Abstract
Chlamydial 60-kDa heat shock proteins (cHsp60s) are known to play a prominent role in the immunopathogenesis of disease. It is also known that several stress-inducing growth conditions, such as heat, iron deprivation, or exposure to gamma interferon, result in the development of persistent chlamydial forms that often exhibit enhanced expression of cHsp60. We have shown previously that the expression of cHsp60 is greatly enhanced in Chlamydia trachomatis serovar E propagated in an iron-deficient medium. The objective of this work was to determine which single cHsp60 or combination of the three cHsp60 homologs encoded by this organism responds to iron limitation. Using monospecific polyclonal peptide antisera that recognize only cHsp60-1, cHsp60-2, or cHsp60-3, we found that expression of cHsp60-2 is responsive to iron deprivation. Overall, our studies suggest that the expression of cHsp60 homologs differs among the mechanisms currently known to induce persistence.
Studies have consistently shown that there is a correlation between the production of chlamydial 60-kDa heat shock protein (cHsp60) antibodies in chlamydia-infected patients and adverse disease consequences. These observations appear to be universal for Chlamydia species and their disease presentations. Early studies of whether cHsp60 plays a role in immunopathogenesis involved analyses of serum antibodies from female patients presenting with Chlamydia trachomatis-associated tubal infertility (7, 52); elevated levels of anti-cHsp60 in the sera of these patients were significantly associated with disease. A separate group of investigators described the contribution of cHsp60 in a guinea pig model of trachoma; an intense mononuclear cell inflammatory response was observed after conjunctival inoculation of cHsp60 following resolution of a primary ocular infection with Chlamydia psittaci GPIC (37). Patients with coronary artery disease often have serological evidence of previous infection by Chlamydia pneumoniae (45), and cHsp60 has been directly identified in human atheromatous tissue (31). Most recently, C. pneumoniae and Chlamydia pecorum cHsp60s have been implicated in urogenital tract disease in koalas (Phascolarctos cinereus), leading to infertility and death; chlamydiae are the most commonly recognized disease agents in the threatened koala population (23, 24).
Studies to determine the role of cHsp60 and immunopathogenesis are still being performed. Recently, workers have examined the initial interactions of cHsp60 with host cells that induce an inflammatory response. For example, cHsp60 interacts with Toll-like receptor 4, which stimulates the proliferation of human vascular smooth muscle cells (47), activates macrophages, and activates endothelial cells (8). The interaction between cHsp60 and Toll-like receptor 4 also leads to apoptosis in primary human trophoblasts, placental fibroblasts, and a trophoblast cell line by both caspase-dependent and -independent pathways (18). cHsp60 and other microbial ligands can also activate mononuclear cells by binding to CD14, the monocyte receptor for lipopolysaccharide (32). Although cHsp60 clearly plays a prominent role in chlamydial pathogenesis, it is not the only molecule involved. The genetically linked protein cHsp10, encoded by the groES gene upstream of groEL-1, is also associated with disease complications (6, 23, 27, 33). Moreover, several studies have demonstrated that genetic predisposition plays a significant role in chronic chlamydial disease (10, 12, 38). Perhaps most interesting is the fact that cHsp60 has been used in a human trial involving women at high risk for C. trachomatis infection; cHsp60 was used to stimulate the patients’ peripheral blood mononuclear cells to produce gamma interferon (IFN-γ), and the results indicated that a protective response against incidental infection developed (11).
Our laboratory is involved in identifying and analyzing C. trachomatis proteins that respond to iron restriction, as well as the mechanisms involved (42, 43, 55); cHsp60 is one of several proteins whose expression increases significantly during iron limitation in vitro (43). It is known that iron sources and the availability of iron fluctuate in menstruating women due to the cyclic pressures of estrogen and progesterone (1, 29); active or persistent C. trachomatis organisms in the reproductive tract are therefore likely to respond to this dynamic environment using transcriptional, translational, or posttranslational mechanisms to alter the production of specific chlamydial proteins. While we are not involved in direct studies of persistent chlamydiae, which have been defined as viable but nonculturable organisms (3), iron deprivation is one of several modes for induction of persistent chlamydiae (22, 39, 43). In women with tubal factor subfertility, cHsp60 is a serological marker for persistence (15) along with chlamydial proteasome/protease-like activity factor (48). However, the results of recent studies with C. psittaci (22) and C. pneumoniae (39) indicate that cHsp60 is not a general marker for persistence.
When the complete sequence of the C. trachomatis serovar D chromosome became available, one of many surprises was that there are three open reading frames (ORF) that code for groEL-related proteins (49). These ORF are located in separate regions of the chromosome and designated as follows: CT110 or groEL-1, encoding cHsp60-1; CT604 or groEL-2, encoding cHsp60-2; and CT755 or groEL-3, encoding cHsp60-3. Only groEL-1 is preceded by groES. Matching cHsp60s in different Chlamydia species appear to be conserved in the sequences that are currently available. For example, the predicted level of amino acid sequence identity between cHsp60-1 in C. trachomatis serovar D and cHsp60-1 in C. pneumoniae AR39 is 91%. However, there are considerable differences between cHsp60-1, cHsp60-2, and cHsp60-3 in a given species or serovariant. In C. trachomatis serovar D, the levels of amino acid identity and similarity between cHsp60-1 and cHsp60-2 are 23 and 19%, respectively; the levels of amino acid identity and similarity between cHsp60-1 and cHsp60-3 are 18 and 20%, respectively; and the levels of amino acid identity and similarity between cHsp60-2 and cHsp60-3 are 17 and 15%, respectively (28, 35, 49).
Although the majority of previous studies clearly involved cHsp60-1, as confirmed by sequence analysis, certain studies, including our study (43), generated new questions concerning the extent to which each cHsp60 responds to a given microenvironment, especially a microenvironment leading to chlamydial persistence. Thus, the purpose of this study was to determine which cHsp60 is iron responsive in C. trachomatis serovar E.
MATERIALS AND METHODS
Bacterial strains, eukaryotic host cells, and growth.
Stock inocula of C. trachomatis serovar E/UW-5CX EB were generated in McCoy cell fibroblasts and titrated to determine their infectivity. Polarized human endometrial epithelial cells (HEC-1B) were used as host cells in iron deprivation experiments and were maintained in Eagle's minimal essential medium containing 2 mM glutamine and 5% (vol/vol) heat-inactivated fetal bovine serum at 37°C. For induction of iron deprivation, chlamydia-infected cultures were allowed to grow until 36 h postinoculation (hpi), and one-half of the samples were exposed to 500 μM Desferal for 30 min and 1 and 2 h.
Escherichia coli LMG194(pBAD/HisA) was used to engineer and overexpress each cHsp60. The recombinants expressing cHsp60-1, cHsp60-2, and cHsp60-3 were designated E. coli LMG194(pJER516), LMG194(pJER517), and LMG194(pJER518), respectively. Each recombinant E. coli was grown in reduced medium (Invitrogen, Carlsbad, CA) containing 0.2% (wt/vol) glucose and 100 μg/ml ampicillin (Sigma-Genosys, The Woodlands, TX) to the mid-log phase (A600, 0.4 to 0.6) at 37°C. Cultures were subsequently centrifuged, washed, and resuspended in prewarmed glucose-free medium. Arabinose was then added to each culture for 4 h of induction. Maximum expression of cHsp60-1 and cHsp60-2 in E. coli LMG194(pJER516) and LMG194(pJER517) required 0.002% (wt/vol) arabinose, whereas maximum expression of cHsp60-3 in E. coli LMG194(pJER518) required 20% (wt/vol) arabinose.
DNA amplification, cloning, and sequence analysis.
The primers used for PCR amplification of the chlamydial groEL genes were designed using the genome sequence of C. trachomatis serovar D (49). The reactions were carried out with an Expand High Fidelity PCR system kit (Roche, Nutley, NJ) in the presence of 0.5 pmol of forward, 0.5 pmol of reverse primer, and 10-fold (1:10 to 1:1,000) dilutions of C. trachomatis serovar E DNA template. After 35 cycles of amplification, the PCR products were cleaned up using a QIAquick PCR purification kit (QIAGEN, Germantown, MD); the sizes and concentrations of the purified products were monitored by agarose gel electrophoresis in the presence of ethidium bromide and by determining the optical density, respectively. All PCRs were done in duplicate to reduce introduction of errant nucleotides. The PCR products were then directionally cloned into the pBAD/HisA vector (Invitrogen, Carlsbad, CA) under the control of the araC promoter with an N-terminal six-histidine tag for recombinant protein detection and used to transform E. coli LMG194 by the traditional CaCl2 method (46). For each groEL gene, the recombinant plasmids from three clones were purified using a Concert nucleic acid purification kit (Invitrogen) and then sequenced to verify in-frame cloning and to determine the complete nucleic acid sequences.
Peptide antibodies.
The predictive amino acid sequences of C. trachomatis serovar E Hsp60-1, Hsp60-2, and Hsp60-3 were aligned using the EditSeq and MegAlign software from DNAStar, Inc. (Madison, WI). Peptides that were 17 to 21 residues long, described by Giles et al. (21), were commercially synthesized, the purity was assessed by analytical high-pressure liquid chromatography and mass spectroscopy, and each peptide was subsequently used to immunize two female New Zealand White rabbits (Sigma-Genosys, The Woodlands, TX). The results of enzyme-linked immunosorbent assays were provided by the manufacturer to ensure reactivity.
The majority of antiserum from each bleed was immediately stored at −20°C upon receipt. One milliliter of antiserum from each bleed was kept at 4°C to determine the Western blot reactivities of the crude preimmune and immune sera against total protein from HEC-1B cells and E. coli LMG194 as controls and arabinose-induced recombinant E. coli LMG194(pJER516), LMG194(pJER517), and LMG194(pJER518). Immune sera exhibiting the most selective reactivity with the intended Hsp60 homolog were then placed on protein A columns (ImmunoPure immobilized protein A; Pierce, Rockford, IL) to purify immunoglobulin G, and Western blotting was performed to determine the degree of monospecificity and the reduction in the cross-reactivity with other E. coli proteins. Antiserum against the peptide from cHsp60-3 required a further step of adsorption against whole cells of arabinose-induced E. coli LMG194(pJER516) expressing cHsp60-1. A monoclonal antibody reagent (Sigma-Genosys, The Woodlands, TX) against the polyhistidine tag was also used in this study.
Protein quantitation, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blotting, and chemiluminescence.
The total protein concentrations of samples were determined using the Micro BCA assay (Pierce, Rockford, IL). Cell pellets were resuspended in lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 0.5 mg/ml lysozyme, 0.1 mg/ml DNase I, 10 mM CaCl2) and subjected to three freeze-thaw cycles. After the final thaw, samples were centrifuged at 8,000 × g for 10 min, and each supernatant was combined with denaturing sample buffer and heated at 100°C for 5 min. The proteins were resolved in small-format 4 to 12% bis-Tris NuPAGE gels (Invitrogen, Carlsbad, CA) and transferred to nitrocellulose membranes for Western blotting. Preliminary separations were conducted in large-format 12.5% polyacrylamide gels loaded with 1 mg of protein to accommodate multiple blots for screening and titrating antisera.
For Western blotting, membranes were blocked with Blotto-plus (5% [wt/vol] dry nonfat milk in phosphate-buffered saline, 0.1% [vol/vol] Tween 20, 10% [vol/vol] heat-inactivated fetal bovine serum), and washing was performed with phosphate-buffered saline containing 0.1% (vol/vol) Tween 20. Various dilutions were examined for the polyclonal peptide antisera generated against each of the cHsp60s, and a monoclonal antibody against the polyhistidine tag (Sigma- Genosys, The Woodlands, TX) was also used as a control. Specific signals were then detected either (i) by a colorimetric assay with an anti-rabbit alkaline phosphatase-conjugated secondary antibody and Western Blue substrate (Promega, Madison, WI) or (ii) by chemiluminescence using an anti-rabbit horseradish peroxidase conjugate, the SuperSignal West (Pierce) solution, and Kodak X-OMAT AR film.
Electron microscopy.
Samples of C. trachomatis-infected polarized HEC-1B cells at 36 hpi were exposed to 500 μM Desferal for 30 min and 1 and 2 h; mock-exposed samples were used as controls. Each sample was immediately washed, fixed, processed, and embedded in Epon-araldite and Lowicryl (Polysciences, Inc.) for high-contrast electron microscopy and immunoelectron microscopy, respectively, as described by Giles et al. (21).
Visualization and image capture were done using a Philips Tecnai 10 transmission electron microscope (FEI Company, Hillsboro, OR) operating at 80 kV.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under the following accession numbers: AY447001 for C. trachomatis serovar E groEL-1, AY447002 for C. trachomatis serovar E groEL-2, and AY447003 for C. trachomatis serovar E groEL-3.
RESULTS
Amplification and nucleotide sequence analysis of C. trachomatis serovar E groEL ORF.
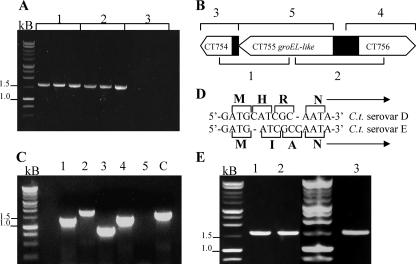
DNA that was the expected size was amplified with all dilutions of the C. trachomatis serovar E groEL DNA templates except the groEL-3/CT755 template (Fig. 1A, lanes 3). Figures 1B and C show the results of a change in primer strategy, and the data revealed that CT755 was present along with the flanking sequences. Further analysis showed that a missing cytosine residue in the initial sequence of CT755 in serovar E at position 25 was responsible for the lack of primer hybridization and amplification (Fig. 1D). As determined by comparison with the previously published sequence of CT755 in serovar D, a frameshift placed the ORF back into frame by insertion of a cytosine residue at position 30 in serovar E. The final results were confirmed using a new set of primers (Fig. 1E). The nucleotide sequences of C. trachomatis serovar E groEL-1, groEL-2, and groEL-3 are 99.7, 98.5, and 99.2% identical to their counterparts in C. trachomatis serovar D, with only 6-, 25-, and 8-bp differences, respectively.
FIG. 1.
PCR amplification of C. trachomatis serovar E groEL. (A) Initial attempt to amplify groEL-1, groEL-2, and groEL-3 (lanes 1, lanes 2, and lanes 3, respectively) using a C. trachomatis serovar E DNA template and primers based on the sequence of C. trachomatis serovar D. (B) Strategy used to amplify groEL-3 and flanking sequences. (C) Result of amplification of groEL-3 and flanking sequences. Lanes 1 through 5 contained areas indicated in panel B, and amplification of groEL-1 was used as a control. (D) Difference in the starting sequences of C. trachomatis serovars D and E. (E) Amplification of C. trachomatis serovar E groEL-1, groEL-2, and groEL-3 with redesigned primers for groEL-3.
Specificity of the peptide antisera.
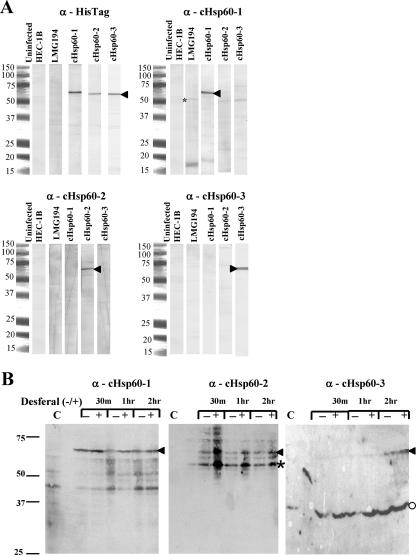
Monospecificity was achieved for anti-cHsp60-1 and anti-cHsp60-2 with purification of immunoglobulin G alone (Fig. 2A). Antiserum against cHsp60-3 initially exhibited faint cross-reactivity with Hsp60-1 that was removed by cross-adsorption against whole cells of arabinose-induced E. coli LMG194(pJER516) expressing cHsp60-1. A control using an anti-histidine monoclonal antibody (Sigma-Genosys, The Woodlands, TX) was included.
FIG. 2.
Specificity of peptide antisera and response of cHsp60-2 to iron deprivation. (A) Samples used for Western blotting included uninfected HEC-1B cells, E. coli LMG194 alone, and arabinose-induced recombinants E. coli LMG194 (pJER516), LMG194 (pJER517), and LMG194 (pJER518), representing cHsp60-1, cHsp60-2 and cHsp60-3, respectively. An anti-His tag monoclonal antibody was used as a control (upper left panel). (B) Samples included uninfected HEC-1B cells (control) (lanes C), cells mock exposed for 30 min and 1 and 2 h, and cells exposed to Desferal for 30 min and 1 and 2 h. One milligram of protein was loaded onto preparative gels (A), whereas 15 μg was loaded into each lane in panel B. Arrowheads indicate the position of cHsp60. The asterisk indicates the position of a major proteolytic product of cHsp60-2, and the circle indicates the position of a cross-reactive protein in HEC-1B cells.
Iron responsiveness of cHsp60s.
Next, the peptide antibodies were used to examine C. trachomatis-infected cells with or without 500 μM Desferal, which were exposed for 30 min and 1 and 2 h, beginning at 36 hpi (Fig. 2B). Preliminary experiments using the standard 50 μM Desferal for 96 h resulted in induction of a persistence-like state (43); these initial experiments indicated that only cHsp60-2 responds to iron deprivation beginning at 36 hpi (data not shown). Therefore, we changed the strategy in a manner analogous to application of heat, cold, or acid shock in other bacteria. Figure 2B confirms that cHsp60-2 is the primary cHsp60 that responds to iron limitation. Notably, cHsp60-2 is a target of proteolysis during cell lysis even in the presence of protease inhibitors (several combinations of inhibitors were tested). The data also showed that cHsp60-1 is strongly expressed, but there was little or no difference between the expression in the absence of Desferal and the expression in the presence of Desferal; cHsp60-3 expression was delayed, but again, there was little difference between the expression in the absence of Desferal and the expression in the presence of Desferal.
Immunoelectron microscopy.
To confirm that C. trachomatis cHsp60-2 is iron responsive, the final experiment involved examination of thin sections by immunoelectron microscopy. Multiple images were captured, saved as TIFF files, and printed; 2-μm square grids were used to enclose 10 to 15 randomly selected squares containing chlamydial reticulate bodies (RB) on prints of each sample, and the gold particles in each box were counted (Fig. 3).
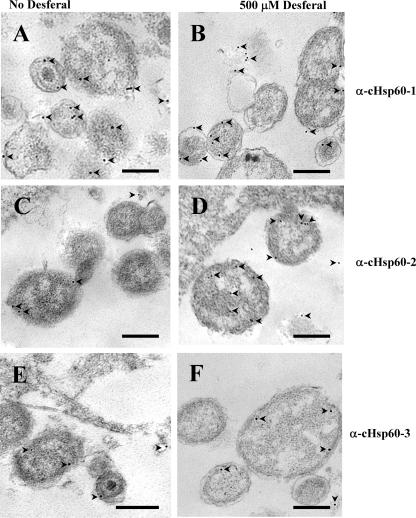
FIG. 3.
Immunolabeling transmission electron microscopy showing the response of cHsp60-2 to iron limitation. Chlamydia-infected HEC-1B cells at 36 hpi were either not exposed to Desferal (A, C, and E) or exposed to 500 μM Desferal (B, D, and F) for 1 h and labeled using a 1:100 (vol/vol) dilution of anti-cHsp60-1 (A and B), a 1:20 (vol/vol) dilution of anti-cHsp60-2 (C and D), or a 1:40 (vol/vol) dilution of anti-cHsp60-3 (E and F). A 15-nm gold-conjugated anti-rabbit serum (Amersham Biosciences) was used at a 1:200 (vol/vol) dilution for visualization. Bars = 0.5 μm. The arrowheads indicate gold particles.
Figures 3A, C, and E show chlamydial RB in HEC-1B cells at 36 hpi (mock exposure); Fig. 3B, D, and F show RB at 36 hpi in cells that were exposed to 500 μM Desferal for 1 h. Consistent with the results of the Western blot analyses, cHsp60-1 was strongly expressed, but there was not a significant difference between mock-exposed chlamydiae and chlamydiae exposed to Desferal (Fig. 3A and B); the numbers of particles in Fig. 3A and B for cHsp60-1 were 19 ± 8 and 18 ± 6, respectively. Likewise, for weakly expressed cHsp60-3 there was no difference between mock exposure and exposure to Desferal; the numbers of gold particles in both Fig. 3E and F were 5 ± 2. However, there was a significant difference (P < 0.001) between expression of cHsp60-2 after mock exposure and expression of cHsp60-2 after exposure to Desferal (Fig. 3C and D), as determined by Student's two-tailed t test; the numbers of gold particles in Fig. 3C and D were 4 ± 2 and 10 ± 3, respectively.
DISCUSSION
In this report we show conclusively that C. trachomatis serovar E cHsp60-2 is the primary cHsp60 that exhibits enhanced expression in response to iron restriction. In a larger context, the specificity of cHsp60-2 expression as a result of iron limitation indicates that the mechanisms for development of chlamydial persistence have unique signatures. This is an emerging concept in the study of chlamydial pathogenesis. The expression of cHsp60s, as determined by either protein expression or transcript analysis, has been examined using several models of persistence (2, 4, 17, 19, 22, 39).
Belland et al. (4) conducted a comprehensive microarray study of C. trachomatis serovar D transcription and compared standard growth and growth of IFN-γ-mediated persistent chlamydiae in HeLa 229 cells. None of the groEL transcripts varied significantly for the first 24 h; however, by 48 hpi, transcription of groEL-1 had increased 2.8-fold due to tryptophan depletion by IFN-γ. Tryptophan is an essential amino acid for C. trachomatis. A separate group of investigators examined transcription using quantitative real-time PCR for three distinct modes of persistence, exposure to IFN-γ, penicillin G, and iron depletion, in C. psittaci growing in HEp-2 cells (22). At 24 hpi, groEL-1 was upregulated only in the penicillin G model of persistence; IFN-γ persistence actually resulted in significant downregulation of groEL-1. Downregulation of groEL-1 was also observed by 48 hpi for C. psittaci persistence induced by iron deprivation; groEL-2 was not examined in this study. Using a different stress environment, Karunakaran and colleagues (28) examined transcription using a heat shock model. HeLa 229 cells were infected with C. trachomatis serovar D for 18 h and subsequently subjected to a 10-min heat pulse at 45°C. mRNA was quantified using a microarray procedure, and the results showed that there was a >5-fold increase in groEL-1 transcripts; the quantities of the groEL-2 and groEL-3 transcripts did not change. In studies of protein expression, expression of C. trachomatis serovar A cHsp60-1, as determined using Western blotting and an anti-cHsp60-1 monoclonal antibody, was enhanced in an in vitro model of IFN-γ-mediated persistence (2). For C. pneumoniae cHsp60-1, there was a twofold increase in expression at 48 hpi with the following three different models of persistence and/or stress: (i) IFN-γ exposure, (ii) iron deprivation, and (iii) heat shock (39).
Our findings are more consistent with results reported by Gerard and colleagues (20). These investigators quantified mRNA for each groEL homolog in C. trachomatis serovar K using real-time reverse transcription-PCR with the following systems: (i) active infection in HEp-2 cells, (ii) persistent infection in human monocytes, and (iii) synovial tissue from patients with Chlamydia-associated arthritis. In active HEp-2 cell infection, all groEL transcripts were present beginning at 8 hpi, and the levels increased throughout chlamydial development; groEL-3 was transcribed at the highest levels. In the monocyte persistence model, the levels of groEL-1 and groEL-3 transcripts were low, whereas the level of the groEL-2 transcripts increased threefold over 3 days as the organisms entered the persistent state. Findings for the synovial tissues also showed that the levels of groEL-2 transcripts were high. Comparisons of our model with this model of C. trachomatis serovar K persistence in monocytes may not be entirely legitimate because our model involves C. trachomatis serovar E, a less invasive organism, in epithelial cells, but the observations are intriguing nonetheless.
From the standpoint of immunopathogenesis, the importance of cHsp60s in disease has been the subject of several excellent reviews (9, 13, 14, 16, 26, 30, 34, 40). Our previous work, performed with the antisera generated in this study, showed that cHsp60-2 and cHsp60-3, but not cHsp60-1, escape from chlamydial inclusions via vesicle eversion, a process that is exacerbated by exposure to azithromycin (21, 44). The vesicles are thought to interact with host cell antigen presentation and to contribute to the inflammatory response. Studies of heat shock proteins, in general, are being performed since heat shock proteins carry antigens and deliver peptides to the major histocompatibility complex, thus priming the adaptive immune response by inducing specific B and T cells in the absence of adjuvants (41). Heat shock proteins also participate in the innate immune response by stimulating the production of chemokines (41). In one study, cHsp60 serum antibodies were shown to be the best predicting factor for tubal factor infertility (51). Mapping of cHsp60-1 peptide epitopes in human sera has been done (50, 54); it may be worthwhile to investigate whether peptides of cHsp60-2 and/or cHsp60-3 contribute to the generation of specific immunoglobulins.
GroEL proteins are essential for bacterial growth and ensure that newly synthesized proteins are functional; expression of GroEL proteins increases in response to a variety of stresses, including heat shock and nutrient deprivation (56). Structural studies of cHsp60s have shown that although the primary amino acid sequences of cHsp60s differ from the primary amino acid sequences of other organisms, amino acid residues involved in binding polypeptides are conserved (28). It is also clear that cHsp60-1 is negatively regulated by the interaction of a negative regulator, HcrA, with a CIRCE element in the operator regions of the groES-groEL-1 and dnaK operons; HrcA does not appear to regulate groEL-2 or groEL-3 (53). Although the results of studies of C. trachomatis serovar K persistence and synovial fluid support the hypothesis that there is regulation of chlamydial groEL-2 at the level of transcription, there appears to be no Fur/DcrA binding site in upstream sequences. There is only a partial Fur box approximately 300 bp downstream in C. trachomatis serovar E groEL-2; determining whether DcrA binds to groEL-2 sequences is part of a separate project in our laboratory. A likely alternative mechanism for enhanced expression of cHsp60-2 may involve small RNAs that regulate genes posttranscriptionally or by stabilization of mRNA; numerous iron-responsive proteins in other bacteria are known to be regulated in this fashion (25). Chlamydiae code for several small RNAs (5). Finally, the increased level of cHsp60-2 might also involve protein stability. For example, when E. coli GroEL is bound to an unfolded substrate in vitro, the complex remains stable at 25°C for more than 2 weeks; at 43°C, the half-life is 1.5 h (36).
Overall, the results of this study and our previous work (21, 43) strongly indicate that cHsp60-2 should be considered a potential mediator of immune-mediated damage, and they mechanistically indicate that not all modes of chlamydial persistence are identical.
Acknowledgments
This work was supported by grant RO1AI40915 from the NIAID, National Institutes of Health, to J.E.R.
Our appreciation is extended to Priscilla B. Wyrick and Robert V. Schoborg for helpful suggestions and advice.
Editor: D. L. Burns
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Andrews, N. C. 2000. Iron homeostasis: insights from genetics and animal models. Nat. Rev. 1:208-216. [DOI] [PubMed] [Google Scholar]
- 2.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc. Natl. Acad. Sci. USA 90:3998-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beatty, W. L., G. I. Byrne, and R. P. Morrison. 1994. Repeated and persistent infection with Chlamydia and the development of chronic inflammation and disease. Trends Microbiol. 2:94-98. [DOI] [PubMed] [Google Scholar]
- 4.Belland, R. J., D. E. Nelson, D. Virok, D. D. Crane, D. Hogan, D. Sturdevant, W. L. Beatty, and H. D. Caldwell. 2003. Transcriptome analysis of chlamydial growth during IFN-γ-mediated persistence and reactivation. Proc. Natl. Acad. Sci. USA 100:15971-15976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belland, R. J., L. Rose, and Y. AbdelRahman. 2006. Promoter mapping of coding and non-coding RNAs using an intergenic microarray for Chlamydia trachomatis, p. 17-20. In M. Chernesky, H. Caldwell, G. Christiansen, I. N. Clarke, B. Kaltenboek, C. Knirsch, C.-C. Kuo, J. Mahony, R. G. Rank, P. Saikku, J. Schachter, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Proceedings of the 11th International Symposium on Human Chlamydial Infections, Niagara-on-the-Lake, Ontario, Canada. International Chlamydia Symposium, San Francisco, CA.
- 6.Betsou, F., J. M. Sueur, and J. Orfila. 2003. Anti-Chlamydia pneumoniae heat shock protein 10 antibodies in asthmatic adults. FEMS Immunol. Med. Microbiol. 35:107-111. [DOI] [PubMed] [Google Scholar]
- 7.Brunham, R. C., I. W. Maclean, B. Binns, and R. W. Peeling. 1985. Chlamydia trachomatis: its role in tubal infertility. J. Infect. Dis. 152:1275-1282. [DOI] [PubMed] [Google Scholar]
- 8.Bulut, Y., E. Faure, L. Thomas, H. Karahashi, K. S. Michelson, O. Equils, S. G. Morrison, R. P. Morrison, and M. Arditi. 2002. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J. Immunol. 168:1435-1440. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, C. R., and R. C. Brunham. 1999. Pathogenesis of Chlamydia induced pelvic inflammatory disease. Sex. Transm. Infect. 75:21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen, C. R., J. Gichui, Rukaria, S. Sinei, L. K. Gaur, and R. C. Brunham. 2003. Immunogenetic correlates for Chlamydia trachomatis-associated tubal infertility. Obstet. Gynecol. 101:438-444. [DOI] [PubMed] [Google Scholar]
- 11.Cohen, C. R., K. M. Koochesfahani, A. S. Meier, C. Shen, K. Karunakaran, B. Ondondo, T. Kinyari, N. R. Mugo, R. Nguti, and R. C. Brunham. 2005. Immunoepidemiologic profile of Chlamydia trachomatis infection: importance of heat shock protein 60 and interferon-gamma. J. Infect. Dis. 4:591-599. [DOI] [PubMed] [Google Scholar]
- 12.Conway, D. J., M. J. Holland, A. E. Campbell, R. L. Bailey, P. Krausa, R. W. Peeling, H. C. Whittle, and D. C. Mabey. 1996. HLA class I and II polymorphisms and trachomatous scarring in a Chlamydia trachomatis-endemic population. J. Infect. Dis. 174:643-646. [DOI] [PubMed] [Google Scholar]
- 13.Darville, T. 2000. Chlamydia spp., p. 229-261. In J. P. Nataro, M. J. Blaser, and S. Cunningham-Rundles (ed.), Persistent bacterial infections. ASM Press, Washington, DC.
- 14.Debattista, J., P. Timms, J. Allan, and J. Allan. 2003. Immunopathogenesis of Chlamydia trachomatis infections in women. Fertil. Steril. 79:1273-1287. [DOI] [PubMed] [Google Scholar]
- 15.den Hartog, J. E., J. A. Land, F. R. M. Stassen, A. G. H. Kessels, and C. A. Bruggeman. 2005. Serological markers of persistent C. trachomatis infections in women with tubal factor subfertility. Hum. Reprod. 20:986-990. [DOI] [PubMed] [Google Scholar]
- 16.Di Felice, V., S. David, F. Cappello, F. Farina, and G. Zummo. 2005. Is chlamydial heat shock protein 60 a risk factor for oncogenesis? Cell. Mol. Life Sci. 62:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreses-Werringloer, U., I. Padubrin, B. Jurgens-Saathoff, A. P. Hudson, H. Zeidler, and L. Kohler. 2000. Persistence of Chlamydia trachomatis is induced by ciprofloxacin and ofloxacin in vitro. Antimicrob. Agents Chemother. 44:3288-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Equils, O., D. Lu, M. Gatter, S. S. Witkin, C. Bertolotto, M. Arditi, J. A. McGregor, C. F. Simmons, and C. J. Hobel. 2006. Chlamydia heat shock protein 60 induces trophoblast apoptosis through TLR4. J. Immunol. 177:1257-1263. [DOI] [PubMed] [Google Scholar]
- 19.Gerard, H. C., L. Kohler, P. J. Branigan, H. Zeilder, H. R. Schumacher, and A. P. Hudson. 1998. Viability and gene expression in Chlamydia trachomatis during persistent infection of cultured human monocytes. Med. Microbiol. Immunol. 187:115-120. [DOI] [PubMed] [Google Scholar]
- 20.Gerard, H. C., J. A. Whittum-Hudson, H. R. Schumacher, and A. P. Hudson. 2004. Differential expression of three Chlamydia trachomatis Hsp60-encoding genes in active vs. persistent infections. Microb. Pathog. 36:35-39. [DOI] [PubMed] [Google Scholar]
- 21.Giles, D. K., J. D. Whittimore, R. W. LaRue, J. E. Raulston, and P. B. Wyrick. 2006. Ultrastructural analysis of chlamydial antigen-containing vesicles everting from the Chlamydia trachomatis inclusion. Microb. Infect. 8:1579-1591. [DOI] [PubMed] [Google Scholar]
- 22.Goellner, S., E. Schubert, E. Liebler-Tenorio, H. Hotzel, H. P. Saluz, and K. Sachse. 2006. Transcriptional response patterns of Chlamydia psittaci in different in vitro models of persistent infection. Infect. Immun. 74:4801-4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins, D. P., S. Hemsley, and P. J. Canfield. 2005. Association of uterine and salpingeal fibrosis with chlamydial Hsp60 and Hsp10 antigen-specific antibodies in Chlamydia-infected koalas. Clin. Diag. Lab. Immunol. 12:632-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson, M., N. White, P. Giffard, and P. Timms. 1999. Epizootiology of Chlamydia infections in two free-range koala populations. Vet. Microbiol. 65:255-264. [DOI] [PubMed] [Google Scholar]
- 25.Kadner, R. J. 2005. Regulation by iron: RNA rules the rust. J. Bacteriol. 187:6870-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalayoglu, M. V., Indrawati, R. P. Morrison, S. G. Morrison, Y. Yuan, and G. I. Byrne. 2000. Chlamydial virulence determinants in atherogenesis: the role of chlamydial lipopolysaccharide and heat shock protein 60 in macrophage-lipoprotein interactions. J. Infect. Dis. 181:S483-S489. [DOI] [PubMed] [Google Scholar]
- 27.Karinen, L., A. Pouta, A. L. Hartikainen, A. Bloigu, M. Paldanius, M. Leinonen, P. Saikku, and M. R. Jarvelin. 2004. Antibodies to Chlamydia trachomatis heat shock proteins Hsp60 and Hsp10 and subfertility in general population at age 31. Am. J. Reprod. Immunol. 52:291-297. [DOI] [PubMed] [Google Scholar]
- 28.Karunakaran, K. P., Y. Noguchi, T. D. Read, A. Cherkasov, J. Kwee, C. Shen, C. C. Nelson, and R. C. Brunham. 2003. Molecular analysis of the multiple GroEL proteins of chlamydiae. J. Bacteriol. 185:1958-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kelver, M. E., A. Kaul, B. Nowicki, W. E. Findley, T. W. Hutchens, and N. Nagamani. 1996. Estrogen regulation of lactoferrin expression in human endometrium. Am. J. Reprod. Immunol. 36:243-247. [DOI] [PubMed] [Google Scholar]
- 30.Kinnunen, A., J. Paavonen, and H. M. Surcel. 2001. Heat shock protein 60 specific T-cell response in chlamydial infections. Scand. J. Immunol. 54:76-81. [DOI] [PubMed] [Google Scholar]
- 31.Kol, A., G. K. Suhkova, A. H. Lichtman, and P. Libby. 1998. Chlamydial heat shock protein 60 localizes in human atheroma and regulates macrophage tumor necrosis factor-α and matrix metalloproteinase expression. Circulation 98:300-307. [DOI] [PubMed] [Google Scholar]
- 32.Kol, A., A. H. Lichtman, R. W. Finberg, P. Libby, and E. A. Kurt-Jones. 2000. Cutting edge: heat shock protein (HSP) 60 activates the innate immune response: CD14 is an essential receptor for HSP60 activation of mononuclear cells. J. Immunol. 164:13-17. [DOI] [PubMed] [Google Scholar]
- 33.LaVerda, D., L. N. Albanese, P. E. Ruther, S. G. Morrison, R. P. Morrison, K. A. Ault, and G. I. Byrne. 2000. Seroreactivity to Chlamydia trachomatis Hsp10 correlates with severity of human genital tract disease. Infect. Immun. 68:303-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LaVerda, D., M. V. Kalayoglu, and G. I. Byrne. 1999. Chlamydial heat shock proteins and disease pathology: new paradigms for old problems? Infect. Dis. Obstet. Gynecol. 7:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 36.Melkani, G. C., G. Zardeneta, and J. A. Mendoza. 2005. On the chaperone activity of GroEL at heat-shock temperature. Int. J. Biochem. Cell. Biol. 37:1375-1385. [DOI] [PubMed] [Google Scholar]
- 37.Morrison, R. P., K. Lyng, and H. D. Caldwell. 1989. Chlamydial disease pathogenesis. Ocular hypersensitivity elicited by a genus-specific 57-kDa protein. J. Exp. Med. 169:663-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mozzato-Chamay, N., O. S. Madhi, O. Jallow, D. C. Mabey, R. L. Bailey, and D. J. Conway. 2000. Polymorphisms in candidate genes and risk of scarring trachoma in a Chlamydia trachomatis-endemic population. J. Infect. Dis. 182:1545-1548. [DOI] [PubMed] [Google Scholar]
- 39.Mukhopadhyay, S., R. D. Miller, E. D. Sullivan, C. Theodoropoulos, S. A. Mathews, P. Timms, and J. T. Summersgill. 2006. Protein expression profiles of Chlamydia pneumoniae in models of persistence versus those of heat shock stress response. Infect. Immun. 74:3853-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peeling, R. W., and D. C. Mabey. 1999. Heat shock protein expression and immunity in chlamydial infections. Infect. Dis. Obstet. Gynecol. 7:72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qazi, K. R., M. R. Qazi, E. Julian, M. Singh, M. Abedi-Valugerdi, and C. Fernandez. 2005. Exposure to mycobacteria primes the immune system for evolutionarily diverse heat shock proteins. Infect. Immun. 73:7687-7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rau, A., S. Wyllie, J. Whittimore, and J. E. Raulston. 2005. Identification of Chlamydia trachomatis genomic sequences recognized by chlamydial divalent cation-dependent regulator A (DcrA). J. Bacteriol. 187:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raulston, J. E. 1997. Response of Chlamydia trachomatis serovar E to iron restriction in vitro and evidence for iron-regulated chlamydial proteins. Infect. Immun. 65:4539-4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raulston, J. E., T. R. Paul, S. T. Knight, and P. B. Wyrick. 1998. Localization of Chlamydia trachomatis heat shock proteins 60 and 70 during infection of a human endometrial epithelial cell line in vitro. Infect. Immun. 66:2323-2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saikku, P., M. Leinonen, K. Mattila, M. R. Ekman, M. S. Nieminen, P. H. Huttunen, and V. Valtonen. 1988. Serological evidence of an association of a novel Chlamydia, TWAR, with chronic coronary heart disease and acute myocardial infarction. Lancet ii:983-986. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 47.Sasu, S., D. LaVerda, N. Qureshi, D. T. Golenbock, and D. Beasley. 2001. Chlamydia pneumoniae and chlamydial heat shock protein 60 stimulate proliferation of human vascular smooth muscle cells via Toll-like receptor 4 and p44/p42 mitogen-activated protein kinase activation. Circ. Res. 89:244-250. [DOI] [PubMed] [Google Scholar]
- 48.Sharma, J., F. Dong, M. Pirbhai, and G. Zhong. 2005. Inhibition of proteolytic activity of a chlamydial proteosome/protease-like factor by antibodies from humans infected with Chlamydia trachomatis. Infect. Immun. 73:4414-4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 50.Sziller, I., S. S. Witkin, M. Zeigert, Z. Csapo, A. Ujhazy, and Z. Papp. 1998. Serological responses of patients with ectopic pregnancy to epitopes of the Chlamydia trachomatis 60kDa heat shock protein. Hum. Reprod. 13:1088-1093. [DOI] [PubMed] [Google Scholar]
- 51.Tiitinen, A., H. M. Surcel, M. Halttunen, S. Birkelund, A. Bloigu, G. Christiansen, P. Koskela, S. G. Morrison, R. P. Morrison, and J. Paavonen. 2006. Chlamydia trachomatis and chlamydial heat shock protein 60-specific antibody and cell-mediated responses predict tubal factor infertility. Hum. Reprod. 21:1533-1538. [DOI] [PubMed] [Google Scholar]
- 52.Toye, B., C. Laferrie, P. Claman, P. Jessamine, and R. Peeling. 1993. Association between antibody to the chlamydial heat shock protein and tubal infertility. J. Infect. Dis. 168:1236-1240. [DOI] [PubMed] [Google Scholar]
- 53.Wilson, A. C., C. C. Wu, J. R. Yates III, and M. Tan. 2005. Chlamydial GroEL autoregulates its own expression through direct interactions with the HrcA repressor protein. J. Bacteriol. 187:7535-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Witkin, S. S., M. Askienazy-Elbhar, J. Henry-Suchet, J. Belaisch-Allart, J. Tort-Grumbacj, and K. Sarjdine. 1998. Circulating antibodies to a conserved epitope of the Chlamydia trachomatis 60 kDa heat shock protein (hsp60) in infertile couples and its relationship to antibodies to C. trachomatis surface antigens and the Escherichia coli and human Hsp60. Hum. Reprod. 13:1175-1179. [DOI] [PubMed] [Google Scholar]
- 55.Wyllie, S., and J. E. Raulston. 2001. Identifying regulators of transcription in an obligate intracellular pathogen: a metal-dependent repressor in Chlamydia trachomatis. Mol. Microbiol. 40:1027-1036. [DOI] [PubMed] [Google Scholar]
- 56.Young, R. A., and T. J. Elliot. 1989. Stress proteins, infection, and immune surveillance. Cell 59:5-8. [DOI] [PubMed] [Google Scholar]