Abstract
Transcriptional profiling and ontology tools were utilized to define the biological pathways of gingival epithelial cells modulated by coculture with the oral commensal Streptococcus gordonii and the opportunistic commensal Fusobacterium nucleatum. Overall, F. nucleatum and S. gordonii perturbed the gingival epithelial cell transcriptome much less significantly than the oral pathogens Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans perturbed the transcriptome, indicating that there was a greater degree of host adaptation by the commensal species (M. Handfield, J. J. Mans, G. Zheng, M. C. Lopez, S. Mao, A. Progulske-Fox, G. Narasimhan, H. V. Baker, and R. J. Lamont, Cell. Microbiol. 7:811-823, 2005). The biological pathways significantly impacted by F. nucleatum and S. gordonii included the mitogen-activated protein kinase (MAPK) and Toll-like receptor signaling pathways. Differential regulation of GADD45 and DUSP4, key components of the MAPK pathway, was confirmed at the protein level by Western blotting. Modulation of the MAPK pathway is likely to affect host cell proliferation and differentiation. In addition, both the MAPK and Toll-like receptor pathways ultimately converge on cytokine gene expression. An enzyme-linked immunosorbent assay of secreted interleukin-6 (IL-6) and IL-8 demonstrated that F. nucleatum induced production of these cytokines, whereas S. gordonii inhibited secretion from the epithelial cells. Stimulation of secretion of proinflammatory cytokines from epithelial cells may reflect the invasive phenotype of F. nucleatum and contribute to the greater pathogenic potential of F. nucleatum than of S. gordonii.
Humans are hosts of a complex and abundant population of microorganisms that colonize the mucosal membranes and skin. This commensal microbiota is an integral component of a complex homeostasis mechanism that impedes the activity of pathogenic microorganisms (6, 24). At optimal composition, commensals can prevent both the attachment and into the multiplication of pathogens and their invasion of epithelial cells and into the circulation. In addition, commensals supply essential nutrients, regulate epithelial development, and contribute to the maturation and maintenance of the immune system (5, 11, 25, 43). Thus, commensal inhabitants have become host adapted during a long evolutionary relationship. Under certain conditions, however, a subset of species can escape host restraint mechanisms and initiate disease, and these species have been called opportunistic commensals. There is increasing evidence that the innate immune system may discriminate among commensals, opportunistic commensals, and overt pathogens, and it has been suggested that this discrimination controls the balance between microbial intrusion and host integrity (21, 40, 50, 51, 55).
More than 700 species, or phylotypes, of bacteria can inhabit the oral cavity (1). Temporally distinct patterns of microbial colonization result in biofilm formation on all surfaces in the oral cavity. On the tooth surfaces, the initial colonizers of the dental plaque biofilm are principally oral streptococci and actinomyces. Establishment of these organisms facilitates the subsequent colonization of additional actinomyces and related gram-positive rods along with gram-negative bacteria, such as Fusobacterium nucleatum. Further maturation is characterized by colonization by gram-negative anaerobes, such as Porphyromonas gingivalis (34, 52). Once colonization of the subgingival area has occurred, organisms shed from the plaque biofilm can interact with host epithelial cells that both have a barrier function and act as sensors of microbial infection (28). While many common oral organisms can adhere to gingival epithelial cells, only a subset of these organisms, including F. nucleatum, Aggregatibacter (Actinobacillus) actinomycetemcomitans, and P. gingivalis, can invade host cells (22, 23, 39, 59). Although it is well established that the bacterial inhabitants of the subgingival crevice are direct precursors of periodontal disease, the oral microbiota includes a spectrum ranging from commensals, such as Streptococcus gordonii, to aggressive pathogens, such as P. gingivalis. Some species, such as F. nucleatum, are located near the center of this spectrum as opportunistic commensals and are frequently found in individuals with good oral health but are also potentially able to contribute to disease (4, 15, 17, 44, 58, 62).
Transcriptional profiling using microarrays provides a way to monitor host cell responses to colonizing microorganisms on a global scale (8, 41). Numerous innate immune factors, for example, have been consistently found to be differentially regulated in host cells infected with pathogenic organisms compared to the regulation in uninfected controls (23, 29, 42). In the oral cavity, the pathogens P. gingivalis and A. actinomycetemcomitans induce widespread changes in the gingival epithelial cell transcriptome that are largely organism specific (23). There have been only a limited number of studies of the transcriptional responses to commensal organisms, and in these studies the focus is almost exclusively on nonoral mucosal ecosystems. For example, a study of the gastrointestinal tract commensal Bacteroides thetaiotaomicron showed it can modulate expression of genes involved in several important intestinal functions, including nutrient absorption, mucosal barrier fortification, xenobiotic metabolism, angiogenesis, and postnatal intestinal maturation (25). Commensal bacterial reconstitution of germfree mice has been shown to up-regulate expression of colonic epithelial cell genes associated with growth, apoptosis, and immune responses (19). In contrast, genes that may participate in extracellular oxidant defense and cellular metabolism were down-regulated by a nonpathogenic bacterial challenge (19). Hence, there is evidence that the host cell transcriptional response can be specific for the infecting organism and that commensals and pathogens can regulate distinct physiological functions in host cells.
In this study we utilized expression microarrays to investigate the transcriptional responses of oral epithelial cells to challenge with the commensal S. gordonii and the opportunistic commensal F. nucleatum. The transcriptional responses induced by these organisms were very similar to each other yet significantly different than the responses reported previously for oral pathogens (23). Among the biological processes altered most significantly in the host cells was the signal transduction pathway associated with mitogen-activated protein kinase (MAPK) and downstream effector molecules, including interleukins. Understanding how the host has adapted to commensals and how barrier cells respond to limit the impact of commensals should provide a mechanistic biological basis for health in the mixed bacterium-human ecosystem of the oral cavity.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
F. nucleatum ATCC 25586 was cultured anaerobically at 37°C in Trypticase soy broth supplemented with yeast extract (1 mg ml−1), hemin (5 μg ml−1), and menadione (1 μg ml−1). S. gordonii DL-1 was cultured anaerobically at 37°C in Todd-Hewitt broth with 0.5% yeast extract and glucose (0.5%).
Epithelial cells.
Human immortalized gingival keratinocytes (HIGK) were originally generated by transfection of primary gingival epithelial cells with E6/E7 from human papillomavirus (45). HIGK were cultured in the presence of 5% CO2 in keratinocyte serum-free medium (Gibco/Invitrogen, Carlsbad, CA) supplemented with 0.05 mM calcium chloride, 200 mM l-glutamine, and an antibiotic-antimycotic (Invitrogen).
Microbe-host cell coculture.
Bacteria in the mid-log phase were harvested and washed by centrifugation and then resuspended in antibiotic-free keratinocyte serum-free medium. HIGK (107 cells) were cocultured with bacteria to obtain a total association (adherent plus invading bacteria) of approximately 100 bacteria per epithelial cell. The numbers of adherent and invading organisms were confirmed in parallel experiments by plate counting (23). After 2 h of incubation at 37°C in the presence of 5% CO2, the HIGK were lysed with Trizol (Invitrogen) prior to RNA extraction. Cocultures were carried out in quadruplicate.
RNA isolation, cDNA-cRNA synthesis, and chip hybridization.
RNA isolation, cDNA synthesis, labeled cRNA synthesis, and chip hybridization were performed as previously described (23). Briefly, total RNA was extracted from Trizol-lysed cells, treated with DNase I, purified, and quantified by using standard methods (QIAGEN, Valencia, CA, and Affymetrix, Santa Clara, CA). cDNA was synthesized by using the Affymetrix protocol (SuperScript double-stranded cDNA synthesis kit; Invitrogen); 5 to 8 μg of total cellular RNA was used as a template to amplify mRNA species for detection. Double-stranded cDNA was purified and used as a template for labeled cRNA synthesis. In vitro transcription was performed using a BioArray high-yield RNA transcript labeling kit (T7; Enzo Life Science, Farmingdale, NY) to incorporate biotinylated nucleotides. cRNA was subsequently fragmented and hybridized on Genechip human genome U133-A oligonucleotide arrays (Affymetrix) with proper controls. RNA samples were not pooled. The microarrays were hybridized for 16 h at 45°C, stained with phycoerythrin-conjugated streptavidin, and washed using the Affymetrix protocol (EukGE-WS2v4) with an Affymetrix fluidics station, and then they were scanned with an Affymetrix GeneChip 3000 scanner.
Microarray data analysis.
Microarray data analysis was performed as previously described (23, 41). Briefly, expression filters were applied to remove Affymetrix controls and probe sets whose signals were not detected in all samples. The signal intensity values in the resulting data set were variance normalized, mean centered, and ranked by their coefficients of variation. Normalization was performed to give equal weight to all probe sets in the analysis, regardless of the order of magnitude of the raw signal intensity. To reduce the confounding effect of background signal variation on the analysis, only the half of the data set exhibiting the most variation across samples was used to perform an unsupervised hierarchical cluster analysis using the Cluster software (16). The resulting heat map and cluster dendrograms were visualized with the Treeview software (16) to determine the extent of characteristic host cell responses to each infection state, defined as identical treatments clustering together. Additional quality control data for the arrays are provided in the supplemental material.
Following the initial assessment of the host cell response to each condition, a supervised analysis was performed to investigate differences in gene regulation among experimental conditions. For this analysis, the raw signal intensities were log transformed for all probe sets that passed the initial expression filters and were correlated using BRB Array Tools (R. Simon and A. Peng-Lam, National Cancer Institute, Rockville, MD). In each supervised analysis, biological replicates were grouped into classes based on their infection states during coculture experiments, and probe sets significant at the P <0.001 level for the class were identified. To test the abilities of the significant probe sets to truly distinguish between the classes, leave-one-out cross-validation (LOOCV) studies were performed. In these LOOCV studies each array was left out in turn and a classifier was derived for the three groups by selecting probe sets significant at a P value of <0.001. The significant probe sets were then used with several prediction models (compound covariate predictor, nearest-neighbor predictor, and support vector machine predictor) to predict the class identity of the array that was left out and not included when the classification model was constructed. The ability of the classifier to correctly predict the class identity of the array that was left out was estimated using Monte Carlo simulations with 2,000 permutations of the data set.
Functional categorization by gene ontology and bioinformatics analyses.
Gene ontology trees were populated using Pathway Express (33), available at http://vortex.cs.wayne.edu/projects.htm.
Immunoblotting.
HIGK were infected with F. nucleatum or S. gordonii as described above, using a time course of 1, 2, or 6 h. Cells were washed three times with phosphate-buffered saline and lysed in radioimmunoprecipitation buffer with proteinase inhibitors (Sigma, St. Louis, MO). Twenty-five micrograms of protein was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA), and blocked with 5% skim milk in Tris-buffered saline-0.1% Tween 20. The membranes were incubated for 1 h with primary antibodies to GADD45α (1:200; Santa Cruz Biotechnology, Santa Cruz, CA), GADD45β (1:200; Santa Cruz Biotechnology), DUSP4 (1:2,000; Abcam, Cambridge, MA), and β-actin (1:10,000; Abcam). After three washes in Tris-buffered saline-0.1% Tween 20, each membrane was reacted with the species-appropriate peroxidase-coupled secondary antibody (1:1,000). Visualization was performed with the enhanced chemiluminescence system (Amersham, Buckinghamshire, United Kingdom). Band intensities were scanned and quantified using the Kodak 1D image analysis software (v.3.6.1).
Detection of cytokines.
Supernatants of HIGK infected with F. nucleatum or S. gordonii were collected and filter sterilized. Interleukin-6 (IL-6) and IL-8 concentrations were determined by an enzyme-linked immunosorbent assay (Quantikine, Minneapolis MN) performed according to the manufacturer's protocol. Experiments were conducted in triplicate.
Microarray data accession numbers.
The array results have been deposited in the GEO repository (http://www.ncbi.nlm.nih.gov/geo/index.cgi) under accession numbers GSM159477 to GSM159488, series GSE6927.
RESULTS
Association of F. nucleatum and S. gordonii with HIGK.
F. nucleatum and S. gordonii exhibited different capacities to invade HIGK. At a concentration of approximately 100 organisms per epithelial cell, 55% of F. nucleatum cells were recovered intracellularly, whereas S. gordonii remained essentially extracellular (99.5%).
Comparison of the transcriptional profiles of HIGK infected with F. nucleatum and HIGK infected with S. gordonii.
All samples of uninfected HIGK, F. nucleatum-infected cells, and S. gordonii-infected cells were used to determine the overall similarity of the transcriptional responses. After elimination of probe sets whose signals were not greater than background levels on all arrays, signal intensity data for the 12,125 probe sets that passed the initial expression filters were included in an unsupervised cluster analysis and supervised class prediction analysis. The unsupervised hierarchical cluster analysis revealed an infection state-dependent host cell transcriptional profile, as biological replicates clustered together (data not shown). In a supervised analysis based on the infection state, at a significance level of P < 0.001, 240 probe sets were differentially expressed. Assuming normality of the data set, the 240 significant genes is 20-fold higher than the 12 probe sets that would be expected by chance at a significance threshold of P < 0.001, given that 12,125 probe sets passed the expression filter. Treeview visualization of the 240 probe sets differentially expressed in the three classes is shown in Fig. 1. The major separation node occurred between uninfected cells and infected organisms, independent of bacterial species, and the differences between the uninfected and infected states exceeded the observable differences between F. nucleatum and S. gordonii, although organism-specific gene signatures could be discerned. Overall, 84% of the probe sets were regulated similarly by F. nucleatum and S. gordonii, which may reflect the degree of adaptation to the host of both these species.
FIG. 1.
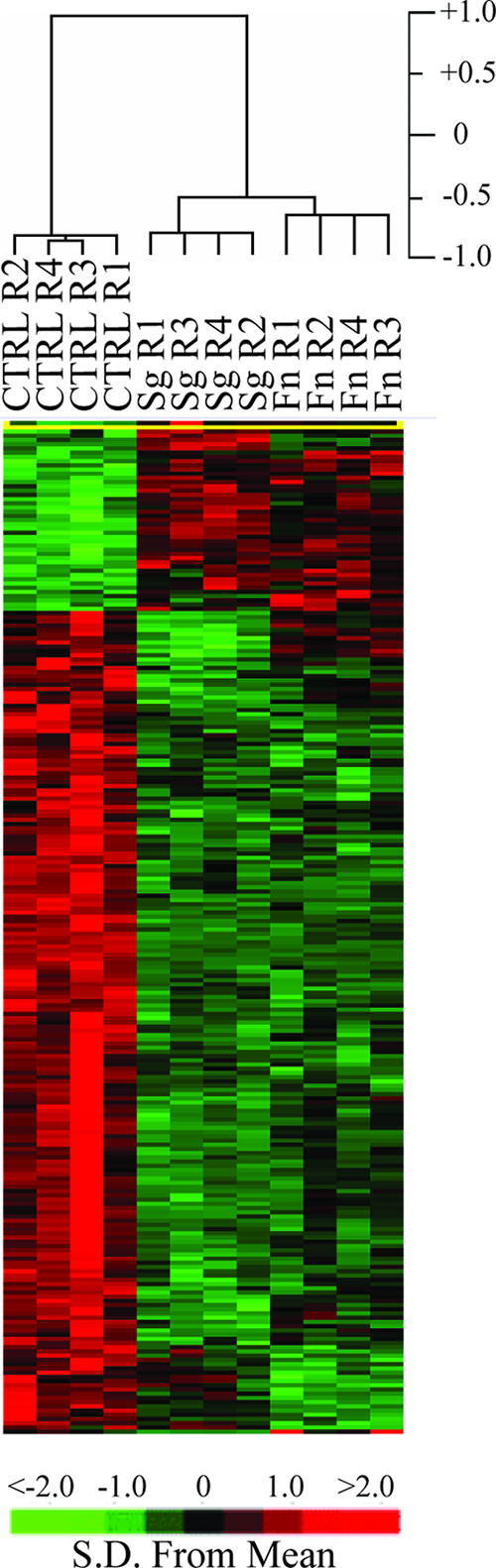
Hierarchical clustering of variance-normalized gene expression data. The expression pattern of the cRNAs analyzed by using microarrays was determined by a supervised analysis of the variance-normalized data set of differentially expressed genes (P < 0.001, BRB ArrayTools) with the algorithm Cluster and was displayed with Treeview. Each row represents an individual gene element on the array, and each column represents the expression states of cRNAs for the challenge condition indicated. Each expression data point represents the relative fluorescence intensity of the cRNA from F. nucleatum-infected cells (columns Fn R1 to Fn R4) or S. gordonii-infected cells (columns Sg R1 to Sg R4) to the fluorescence intensity of the cRNA from uninfected cells (columns CTRL R1 to CTRL R4). The distance matrix used to determine the relatedness of samples through gene expression space was 1 − Pearson's correlation coefficient. The cluster is subdivided into three groups consisting of genes that were repressed (green), genes that were induced (red), and genes whose expression did not change (black). The variation in gene expression for a given gene is expressed as the distance from the mean observation for that gene according to the color scale below the heat map.
Ontology analysis.
While it is becoming increasingly clear that the predictive power of regulation of individual genes is limited due to the extensive interconnectivity among regulatory networks, the assembly of regulated genes into biologically relevant pathways by ontology analysis has greater biological resolution. Therefore, in order to mine the array data for biologically relevant information, an ontology analysis of known metabolic pathways was performed using individual comparisons of the F. nucleatum-infected state or the S. gordonii-infected state with the corresponding baseline uninfected state. A pairwise comparison was performed by using the ontology algorithms described above. For this analysis, signal intensities were renormalized across all samples, and supervised analyses were repeated as described above. In F. nucleatum-infected HIGK, 11,909 genes passed the initial expression filters, while 11,835 genes were analyzed for S. gordonii-infected cells. At a significance level of P < 0.001, 145 genes were differentially expressed in the F. nucleatum-infected cells compared to the expression in uninfected controls, whereas 268 genes were differentially expressed in the S. gordonii-infected cells. At a less stringent significance level, P < 0.05, class prediction analysis revealed 1,917 F. nucleatum-regulated genes and 2,910 S. gordonii-regulated genes. The abilities of probe sets with significance at P < 0.001 or at P < 0.05 to correctly identify treatment groups were confirmed by LOOCV analysis. The classifiers performed flawlessly and correctly predicted the treatment group with 100% accuracy. To populate biological pathways to the maximal extent and thus enhance their predictive power, probe sets at a significance level of P < 0.05 were analyzed by the Pathway Express algorithm (12-14, 31-33). Table 1 shows the most impacted epithelial pathways generated in this analysis in order of their impact factors. Pathways impacted by F. nucleatum or S. gordonii showed considerable overlap, and the MAPK pathway and the Toll-like receptor signaling pathway were two of the top three most affected pathways for both organisms. Thus, recognition of and response to these oral commensals by gingival epithelial cells involve the “classical” system of microbe-associated molecular pattern-pattern recognition receptor binding and signaling propagation through MAPK pathways. Coupled with the absence of a danger signal (46), this interaction may maintain a physiologic balance between the host and the organism.
TABLE 1.
Ontology analysis of epithelial cell pathways impacted by infection with F. nucleatum or S. gordoniia
| Impacted pathwayb | Impact factorc | No. of input genes/no. of pathway genesd |
|---|---|---|
| F. nucleatum-infected cells | ||
| Phosphatidylinositol signaling pathway | 178.648 | 11/79 |
| MAPK signaling pathway | 173.395 | 43/273 |
| Toll-like receptor signaling pathway | 105.509 | 13/91 |
| Regulation of actin cytoskeleton | 91.253 | 31/206 |
| Cell cycle | 75.558 | 20/112 |
| Wnt signaling pathway | 69.173 | 26/147 |
| Cytokine-cytokine receptor interaction | 61.258 | 24/256 |
| Focal adhesion | 55.455 | 25/194 |
| S. gordonii-infected cells | ||
| MAPK signaling pathway | 248.404 | 57/273 |
| Toll-like receptor signaling pathway | 179.365 | 20/91 |
| Phosphatidylinositol signaling pathway | 172.608 | 9/79 |
| Cell cycle | 95.198 | 30/112 |
| Regulation of actin cytoskeleton | 93.062 | 39/206 |
| Wnt signaling pathway | 74.227 | 31/147 |
| Cytokine-cytokine receptor interaction | 72.52 | 29/256 |
| Focal adhesion | 72.117 | 37/194 |
| Tight junction | 64.433 | 21/119 |
| Jak-STAT signaling pathway | 56.993 | 25/153 |
| Apoptosis | 52.94 | 20/84 |
The epithelial cell pathways were determined by Pathway Express (33).
Kyoto Encyclopedia of Genes and Genomes pathways (http://www.genome.jp/kegg/).
The impact factor measures the pathways most affected by changes in gene expression by considering the proportion of differentially regulated genes, the perturbation factors of all pathway genes, and the propagation of these perturbations throughout the pathway. Only pathways with an impact factor greater than 50 are included.
Number of regulated genes in a pathway/total number of genes currently mapped to this pathway.
Correlation between mRNA and protein levels in the MAPK pathway.
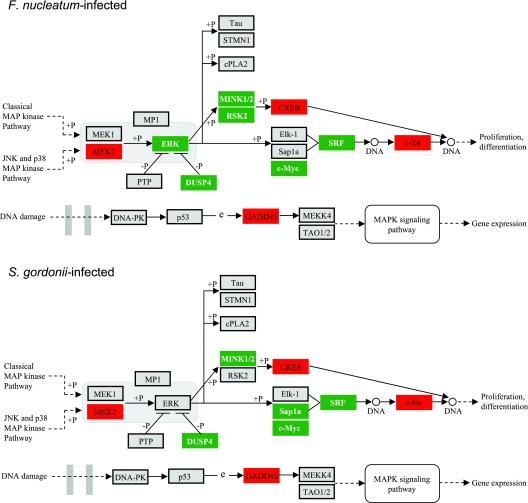
As the MAPK pathway was significantly (P < 0.001) impacted at the transcriptional level by both organisms, we investigated the correlation between mRNA and protein levels for three of the most statistically significantly regulated genes. Genes encoding GADD45α and GADD45β were transcriptionally up-regulated following F. nucleatum infection (P = 0.0005 and P < 10−6, respectively), whereas expression of the gene encoding DUSP4 (one of several MAPK phosphatase family members) was down-regulated (P < 10−6). S. gordonii up-regulated GADD45β gene expression (P < 10−6), down-regulated DUSP4 gene expression (P < 10−6), and had no detectable effect on GADD45α gene expression. The protein expression data obtained by Western blotting (Fig. 2) were consistent with the transcriptional data. Both F. nucleatum and S. gordonii decreased the levels of DUSP4; F. nucleatum up-regulated GADD45α and GADD45β expression; and S. gordonii infection resulted in an increase in the GADD45β level. In general, the amount of regulated protein continued to change over a 6-h period, corroborating the predictive power of the transcriptional “snapshot” for the longer-term phenotype of the epithelial cells, at least for these genes. The roles of these genes in signal transduction through the MAPK pathway and their impact on cell physiology are shown diagrammatically in Fig. 3.
FIG. 2.
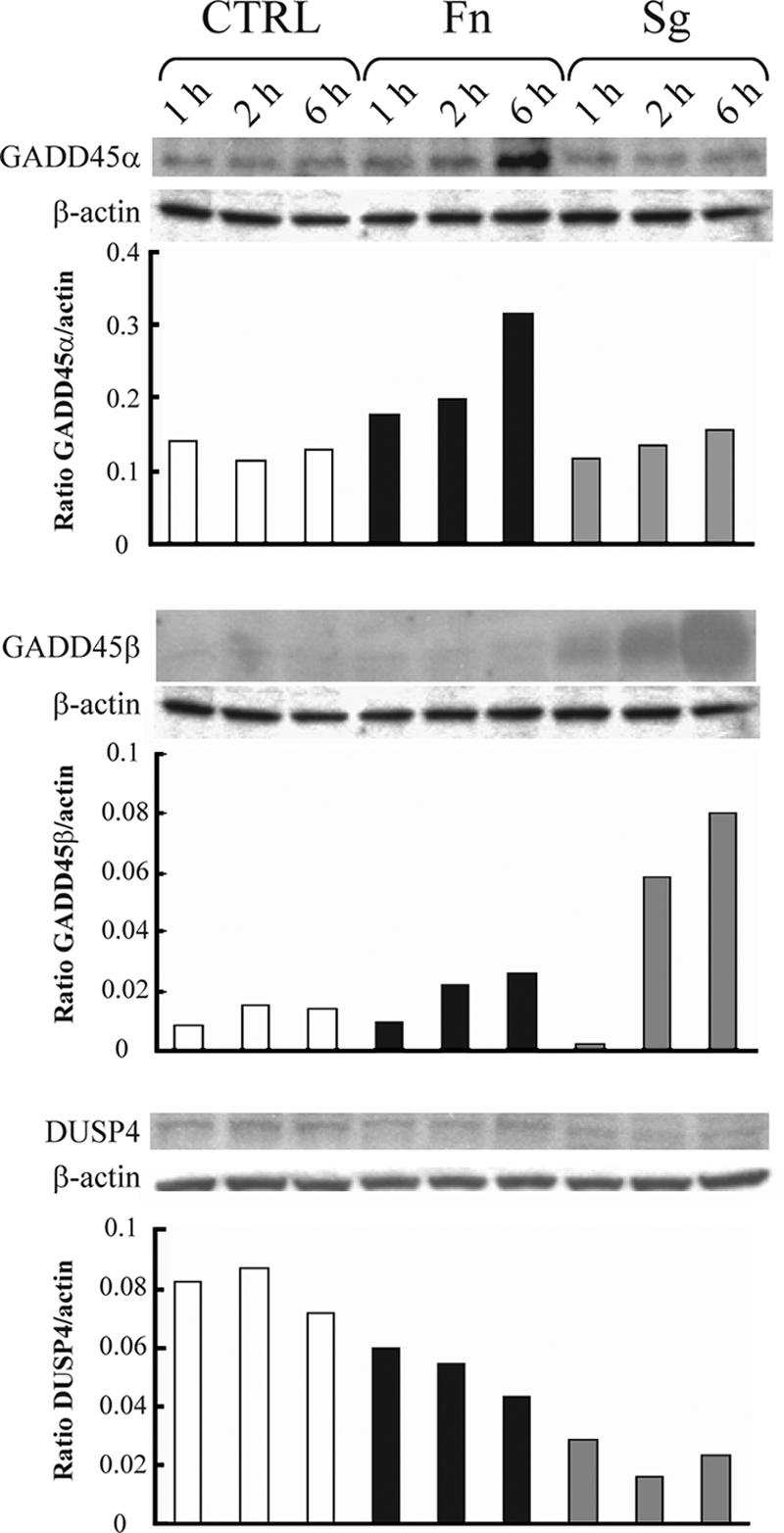
Western immunoblots of HIGK infected with F. nucleatum (Fn) or S. gordonii (Sg) and uninfected controls (CTRL) for 1, 2, or 6 h. The blots were probed with antibodies to GADD45α (upper panel), GADD45β (middle panel), and DUSP4 (lower panel) and then stripped and reprobed with antibodies to β-actin. The graphs show the results of densitometric analyses of the ratio of test protein band intensity to β-actin band intensity.
FIG. 3.
MAPK-related pathways containing F. nucleatum and S. gordonii differentially regulated genes at P < 0.05, adapted from Pathway Express and using the Kyoto Encyclopedia of Genes and Genomes nomenclature (see text for details). Red indicates up-regulation, green indicates down-regulation, and gray indicates no change in expression. +P indicates phosphorylation, and −P indicates dephosphorylation. An arrow indicates a molecular interaction resulting in activation, and a line without an arrowhead indicates a molecular interaction resulting in inhibition. e, expression.
Correlation between mRNA and secreted cytokines.
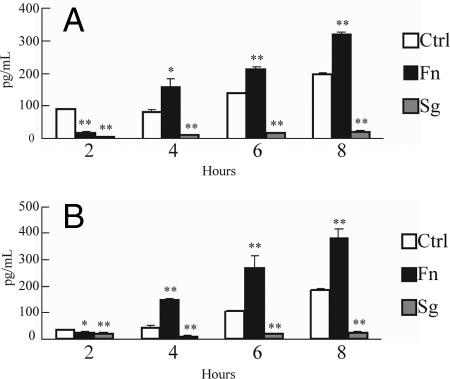
Both the MAPK and Toll-like pathways can converge on expression of cytokines. While recognizing that control of cytokine secretion is hierarchical and can occur at the transcriptional and posttranscriptional levels, we assessed the phenotypic significance of the ontology analysis by directly assaying at the protein level the proinflammatory cytokines IL-6 and IL-8. Secretion of IL-6 and IL-8 following infection with F. nucleatum or S. gordonii was monitored over time (Fig. 4). After 4 h of coculture, F. nucleatum stimulated secretion of IL-8, which is consistent with previous reports (9, 26, 36), and of IL-6. In contrast, S. gordonii reduced the levels of secreted IL-6 and IL-8 compared to the levels in uninfected control cells over an 8-h incubation. Inhibition of proinflammatory cytokine secretion by S. gordonii may eliminate initiation of a potentially destructive immune response and contribute to the maintenance of oral health.
FIG. 4.
Enzyme-linked immunosorbent assay of IL-6 (A) and IL-8 (B) accumulation in HIGK supernatants following coculture with F. nucleatum or S. gordonii for 2, 4, 6, or 8 h. Ctrl, uninfected control. The error bars indicate standard deviations (n = 3). One asterisk, P < 0.05; two asterisks, P < 0.001 (as determined by a t test).
DISCUSSION
The epithelial cells that line the gingival crevice form the initial interactive interface between the host and subgingival bacteria. The bacterial inhabitants of this area are diverse and have a spectrum of pathogenic potentials, ranging from commensal (such as S. gordonii) through opportunistic commensal (such as F. nucleatum) to overtly pathogenic (such as P. gingivalis and A. actinomycetemcomitans). The outcome of the bacterium-epithelial cell interaction, in which the host cells distinguish the infecting bacteria and tailor a response while the bacteria attempt to manipulate host cell responses, is an important component in establishing pathogenic potential. Transcriptional profiling provides insights into these extremely complex systems. In a previous study Handfield et al. (23) reported that HIGK responses to the pathogens P. gingivalis and A. actinomycetemcomitans reverberated throughout the transcriptome and impacted biological pathways, including cell development and morphogenesis pathways. In contrast, the HIGK response to the less pathogenic organisms F. nucleatum and S. gordonii was much more restrained. The similar transcriptional responses to F. nucleatum and S. gordonii provide experimental support for the concept that commensals and opportunistic commensals have developed a more balanced evolutionary relationship with the host than pathogens have developed (17, 24). Future studies will be directed toward trying to identify host cell genes and pathways that can be used to predict a commensal or pathogenic challenge. Such genes and pathways could be used as markers to assess the pathogenic potentials of the hundreds of species or complex mixtures of subgingival bacteria for which no virulence information or culture methods are available (38). An additional distinction between the gingival cell responses to more pathogenic and less pathogenic organisms was that the core common transcriptional responses to P. gingivalis and A. actinomycetemcomitans were very limited (around 15% of the regulated genes), and organism-specific responses predominated (23). However, the core transcriptional responses to F. nucleatum and S. gordonii were more extensive, with 84% of the genes similarly regulated by these two organisms, including the genes phenotypically validated and described here. Considering that F. nucleatum possesses an outer membrane while S. gordonii does not and that F. nucleatum is orders of magnitude more invasive than S. gordonii, it appears that transcriptional responses are not strictly dependent on the physical nature of the bacterial microbe-associated molecular patterns or on the extra- or intracellular location of the bacteria.
One pathway that was significantly impacted by F. nucleatum (P = 0.00825) and S. gordonii (P = 0.00437) involves MAPK signal transduction. This signaling pathway, which is evolutionarily conserved, connects cell surface receptors to regulatory targets within cells. MAPK signaling is involved in a vast array of physiological processes, including cell growth, migration, proliferation, differentiation, survival, development, and innate immunity (37, 49, 64). Consistent with these diverse roles, a variety of stimuli can activate MAPK pathways; these stimuli include growth factors, cytokines, ligands for G protein-coupled receptors, transforming agents, environmental stress, and viral and bacterial ligands. In the MAPK pathway, expression of GADD45α and GADD45β was up-regulated by F. nucleatum infection, whereas expression of DUSP4 was down-regulated. Similarly, S. gordonii up-regulated GADD45β expression and down-regulated DUSP4 expression. Regulation occurred at both the mRNA and protein levels. The GADD45 gene was originally identified as a gene that is rapidly induced by agents that cause DNA damage (18, 48). Transcriptional regulation of the GADD45 gene is mediated by both p53-dependent and p53-independent mechanisms (56), and GADD45 family members (α, β, and γ) are involved in the activation of the p38 and JNK pathways through MEKK4. Up-regulation of GADD45 expression may ultimately converge on growth arrest and on the activation of the nuclear transcription factor NF-κB (47, 56, 57, 63). The dual-specificity phosphatase DUSP4 is involved in the inactivation of the MAPKs by dephosphorylating both Thr and Tyr residues of ERK1 and ERK2 (30). One role of the DUSP family of phosphatases may be in the postinduction repression of MAPK activity (60). Hence, both up-regulation of GADD45 expression and down-regulation of DUSP4 expression by F. nucleatum or S. gordonii engender the concordant phenotype of enhanced information flow through the MAPK pathway. Together, these results provide a mechanistic framework for previous reports which showed that F. nucleatum and S. gordonii activated MAPK and NF-κB in gingival epithelial cells (7, 9, 26, 27, 35, 61).
One of the downstream targets of MAPK signaling is the production of cytokines. F. nucleatum induced IL-6 and IL-8 secretion, whereas S. gordonii repressed secretion of both of these proinflammatory cytokines. Differences in the cytokine secretion profiles may be related to differential regulation of individual components of the MAPK pathway, such as the ability of F. nucleatum to up-regulate both GADD45α expression and GADD45β expression. In addition, F. nucleatum, but not S. gordonii, down-regulated expression of RSK2, whereas S. gordonii, but not F. nucleatum, down-regulated expression of Sap1a. RSK2 is a serine/threonine kinase that may play a role in mediating the growth factor- and stress-induced activation of the transcription factor CREB. SAP1a is a nuclear protein that stimulates transcription via the c-fos serum response element and also via an Ets binding site independent of the serum response factor. However, although RSK2 and Sap1a are involved in distinct aspects of MAPK signaling, their precise physiological roles are not known. The induction of expression of proinflammatory cytokines in gingival tissues and the subsequent inflammatory tissue damage are considered contributory factors in the pathogenesis of periodontal disease (10, 20, 53). Hence, stimulation of IL-6 and IL-8 by F. nucleatum may be one property that increases the pathogenicity of this organism compared to the pathogenicity of S. gordonii. Indeed, a general hyporesponsiveness to commensals may be advantageous in order to limit tissue destruction that might occur if a strong proinflammatory response were induced. Furthermore, it has been proposed that commensal species can “program” host cells to limit subsequent responses to more pathogenic organisms (2, 54) Conversely, in gingival epithelial cells, prior infection with the pathogen P. gingivalis can paralyze the local chemokine response to F. nucleatum (9). The extent to which challenge with S. gordonii can modulate subsequent host cell responses to other organisms is currently being investigated. Of possibly greater in vivo relevance is the potential of a complex microbial community to impact the transcriptional profile and phenotypic responses of host cells in a manner distinct from the summed activities of its constituents. Such community-based responses could partially explain apparent discrepancies that are observed between certain oral clinical manifestations and the putative pathogenic potentials of causative microbial species. For example, it is generally recognized that clinical cases of localized aggressive periodontitis that are associated with proapoptotic and proinflammatory A. actinomycetemcomitans do not result in significant inflammation or gingival destruction (3). It is tempting to speculate that the anti-inflammatory nature of certain commensal species has the potential to restrain the proinflammatory capability of pathogenic species.
Supplementary Material
Acknowledgments
This study was supported by grants DE11111 (R.J.L.), DE16715 (M.H.), and T32 DE7200 (J.J.M.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 16 February 2007.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abreu, M. T., P. Vora, E. Faure, L. S. Thomas, E. T. Arnold, and M. Arditi. 2001. Decreased expression of Toll-like receptor-4 and MD-2 correlates with intestinal epithelial cell protection against dysregulated proinflammatory gene expression in response to bacterial lipopolysaccharide. J. Immunol. 167:1609-1616. [DOI] [PubMed] [Google Scholar]
- 3.Armitage, G. C. 2004. Periodontal diagnoses and classification of periodontal diseases. Periodontol. 2000 34:9-21. [DOI] [PubMed] [Google Scholar]
- 4.Avila-Campos, M. J., C. T. Sacchi, A. M. Whitney, A. G. Steigerwalt, and L. W. Mayer. 1999. Arbitrarily primed-polymerase chain reaction for identification and epidemiologic subtyping of oral isolates of Fusobacterium nucleatum. J. Periodontol. 70:1202-1208. [DOI] [PubMed] [Google Scholar]
- 5.Boman, H. G. 2000. Innate immunity and the normal microflora. Immunol. Rev. 173:5-16. [DOI] [PubMed] [Google Scholar]
- 6.Casadevall, A., and L. A. Pirofski. 2000. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect. Immun. 68:6511-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, W. O., and B. A. Dale. 2004. Innate immune response of oral and foreskin keratinocytes: utilization of different signaling pathways by various bacterial species. Infect. Immun. 72:352-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings, C. A., and D. A. Relman. 2000. Using DNA microarrays to study host-microbe interactions. Emerg. Infect. Dis. 6:513-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darveau, R. P., C. M. Belton, R. A. Reife, and R. J. Lamont. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66:1660-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dixon, D. R., B. W. Bainbridge, and R. P. Darveau. 2004. Modulation of the innate immune response within the periodontium. Periodontol. 2000 35:53-74. [DOI] [PubMed] [Google Scholar]
- 11.Dixon, D. R., R. A. Reife, J. J. Cebra, and R. P. Darveau. 2004. Commensal bacteria influence innate status within gingival tissues: a pilot study. J. Periodontol. 75:1486-1492. [DOI] [PubMed] [Google Scholar]
- 12.Draghici, S., P. Khatri, P. Bhavsar, A. Shah, S. A. Krawetz, and M. A. Tainsky. 2003. Onto-Tools, the toolkit of the modern biologist: Onto-Express, Onto-Compare, Onto-Design and Onto-Translate. Nucleic Acids Res. 31:3775-3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Draghici, S., P. Khatri, R. P. Martins, G. C. Ostermeier, and S. A. Krawetz. 2003. Global functional profiling of gene expression. Genomics 81:98-104. [DOI] [PubMed] [Google Scholar]
- 14.Draghici, S., S. Sellamuthu, and P. Khatri. 2006. Babel's tower revisited: a universal resource for cross-referencing across annotation databases. Bioinformatics 22:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzink, J. L., S. S. Socransky, and A. D. Haffajee. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316-323. [DOI] [PubMed] [Google Scholar]
- 16.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, Z., and A. Weinberg. 2006. Role of bacteria in health and disease of periodontal tissues. Periodontol. 2000 40:50-76. [DOI] [PubMed] [Google Scholar]
- 18.Fornace, A. J., Jr., I. Alamo, Jr., and M. C. Hollander. 1988. DNA damage-inducible transcripts in mammalian cells. Proc. Natl. Acad. Sci. USA 85:8800-8804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushima, K., H. Ogawa, K. Takahashi, H. Naito, Y. Funayama, T. Kitayama, H. Yonezawa, and I. Sasaki. 2003. Non-pathogenic bacteria modulate colonic epithelial gene expression in germ-free mice. Scand. J. Gastroenterol. 38:626-634. [DOI] [PubMed] [Google Scholar]
- 20.Graves, D. T. 1999. The potential role of chemokines and inflammatory cytokines in periodontal disease progression. Clin. Infect. Dis. 28:482-490. [DOI] [PubMed] [Google Scholar]
- 21.Han, D. C., G. T. Huang, L. M. Lin, N. A. Warner, J. S. Gim, and A. Jewett. 2003. Expression of MHC class II, CD70, CD80, CD86 and pro-inflammatory cytokines is differentially regulated in oral epithelial cells following bacterial challenge. Oral Microbiol. Immunol. 18:350-358. [DOI] [PubMed] [Google Scholar]
- 22.Han, Y. W., W. Shi, G. T. Huang, S. Kinder Haake, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handfield, M., J. J. Mans, G. Zheng, M. C. Lopez, S. Mao, A. Progulske-Fox, G. Narasimhan, H. V. Baker, and R. J. Lamont. 2005. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell. Microbiol. 7:811-823. [DOI] [PubMed] [Google Scholar]
- 24.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 25.Hooper, L. V., M. H. Wong, A. Thelin, L. Hansson, P. G. Falk, and J. I. Gordon. 2001. Molecular analysis of commensal host-microbial relationships in the intestine. Science 291:881-884. [DOI] [PubMed] [Google Scholar]
- 26.Huang, G. T., D. Kim, J. K. Lee, H. K. Kuramitsu, and S. K. Haake. 2001. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect. Immun. 69:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang, G. T., H. B. Zhang, H. N. Dang, and S. K. Haake. 2004. Differential regulation of cytokine genes in gingival epithelial cells challenged by Fusobacterium nucleatum and Porphyromonas gingivalis. Microb. Pathog. 37:303-312. [DOI] [PubMed] [Google Scholar]
- 28.Ismail, A. S., and L. V. Hooper. 2005. Epithelial cells and their neighbors. IV. Bacterial contributions to intestinal epithelial barrier integrity. Am. J. Physiol. Gastrointest. Liver Physiol. 289:G779-G784. [DOI] [PubMed] [Google Scholar]
- 29.Jenner, R. G., and R. A. Young. 2005. Insights into host responses against pathogens from transcriptional profiling. Nat. Rev. Microbiol. 3:281-294. [DOI] [PubMed] [Google Scholar]
- 30.Keyse, S. M. 1995. An emerging family of dual specificity MAP kinase phosphatases. Biochim. Biophys. Acta 1265:152-160. [DOI] [PubMed] [Google Scholar]
- 31.Khatri, P., P. Bhavsar, G. Bawa, and S. Draghici. 2004. Onto-Tools: an ensemble of web-accessible, ontology-based tools for the functional design and interpretation of high-throughput gene expression experiments. Nucleic Acids Res. 32:W449-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khatri, P., S. Draghici, G. C. Ostermeier, and S. A. Krawetz. 2002. Profiling gene expression using onto-express. Genomics 79:266-270. [DOI] [PubMed] [Google Scholar]
- 33.Khatri, P., S. Sellamuthu, P. Malhotra, K. Amin, A. Done, and S. Draghici. 2005. Recent additions and improvements to the Onto-Tools. Nucleic Acids Res. 33:W762-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kolenbrander, P. E., R. J. Palmer, Jr., A. H. Rickard, N. S. Jakubovics, N. I. Chalmers, and P. I. Diaz. 2006. Bacterial interactions and successions during plaque development. Periodontol. 2000 42:47-79. [DOI] [PubMed] [Google Scholar]
- 35.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kappaB transcription factor family. J. Immunol. 168:316-324. [DOI] [PubMed] [Google Scholar]
- 36.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuida, K., and D. M. Boucher. 2004. Functions of MAP kinases: insights from gene-targeting studies. J. Biochem. (Tokyo) 135:653-656. [DOI] [PubMed] [Google Scholar]
- 38.Kumar, P. S., A. L. Griffen, M. L. Moeschberger, and E. J. Leys. 2005. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J. Clin. Microbiol. 43:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee, J., J. H. Mo, K. Katakura, I. Alkalay, A. N. Rucker, Y. T. Liu, H. K. Lee, C. Shen, G. Cojocaru, S. Shenouda, M. Kagnoff, L. Eckmann, Y. Ben-Neriah, and E. Raz. 2006. Maintenance of colonic homeostasis by distinctive apical TLR9 signalling in intestinal epithelial cells. Nat. Cell Biol. 8:1327-1336. [DOI] [PubMed] [Google Scholar]
- 41.Mans, J. J., H. V. Baker, D. Oda, R. J. Lamont, and M. Handfield. 2006. Distinctive characteristics of transcriptional profiles from two epithelial cell lines upon interaction with Actinobacillus actinomycetemcomitans. Oral Microbiol. Immunol. 21:261-267. [DOI] [PubMed] [Google Scholar]
- 42.Mans, J. J., R. J. Lamont, and M. Handfield. 2006. Microarray analysis of human epithelial cell responses to bacterial interaction. Infect. Disord. Drug Targets 6:299-309. [DOI] [PubMed] [Google Scholar]
- 43.Mazmanian, S. K., C. H. Liu, A. O. Tzianabos, and D. L. Kasper. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107-118. [DOI] [PubMed] [Google Scholar]
- 44.Moore, W. E., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 45.Oda, D., L. Bigler, P. Lee, and R. Blanton. 1996. HPV immortalization of human oral epithelial cells: a model for carcinogenesis. Exp. Cell Res. 226:164-169. [DOI] [PubMed] [Google Scholar]
- 46.Ogura, Y., F. S. Sutterwala, and R. A. Flavell. 2006. The inflammasome: first line of the immune response to cell stress. Cell 126:659-662. [DOI] [PubMed] [Google Scholar]
- 47.Papa, S., F. Zazzeroni, C. Bubici, S. Jayawardena, K. Alvarez, S. Matsuda, D. U. Nguyen, C. G. Pham, A. H. Nelsbach, T. Melis, E. De Smaele, W. J. Tang, L. D'Adamio, and G. Franzoso. 2004. Gadd45 beta mediates the NF-kappa B suppression of JNK signalling by targeting MKK7/JNKK2. Nat. Cell Biol. 6:146-153. [DOI] [PubMed] [Google Scholar]
- 48.Papathanasiou, M. A., N. C. Kerr, J. H. Robbins, O. W. McBride, I. Alamo, Jr., S. F. Barrett, I. D. Hickson, and A. J. Fornace, Jr. 1991. Induction by ionizing radiation of the gadd45 gene in cultured human cells: lack of mediation by protein kinase C. Mol. Cell. Biol. 11:1009-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qi, M., and E. A. Elion. 2005. MAP kinase pathways. J. Cell Sci. 118:3569-3572. [DOI] [PubMed] [Google Scholar]
- 50.Rakoff-Nahoum, S., and R. Medzhitov. 2006. Role of the innate immune system and host-commensal mutualism. Curr. Top Microbiol. Immunol. 308:1-18. [DOI] [PubMed] [Google Scholar]
- 51.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 52.Rosan, B., and R. J. Lamont. 2000. Dental plaque formation. Microbes Infect. 2:1599-1607. [DOI] [PubMed] [Google Scholar]
- 53.Seymour, G. J., and E. Gemmell. 2001. Cytokines in periodontal disease: where to from here? Acta Odontol. Scand. 59:167-173. [DOI] [PubMed] [Google Scholar]
- 54.Steidler, L., W. Hans, L. Schotte, S. Neirynck, F. Obermeier, W. Falk, W. Fiers, and E. Remaut. 2000. Treatment of murine colitis by Lactococcus lactis secreting interleukin-10. Science 289:1352-1355. [DOI] [PubMed] [Google Scholar]
- 55.Strober, W. 2006. Immunology. Unraveling gut inflammation. Science 313:1052-1054. [DOI] [PubMed] [Google Scholar]
- 56.Takekawa, M., and H. Saito. 1998. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell 95:521-530. [DOI] [PubMed] [Google Scholar]
- 57.Takekawa, M., K. Tatebayashi, F. Itoh, M. Adachi, K. Imai, and H. Saito. 2002. Smad-dependent GADD45beta expression mediates delayed activation of p38 MAP kinase by TGF-beta. EMBO J. 21:6473-6482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanner, A., and H. Bouldin. 1989. The microbiota of early periodontitis lesions in adults. J. Clin. Periodontol. 16:467-471. [DOI] [PubMed] [Google Scholar]
- 59.Tribble, G. D., S. Mao, C. E. James, and R. J. Lamont. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl. Acad. Sci. USA 103:11027-11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wadgaonkar, R., J. W. Pierce, K. Somnay, R. L. Damico, M. T. Crow, T. Collins, and J. G. Garcia. 2004. Regulation of c-Jun N-terminal kinase and p38 kinase pathways in endothelial cells. Am. J. Respir. Cell Mol. Biol. 31:423-431. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe, K., O. Yilmaz, S. F. Nakhjiri, C. M. Belton, and R. J. Lamont. 2001. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect. Immun. 69:6731-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ximenez-Fyvie, L. A., A. D. Haffajee, and S. S. Socransky. 2000. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J. Clin. Periodontol. 27:648-657. [DOI] [PubMed] [Google Scholar]
- 63.Yang, J., H. Zhu, T. L. Murphy, W. Ouyang, and K. M. Murphy. 2001. IL-18-stimulated GADD45 beta required in cytokine-induced, but not TCR-induced, IFN-gamma production. Nat. Immunol. 2:157-164. [DOI] [PubMed] [Google Scholar]
- 64.Yang, S. H., A. D. Sharrocks, and A. J. Whitmarsh. 2003. Transcriptional regulation by the MAP kinase signaling cascades. Gene 320:3-21. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.