2 film using rope wax, and a Planmeca Prostyle Intra X-ray unit was placed at a right angle to the film. Each jaw was radiographed with an exposure time of 0.05 s at a setting of 70 kV and 8 mA. Radiographs were analyzed to determine alveolar bone height, and decreased bone level (i.e., resorption) was the primary outcome parameter of the study. Radiographs projected at a magnification of ×5 were used to obtain linear measurements from the cementoenamel junction to the bone height at mesial and distal interproximal surfaces (two sites per tooth) of each of the two molars and one premolar in each quadrant (19, 31). To ensure comparability of the results, the measurements were determined by investigators blinded to the group designation and were calibrated on a routine basis using a set of standard radiographs from the rats. The sums of alveolar bone resorption (in millimeters) were tabulated and analyzed for intra- and intergroup comparisons (19).
Statistical analyses.
The alveolar bone resorption and IgG antibody data were expressed as means ± standard deviations. The statistical significance of differences between the groups was determined using analysis of variance and the Holm-Sidak post hoc multiple-comparison test (SigmaStat 3.0; SYSTAT Software Inc., Chicago, IL) for normally distributed data. Data that were determined to be nonnormally distributed were analyzed using Kruskal-Wallis analysis of variance for ranks and multiple comparisons adjusted by Dunn's method (SigmaStat 3.0).
RESULTS
Oral monomicrobial infections.
We examined all rats before monomicrobial infection (P. gingivalis, T. denticola, or T. forsythia) or polymicrobial infection (P. gingivalis, T. denticola, and T. forsythia or P. gingivalis, T. denticola, T. forsythia, and F. nucleatum) using bacterium-specific primers in PCR, and all rats were consistently negative for these human periodontal pathogens. We standardized the bacterial inocula and infection protocol (i.e., number of infections) for successful P. gingivalis colonization and infection previously (19); however, T. denticola and T. forsythia, which are fastidious oral pathogens, required six infections during a 16-week monomicrobial infection protocol. Oral microbial samples collected in the weeks after infection (six times) were examined by PCR using appropriate bacterium-specific primers. The PCR results demonstrated that amplicons that were the appropriate sizes for P. gingivalis (600 bp), T. denticola (860 bp), and T. forsythia (426 bp) were present in DNA isolated from rat oral microbial samples following infection with the single-microbe inocula (Table 2). In the three monomicrobial infections, P. gingivalis was found to consistently colonize all rats (n = 7) at three sampling times during the infection period. T. denticola colonized all rats (n = 5) at the time of the fifth sampling, but bacterial DNA was detected at only three of six sampling times. Similarly, T. forsythia colonized all rats (n = 4) at the fifth and sixth sampling times. None of the control uninfected rats were positive for amplicons of P. gingivalis, T. denticola, and T. forsythia when appropriate PCR primers were used.
TABLE 2.
Distribution of oral microbial samples positive for monomicrobial and polymicrobial infections as determined by PCR
| Bacterial infection | Total no. of rats | No. of oral microbial samples positive fora:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
P. gingivalis
|
T. denticola
|
T. forsythia
|
F. nucleatum
|
||||||||||||||||||
| 1 wk | 2 wk | 3 wk | 4 wk | 1 wk | 2 wk | 3 wk | 4 wk | 5 wk | 6 wk | 1 wk | 2 wk | 3 wk | 4 wk | 5 wk | 6 wk | 1 wk | 2 wk | 3 wk | 4 wk | ||
| Monomicrobial infectionsb | |||||||||||||||||||||
| P. ginigvalis 381 | 8c | 7 | 7 | 7 | 3 | ||||||||||||||||
| T. denticola ATCC 35404 | 5 | −d | − | − | 4 | 5 | 4 | ||||||||||||||
| T. forsythia ATCC 49307 | 5c | 1 | − | 1 | 3 | 4 | 4 | ||||||||||||||
| Control uninfected rats | 10 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| Polymicrobial infections | |||||||||||||||||||||
| Expt I (P. ginigvalis + T. denticola + T. forsythia) | 10 | 10 | 6 | 7 | 9 | 10 | 5 | 7 | 8 | 10 | 5 | 8 | 10 | ||||||||
| Expt I (P. ginigvalis + T. denticola + T. forsythia + F. nucleatum) | 11 | 9 | 6 | 7 | 7 | 8 | 5 | 7 | 7 | 6 | 4 | 7 | 8 | 11 | 11 | 7 | 10 | ||||
| Expt II (P. ginigvalis + T. denticola + T. forsythia) | 11 | 10 | 4 | 9 | 11 | 1 | − | − | 11 | 9 | 5 | 10 | 9 | ||||||||
| Control uninfected rats | 8 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||
Total numbers of oral microbial samples that were collected in subsequent weeks (1, 3, 5, 7, 9, and 11 weeks [Fig. 1]) following monomicrobial or polymicrobial infection and were positive as determined by PCR analysis. Control uninfected rat microbial samples were collected periodically and examined for P. gingivalis, T. denticola, T. forsythia, and F. nucleatum using bacterium-specific primers, and all rats were negative for all the species.
Rats were infected for 4 weeks (P. gingivalis) or 6 weeks (T. denticola and T. forsythia) alternatively, and oral microbial samples were collected in the following weeks and analyzed using appropriate bacterium-specific PCR primers along with positive and negative controls.
One rat in each group infected with P. gingivalis or T. forsythia was removed before the end of the experiment due to tumor growth in the neck region.
−, oral microbial samples were negative as determined by PCR analysis.
Oral polymicrobial infection.
Following demonstration of monomicrobial infections with the three periodontal pathogens, we performed experiments to develop a rat model of polymicrobial periodontal disease using P. gingivalis, T. denticola, and T. forsythia as members of a prototype consortium and examined the colonization and infection characteristics and synergistic virulence interactions of these organisms in the rat oral cavity. F. nucleatum was included to examine its potential to enhance “red complex”-mediated polymicrobial periodontal disease in the rat model.
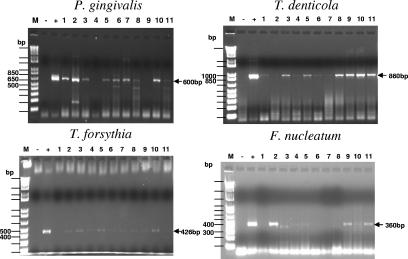
The polymicrobial infection PCR data were derived from two independent experiments with 10 and 11 rats in each study. Figure 2 shows DNA PCR results which demonstrated that amplicons that were the appropriate sizes for P. gingivalis (600 bp), T. denticola (860 bp), T. forsythia (426 bp), and F. nucleatum (360 bp) were obtained from rat oral microbial samples following the third or fourth polymicrobial infection. After polymicrobial infection (P. gingivalis, T. denticola, and T. forsythia), P. gingivalis was detected in 60 to 100% of the rats (experiment I) at the four sampling times (Table 2). Oral microbial samples from all rats were positive for P. gingivalis, and 8 of 10 rats were positive at all three sampling times (individual rat data are not shown). Similarly, T. denticola colonized all rats, although T. denticola was detected in 50 to 100% of the samples collected throughout the study (experiment I). T. denticola DNA was routinely detected at two of four sampling times (experiment II). T. forsythia was also detected in all rats, generally at all four sampling times throughout the infection period. These polymicrobial PCR results indicated that all three periodontal pathogens comprising the polymicrobial consortium were able to colonize the rats during the 12-week study period.
FIG. 2.
PCR analysis of oral microbial samples from rats infected with a polymicrobial inoculum (P. gingivalis, T. denticola, and T. forsythia or P. gingivalis, T. denticola, T. forsythia, and F. nucleatum). Agarose (1.5%) gels contained PCR products from reactions with P. gingivalis, T. denticola, T. forsythia, and F. nucleatum primers. Lane M, 1 Kb Plus DNA ladder; lane −, negative control containing the appropriate bacterial primer but no target DNA; lane +, positive control; lanes 1 to 11, oral microbial samples from individual infected rats. For P. gingivalis 381, a 600-bp amplicon was obtained and 9 of 11 infected rats were positive for P. gingivalis after the third infection. For T. denticola ATCC 35404, a 860-bp amplicon was obtained and all 11 infected rats were positive for T. denticola after the fourth infection (a few bands were faint). For T. forsythia ATCC 43037, a 426-bp amplicon was obtained and 9 of 11 infected rats were positive for T. forsythia after the fourth infection (a few bands were faint). For F. nucleatum ATCC 49256, a 360-bp amplicon was obtained and 7 of 11 infected rats were positive for F. nucleatum after the third infection (a few bands were faint).
For the polymicrobial consortium that included F. nucleatum (P. gingivalis, T. denticola, T. forsythia, and F. nucleatum), P. gingivalis, T. denticola, T. forsythia, and F. nucleatum were observed in 80% of the samples during the infection period (Table 2). During the sampling periods F. nucleatum was detected in 64 to 100% of the rats and P. gingivalis was detected in 55 to 82% of the rats. Similarly, T. denticola and T. forsythia colonized all rats and were detected in 40 to 75% of the rats at all sampling times.
Serum IgG antibody in oral infections.
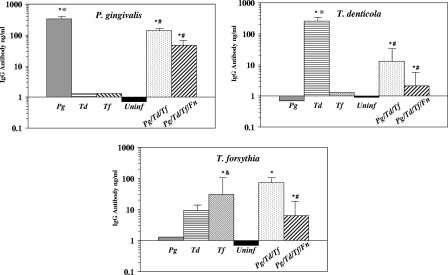
To obtain additional documentation of oral infection with P. gingivalis, T. denticola, and T. forsythia, we evaluated the levels of specific IgG antibody in rat sera collected at the end of the 12- to 16-week infection period (Fig. 3). All rats in the P. gingivalis-infected group had significantly elevated levels of IgG antibody (P < 0.001) compared to the levels in uninfected control rats. Similarly, all rats infected with T. denticola or T. forsythia produced levels of IgG antibody that were significantly greater (P < 0.001) than the levels in uninfected control rats. Of the three species, P. gingivalis induced the highest IgG levels (328 ± 163), while infection with T. forsythia alone induced the lowest levels of antibody (30 ± 19). Generally, the detection and levels of antibody in the serum paralleled the frequency of detection of the individual bacteria in the oral microbial samples.
FIG. 3.
Serum IgG antibody levels in serum from rats (collected at end of a 12- to 16-week infection) following monomicrobial infection (n = 4 to 9) or polymicrobial infection (n = 11). The graphs show the results for IgG antibody reactive with each of the three species of bacteria. The bars indicate the mean antibody levels in serum from rats orally infected with the individual bacteria or with polymicrobial consortia or from uninfected controls. The error bars indicate one standard deviation from the mean. An asterisk indicates that a value is significantly different (P < 0.001) than the value for uninfected controls or for antibody in serum from rats infected with a different microorganism. A number sign indicates that values for the monomicrobial and polymicrobial infection groups are significantly different (P < 0.001 to P < 0.02). An at symbol indicates that the antibody levels are significantly greater (P < 0.001) than the responses to T. forsythia in the rats infected with one species. An ampersand indicates that the antibody level is significantly greater (P < 0.01) than the level of antibody to T. denticola in rats infected with T. forsythia. Pg, P. gingivalis; Td, T. denticola; Tf, T. forsythia; Fn, F. nucleatum; Uninf, uninfected.
To document the polymicrobial infection with the bacterial consortium, we evaluated specific IgG immune responses to the individual pathogens (Fig. 3). All rats in the polymicrobial infection groups had elevated levels of serum IgG antibodies to P. gingivalis and T. forsythia compared to the levels in uninfected control rats. Seventy-three percent of the rats (8 of 11) produced detectable serum antibody to T. denticola, and the overall group IgG antibody levels were significantly elevated compared to the levels in uninfected rats. The IgG antibody levels for the polymicrobial infection groups (with or without F. nucleatum) were significantly lower than the levels in rats infected with P. gingivalis alone (328 ± 163 versus 141 ± 67 or 48 ± 25; P < 0.001), with T. denticola alone (255 ± 226 versus 13 ± 19 or 2.3 ± 3.0; P < 0.004), or with T. forsythia alone (30 ± 19 versus 6.4 ± 3.5; P < 0.02) (Fig. 3). Only the levels of antibody to T. forsythia in a polymicrobial infection without F. nucleatum were elevated compared to the levels in rats infected with T. forsythia alone (72 ± 26 versus 30 ± 19; P < 0.008). F. nucleatum did not increase the serum IgG antibody levels in the context of a polymicrobial infection of rats.
Alveolar bone resorption.
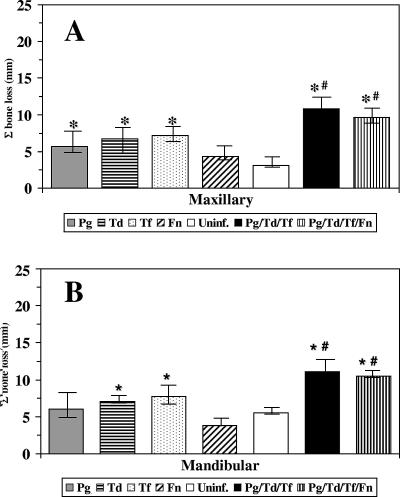
The ability to confirm a monomicrobial or polymicrobial infection with the oral pathogens enabled us to examine the effect on maxillary and mandibular alveolar bone resorption as the primary outcome in addressing the potential virulence synergism of oral microbial consortia in periodontal disease progression in rats. Monomicrobial infections with P. gingivalis, T. denticola, and T. forsythia elicited significant maxillary and mandibular alveolar bone resorption compared to the results for uninfected control rats (Fig. 4A and B). In contrast, infection with F. nucleatum alone did not induce significant maxillary or mandibular bone resorption compared to the results for uninfected control rats. The data show that P. gingivalis, T. denticola, and T. forsythia each induced significant alveolar bone resorption when they were used alone in this model. Importantly, Fig. 4 also shows that oral infection of rats with the polymicrobial consortium containing P. gingivalis, T. denticola, and T. forsythia with or without F. nucleatum induced significantly increased maxillary (P < 0.001) and mandibular (P < 0.001) alveolar bone resorption compared to the results observed with any of the monomicrobial infections. Inclusion of F. nucleatum in the polymicrobial challenge infection did not result in synergism with the P. gingivalis-T. denticola-T. forsythia consortium to induce enhanced alveolar bone resorption.
FIG. 4.
Maxillary and mandibular alveolar bone resorption in rats following monomicrobial infection with P. gingivalis, T. denticola, or T. forsythia or infection with the polymicrobial consortium containing P. gingivalis, T. denticola, and T. forsythia with or without F. nucleatum. Each bar indicates the mean alveolar bone resorption for two sites per tooth and three teeth in each quadrant for 4 to 11 rats per group. The error bars indicate one standard deviation from the mean. An asterisk indicates that a value is significantly different from the value for uninfected rats (P < 0.01), and a number sign indicates that there is a significant difference between a monomicrobial infection and a polymicrobial infection (P < 0.01). (A) Maxillary alveolar bone resorption in rats. (B) Mandibular alveolar bone resorption in rats. Pg, P. gingivalis; Td, T. denticola; Tf, T. forsythia; Fn, F. nucleatum; Uninf., uninfected.
DISCUSSION
Using an experimental polymicrobial periodontal disease model, in this study we explicitly documented colonization and infection by a consortium of oral microorganisms, induction of a specific systemic IgG immune response to the infecting pathogens, and stimulation of maxillary and mandibular alveolar bone resorption in rats. We also documented, for the first time, that synergistic periodontal disease is initiated by a polymicrobial consortium consisting of P. gingivalis, T. denticola, and T. forsythia, compared to monomicrobial infections with any of the individual bacteria. This consortium has been routinely identified as a hallmark of the pathogenic biofilm in deep periodontal pockets in chronic adult periodontitis (30, 40, 41). However, the previous observations generally were based on an epidemiologic approach showing an association among these species with no direct data demonstrating altered virulence attributable to the consortium infection. Synergy of infectious agents is a concept that has been reported in the medical literature for several decades, and the interactions include pathogenic interactions between anaerobes and aerobes, between bacteria and viruses, and between bacteria and parasites (3, 5-7, 28, 32). Oral bacterial synergism in the progression from periodontal health to disease has been suggested in the dental research literature for nearly three decades, but no in vivo studies have documented bacterial synergism due to the inherent complexity of subgingival biofilms (5, 6, 30, 38).
Socransky and colleagues (41) published seminal data which showed the presence and levels of 40 subgingival taxa in 13,261 human subgingival plaque samples from 185 subjects and which were obtained using whole genomic DNA probes and DNA-DNA checkerboard hybridization. These authors suggested that five major complexes (“red complex,” “orange complex,” “yellow complex,” “green complex,” and “violet complex”) were consistently observed in subgingival plaque samples obtained from healthy subjects or subjects with adult periodontitis. One complex (the “red complex”) contained P. gingivalis, T. forsythia, and T. denticola as specific bacterial components; the levels of these bacteria were elevated in patients with adult periodontitis and positively correlated with pocket depth and bleeding on probing. Several species in the complexes are closely associated with each other, and the species belonging to the “red complex” are generally not detected in the absence of members of the “orange complex,” represented by F. nucleatum as a bridging species in the progression of subgingival biofilm maturation. Such relationships can be explained through antagonistic and synergistic physiologic mechanisms, as well as environmental selection.
Increasing clinical evidence supports the concept that bacterial interactions and selected bacterial consortia are essential for both the maintenance of oral health and periodontal disease progression. The ability of bacteria to act in concert to enhance pathogenicity (i.e., synergy) has been suggested to be due to cognate interactions (bacterial coaggregation), physiologic dependency (metabolic nutritional interrelationships), microbial inhibitory mechanisms (bacteriocin and lantibiotics), gene regulation, and products that undermine the host protective response or exacerbate the inflammatory response (5-7, 28, 32). Accordingly, we have initiated in vivo studies to obtain an understanding of the biologic mechanisms contributing to the virulence of the bacterial consortia. We demonstrated previously that mixed infections with P. gingivalis and F. nucleatum resulted in phlegmonous lesions that were larger than the lesions observed with a monomicrobial infection, although it appeared that lesion spread could be significantly decreased by creating a coaggregating environment (13). Chen et al. (8) demonstrated that a mixed infection with P. gingivalis and A. actinomycetemcomitans had a synergistic effect on pathogenicity. We examined the synergistic virulence of T. denticola and P. gingivalis as a mixed infection and observed that at low P. gingivalis challenge doses, T. denticola significantly enhanced the virulence of P. gingivalis compared with the virulence when P. gingivalis was used alone (18). Furthermore, T. forsythia produced relatively small localized abscesses at the sites of monoinfection, and coinfection with P. gingivalis or F. nucleatum produced larger lesions, suggesting that there was pathogenic microbial synergism (42, 44). However, periodontopathic bacterial synergistic virulence has not been examined in periodontal disease models.
A range of putative virulence determinants, including proteinases, have been identified in P. gingivalis, T. denticola, and T. forsythia, although there is little understanding of their contribution to pathogenesis in polymicrobial infections in an experimental periodontal disease model. Our previous studies also showed that when F. nucleatum alone was used for infection, it was a strong inducer of osteoclasts, leading to significant murine calvarial bone resorption compared with the resorption observed with P. gingivalis and C. rectus (46). In the present report we describe our ability to orally infect rats with individual members of the “red complex” consortium and to demonstrate for the first time an oral polymicrobial infection with this pathogenic consortium. This was verified by using molecular approaches to document colonization and infection, by identifying the induction of serum IgG antibodies specific for the species, and by showing that infection-induced alveolar bone resorption occurred.
Monomicrobial oral infections in rats suggested that P. gingivalis had a greater ability to colonize and infect the oral cavity when four alternate weekly infection schedules (20 inoculations) during the 12-week study were used to establish a stable infection. In contrast, Yoshida-Minami et al. (45) infected rats 42 times with a clinical isolate of P. gingivalis to demonstrate alveolar bone loss, but they were unable to recover P. gingivalis throughout the study. We have also shown that infecting rats 15 times with P. gingivalis over a similar interval resulted in consistent detection of the microorganism in oral microbial samples (19). In contrast, it appeared that it was more difficult for T. denticola alone or T. forsythia alone to stably colonize and infect the rat oral cavity, and six infections using a similar alternate week infection schedule were required to establish a stable oral infection.
While differences in alveolar bone resorption levels were observed following with infection P. gingivalis alone, with T. denticola alone, and with T. forsythia alone due to differences in the infection regimen and experimental interval, we could not easily compare the levels of alveolar bone resorption obtained with these individual bacteria. Historically, P. gingivalis has been most frequently associated with human periodontitis and with a plethora of potential virulence determinants (15, 16) and has often been purported to be the major player in induction of alveolar bone resorption (40, 41). Nevertheless, both T. denticola and T. forsythia individually induced significant levels of alveolar bone resorption in rats. These observations were of interest due to parallel studies that we are conducting on polymicrobial infections using a murine calvarial model of inflammation and bone resorption. In particular, we observed that localized P. gingivalis infection had the greatest effect on transcriptome profiles in inflamed soft tissues, while both T. denticola and T. forsythia had greater effects on altering gene expression patterns in the underlying calvarial bone (10). Moreover, the P. gingivalis infection appeared to generally down-regulate genes related to host inflammatory and defense responses (10). Importantly, in testing our hypothesis, oral infection with the polymicrobial consortium containing P. gingivalis, T. denticola, and T. forsythia with or without F. nucleatum significantly increased maxillary and mandibular alveolar bone resorption compared to the resorption observed with any of the monomicrobial infections. This clearly supported the hypothesis that there is in vivo synergism in activation of the alveolar bone resorption process in rats.
The serum IgG antibody data for monomicrobial infections indicated that P. gingivalis is highly effective in colonization and/or is highly antigenic in rats compared to both T. forsythia and T. denticola. In addition, the levels of IgG antibodies to individual species were generally higher in the rats infected with one microbe than in the rats infected with multiple microbes. The antibody responses demonstrated that there is substantial specificity for each of the infecting species. However, we observed an increase in the level of serum IgG antibody to T. denticola following infection with T. forsythia alone. While the level of this “nonspecific” IgG antibody was greater than the level in uninfected controls, it was approximately fivefold lower than the homologous IgG antibody response to T. forsythia. This could be interpreted in different ways; these bacteria could share some epitopes, or alternatively, the increased oral inflammation and disease process initiated by T. forsythia could enable a normal oral microbe resident of rats to induce a cross-reacting response. Our microbiological data demonstrated unequivocally that T. denticola is not normally present in the oral microbiota of rats. The polymicrobial oral infections elicited somewhat different profiles for serum IgG antibodies in rats. First, P. gingivalis still elicited the highest level of IgG antibody, while the levels of T. denticola antibodies were low and only about three-quarters of the rats exhibited a response greater than the response of uninfected controls. Moreover, in general, the levels of serum IgG antibodies to all the bacteria in the consortium infections were lower than the levels detected in monomicrobial infections. This difference occurred in the presence of enhanced maxillary and mandibular alveolar bone resorption, suggesting that the host response to the polymicrobial infection was altered by the bacterial consortium. The altered responses could have been due to a lower colonization capacity of the individual species in the polymicrobial consortium and/or to a decreased ability to expand in the oral cavity during the infection, thus resulting in a lower level of antigenic challenge. However, as observed from the clinical presentation of the rats, significantly enhanced alveolar bone resorption occurred with the consortia, favoring the interpretation that polymicrobial infections may actually modulate the adaptive host responses, leading to more effective evasion of protective immune responses.
The literature is still somewhat limited with respect to polymicrobial infections as related to microbial synergism in pathogenic outcomes, as well as alterations in host responses to polymicrobial infections and to individual pathogens. A recent study of a polymicrobial infection in a mouse model of type 2 diabetes demonstrated that Escherichia coli exhibited strong synergy with Bacteroides fragilis, but not with Clostridium perfringens, in gastrointestinal infections. C. perfringens and B. fragilis exhibited moderate synergy with each other, but only in young mice (28). Our results demonstrated that that there was clear synergy for virulence in alveolar bone resorption in rats following infection with the pathogenic consortium.
Recent in vitro observations showed that there was microbial synergism for biofilm formation between P. gingivalis and T. denticola, between P. gingivalis and T. forsythia, between T. denticola and T. forsythia, and between T. forsythia and F. nucleatum (26, 37, 43). The molecular mechanisms for in vivo synergistic polymicrobial periodontal disease induction may be related to enhancement of expression of the virulence of individual bacteria by the cooperative abilities of the other members of a consortium. Alternatively, the synergism may actually reflect a combination of similar virulence determinants in the bacteria that act in concert to enhance inflammatory alveolar bone resorptive processes. Finally, the increased disease may be due to altered host defense processes that are marginalized by the polymicrobial interactions for triggering host cell responses.
Our findings clearly demonstrated that (i) monomicrobial and polymicrobial colonization and infection by human oral pathogens occurred in the rat oral cavity, (ii) induction of specific serum IgG antibody responses reflected the oral infection and demonstrated that there were variations in infection and/or the immunogenicity of the species, (iii) induction of alveolar bone resorption in rats by P. gingivalis, T. denticola, and T. forsythia and synergistic virulence with a polymicrobial consortium occurred, and (iv) a polymicrobial periodontal disease model in rats was created. The mechanisms of interaction among the species in the subgingival sulcus, which enhance the physiological competitiveness of the organisms in the complex biofilm, integrate the expression of virulence, and contribute to progressing periodontitis, remain undefined. This rat model system should provide an opportunity for further studies to clarify the characteristics and alterations of the host response profiles in gingival tissues that are related to alveolar bone resorption in response to oral polymicrobial infections.
Acknowledgments
This work was supported by USPHS research grant DE-014896 from the National Institute of Dental and Craniofacial Research and by grant RR-020145 from the National Center for Research Resources.
We thank Purnima Kumar for technical assistance with PCR analysis and Howard Kuramitsu (University of Buffalo) for a critical review of the manuscript.
We have no financial conflict of interest.
Editor: V. J. DiRita
Footnotes
Published ahead of print on 8 January 2007.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asai, Y., T. Jinno, H. Igarashi, Y. Ohyama, and T. Ogawa. 2002. Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J. Clin. Microbiol. 40:3334-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakaletz, L. O. 2004. Developing animal models for polymicrobial diseases. Nat. Rev. Microbiol. 2:552-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, P. J., R. T. Evans, and D. C. Roopenian. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035-1040. [DOI] [PubMed] [Google Scholar]
- 5.Brogden, K. A. 2002. Polymicrobial diseases of animals and humans, p. 3-20. In K. Brogden and J. Guthmiller (ed.), Polymicrobial diseases. ASM Press, Washington, DC. [PubMed]
- 6.Brogden, K. A., J. M. Guthmiller, and C. E. Taylor. 2005. Human polymicrobial infections. Lancet 365:253-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brook, I., and R. I. Walker. 1985. Interaction between penicillin, clindamycin or metronidazole and gentamicin against species of clostridia and anaerobic and facultatively anaerobic gram-positive cocci. J. Antimicrob. Chemother. 15:31-37. [DOI] [PubMed] [Google Scholar]
- 8.Chen, P. B., L. B. Davern, J. Katz, J. H. Eldridge, and S. M. Michalek. 1996. Host responses induced by co-infection with Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans in a murine model. Oral Microbiol. Immunol. 11:274-281. [DOI] [PubMed] [Google Scholar]
- 9.Conrads, G., S. E. Gharbia, K. Gulabivala, F. Lampert, and H. N. Shah. 1997. The use of a 16S rDNA directed PCR for the detection of endodontopathogenic bacteria. J. Endod. 23:433-438. [DOI] [PubMed] [Google Scholar]
- 10.Ebersole, J. L., A. Meka, A. Stromberg, C. Saunders, and L. Kesavalu. 2005. Host gene expression in local tissues in response to periodontal pathogens. Oral Biosci. Med. 2:175-184. [Google Scholar]
- 11.Ellen, R. P., and V. B. Galimanas. 2005. Spirochetes at the forefront of periodontal infections. Periodontol. 2000 38:13-32. [DOI] [PubMed] [Google Scholar]
- 12.Evans, R. T., B. Klausen, N. S. Ramamurthy, L. M. Golub, C. Sfintescu, and R. J. Genco. 1992. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch. Oral Biol. 37:813-819. [DOI] [PubMed] [Google Scholar]
- 13.Feuille, F., J. L. Ebersole, L. Kesavalu, M. J. Steffen, and S. C. Holt. 1996. Mixed infection with Porphyromonas gingivalis and Fusobacterium nucleatum in a murine lesion model: potential synergistic effects on virulence. Infect. Immun. 64:2094-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fine, D. H., P. Goncharoff, H. Schreiner, K. M. Chang, D. Furgang, and D. Figurski. 2001. Colonization and persistence of rough and smooth colony variants of Actinobacillus actinomycetemcomitans in the mouths of rats. Arch. Oral Biol. 46:1065-1078. [DOI] [PubMed] [Google Scholar]
- 15.Holt, S. C., and J. L. Ebersole. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72-122. [DOI] [PubMed] [Google Scholar]
- 16.Holt, S. C., L. Kesavalu, S. Walker, and C. A. Genco. 1999. Virulence factors of Porphyromonas gingivalis. Periodontol. 2000 20:168-238. [DOI] [PubMed] [Google Scholar]
- 17.Kesavalu, L., J. L. Ebersole, R. L. Machen, and S. C. Holt. 1992. Porphyromonas gingivalis virulence in mice: induction of immunity to bacterial components. Infect. Immun. 60:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kesavalu, L., S. C. Holt, and J. L. Ebersole. 1998. Virulence of a polymicrobic complex, Treponema denticola and Porphyromonas gingivalis, in a murine model. Oral Microbiol. Immunol. 13:373-377. [DOI] [PubMed] [Google Scholar]
- 19.Kesavalu, L., B. Vasudevan, B. Raghu, E. Browning, D. Dawson, J. M. Novak, M. C. Correll, M. J. Steffen, A. Bhattacharya, G. Fernandes, and J. L. Ebersole. 2006. Omega-3 fatty acid effect on alveolar bone loss in rats. J. Dent. Res. 85:648-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kesavalu, L., S. G. Walker, S. C. Holt, R. R. Crawley, and J. L. Ebersole. 1997. Virulence characteristics of oral treponemes in a murine model. Infect. Immun. 65:5096-5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klausen, B., R. T. Evans, N. S. Ramamurthy, L. M. Golub, C. Sfintescu, J. Y. Lee, G. Bedi, J. J. Zambon, and R. J. Genco. 1991. Periodontal bone level and gingival proteinase activity in gnotobiotic rats immunized with Bacteroides gingivalis. Oral Microbiol. Immunol. 6:193-201. [DOI] [PubMed] [Google Scholar]
- 22.Kolenbrander, P. E. 2000. Oral microbial communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 23.Kolenbrander, P. E., K. D. Parrish, R. N. Andersen, and E. P. Greenberg. 1995. Intergeneric coaggregation of oral Treponema spp. with Fusobacterium spp. and intrageneric coaggregation among Fusobacterium spp. Infect. Immun. 63:4584-4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuramitsu, H. K. 2003. Molecular genetic analysis of the virulence of oral bacterial pathogens: an historical perspective. Crit. Rev. Oral Biol. Med. 14:331-344. [DOI] [PubMed] [Google Scholar]
- 26.Kuramitsu, H. K., W. Chen, and A. Ikegami. 2005. Biofilm formation by the periodontopathic bacteria Treponema denticola and Porphyromonas gingivalis. J. Periodontol. 76:2047-2051. [DOI] [PubMed] [Google Scholar]
- 27.Maiden, M. F. J., C. Pham, and S. Kashket. 2004. Glucose toxicity effect and accumulation of methylglyoxal by the periodontal anaerobe Bacteroides forsythus. Anaerobe 10:27-32. [DOI] [PubMed] [Google Scholar]
- 28.Mastropaolo, M. D., N. P. Evans, M. K. Byrnes, A. M. Stevens, J. L. Robertson, and S. B. Melville. 2005. Synergy in polymicrobial infections in a mouse model of type 2 diabetes. Infect. Immun. 73:6055-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meurman, J. H., J. Wahlfors, A. Korhonen, P. Alakuijala, P. Vaisanen, H. Torkko, and J. Janne. 1997. Identification of Bacteroides forsythus in subgingival dental plaque with the aid of a rapid PCR method. J. Dent. Res. 76:1376-1380. [DOI] [PubMed] [Google Scholar]
- 30.Nishihara, T., and T. Koseki. 2004. Microbial etiology of periodontitis. Periodontol. 2000 36:14-26. [DOI] [PubMed] [Google Scholar]
- 31.Reed, B. E., and A. M. Polson. 1984. Relationships between bitewing and periapical radiographs in assessing crestal alveolar bone levels. J. Periodontol. 55:22-27. [DOI] [PubMed] [Google Scholar]
- 32.Rotstein, O. D., T. L. Pruett, and R. L. Simmons. 1985. Mechanisms of microbial synergy in polymicrobial surgical infections. Rev. Infect. Dis. 7:151-170. [DOI] [PubMed] [Google Scholar]
- 33.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 34.Sakamoto, M., M. Umeda, I. Ishikawa, and Y. Benno. 2000. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 44:643-652. [DOI] [PubMed] [Google Scholar]
- 35.Schreiner, H. C., K. Sinatra, J. B. Kaplan, D. Furgang, S. C. Kachlany, P. J. Planet, B. A. Perez, D. H. Figurski, and D. H. Fine. 2003. Tight-adherence genes of Actinobacillus actinomycetemcomitans are required for virulence in a rat model. Proc. Natl. Acad. Sci. USA 100:7295-7300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma, A., S. Inagaki, K. Honma, C. Sfintescu, P. J. Baker, and R. T. Evans. 2005. Tannerella forsythia-induced alveolar bone loss in mice involves leucine-rich-repeat BspA protein. J. Dent. Res. 84:462-467. [DOI] [PubMed] [Google Scholar]
- 37.Sharma, A., S. Inagaki, W. Sigurdson, and H. K. Kuramitsu. 2005. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol. Immunol. 20:39-42. [DOI] [PubMed] [Google Scholar]
- 38.Simonson, L. G., K. T. McMahon, D. W. Childers, and H. E. Morton. 1992. Bacterial synergy of Treponema denticola and Porphyromonas gingivalis in a multinational population. Oral Microbiol. Immunol. 7:111-112. [DOI] [PubMed] [Google Scholar]
- 39.Slade, G. D., and J. D. Beck. 1999. Plausibility of periodontal disease estimates from NHANES III. J. Public Health Dent. 59:67-72. [DOI] [PubMed] [Google Scholar]
- 40.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38:135-187. [DOI] [PubMed] [Google Scholar]
- 41.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 42.Takemoto, T., H. Kurihara, and G. Dahlen. 1997. Characterization of Bacteroides forsythus isolates. J. Clin. Microbiol. 35:1378-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada, M., A. Ikegami, and H. K. Kuramitsu. 2005. Synergistic biofilm formation by Treponema denticola and Porphyromonas gingivalis. FEMS Microbiol. Lett. 250:271-277. [DOI] [PubMed] [Google Scholar]
- 44.Yoneda, M., T. Hirofuji, H. Anan, A. Matsumoto, T. Hamachi, K. Nakayama, and K. Maeda. 2001. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: involvement of gingipains in a synergistic effect. J. Periodontal Res. 36:237-243. [DOI] [PubMed] [Google Scholar]
- 45.Yoshida-Minami, I., A. Suzuki, K. Kawabata, A. Okamoto, Y. Nishihara, T. Minami, S. Nagashima, I. Morisaki, and T. Ooshima. 1997. Alveolar bone loss in rats infected with a strain of Prevotella intermedia and Fusobacterium nucleatum isolated from a child with prepubertal periodontitis. J. Periodontol. 68:12-17. [DOI] [PubMed] [Google Scholar]
- 46.Zubery, Y., C. R. Dunstan, B. M. Story, L. Kesavalu, J. L. Ebersole, S. C. Holt, and B. F. Boyce. 1998. Bone resorption caused by three periodontal pathogens in vivo in mice is mediated in part by prostaglandin. Infect. Immun. 66:4158-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]