Abstract
PtlH is an essential component of the Ptl system, the type IV transporter responsible for secretion of pertussis toxin (PT) across the outer membrane of Bordetella pertussis. The nine Ptl proteins are believed to interact to form a membrane-spanning apparatus through which the toxin is secreted. In this study, we monitored the subcellular localization of PtlH in strains of B. pertussis lacking PT, lacking other Ptl proteins, or from which ATP has been depleted in order to gain insight into the requirements for assembly of PtlH with the remainder of the Ptl transporter complex that is thought to be tightly embedded in the membrane. We found that PtlH is exclusively localized to the inner membrane fraction of the cell in a wild-type strain of B. pertussis. In contrast, PtlH localized to both the cytoplasmic and inner membrane fractions of a mutant strain of B. pertussis that does not produce PT. In comparison to how it localized in wild-type strains of B. pertussis, PtlH exhibited aberrant localization in strains lacking PtlD, PtlE, PtlF, and PtlG. We also found that localization of PtlH was perturbed in B. pertussis strains that were treated with carbonyl cyanide m-chlorophenylhydrazone and sodium arsenate, which are capable of depleting cellular ATP levels, and in strains of B. pertussis that produce an altered form of PtlH that lacks ATPase activity. When taken together, these results indicate that tight association of PtlH with the membrane, likely through interactions with components of the transporter-PT complex, requires the toxin substrate, a specific subset of the Ptl proteins, and ATP. Based on these data, a model for the assembly of the Ptl transporter-PT complex is presented.
Pertussis toxin (PT) is an essential virulence factor that is secreted by Bordetella pertussis (40), the causative agent of pertussis (whooping cough). PT is a large, complex, oligomeric protein that interferes with signaling pathways in host cells by ADP-ribosylating a family of GTP-binding regulatory proteins (16, 18). The toxin, which belongs to the AB class of bacterial toxins, is composed of five different subunits, S1, S2, S3, S4, and S5, found in a ratio of 1:1:1:2:1 (24, 25, 37). The enzymatically active S1 subunit (the A component) sits atop a ring formed by the remaining subunits that make up the binding component, or the B oligomer, of the toxin (33).
Secretion of PT from the bacterium requires a transport system that comprises nine accessory proteins known as the Ptl proteins (41). The genes encoding the transport system are located directly downstream from the genes encoding the toxin structural subunits (Fig. 1A). The Ptl transport apparatus is a member of the type IV family of secretion systems that are involved in the transport of proteins and/or DNA from bacterial cells (8, 42). The Ptl system displays considerable homology to other type IV transporters, including the prototypic type IV transporter, the VirB/D4 system of Agrobacterium tumefaciens, which has been studied extensively. Because of the similarities between these systems, much can be surmised concerning the general architecture of the Ptl transporter based on what is known about the VirB/D4 system. However, extrapolations from the VirB/D4 system to the Ptl system have certain limitations because distinct differences exist between the Ptl and VirB/VirD4 systems. In particular, these two systems differ dramatically in regards to their substrates and the locations within the bacteria at which that the substrates are believed to initially interact with the transporters. The substrates for the VirB/D4 transporter are cytoplasmic proteins, certain of which are covalently bound to DNA. These substrates are thought to interact with the VirB/D4 transporter at the cytoplasmic surface of the inner membrane through initial interaction with VirD4, a protein that is lacking in the Ptl system (2, 7). In contrast, the substrate for the Ptl system, PT, is an oligomeric protein that is transported via a Sec-like system to the bacterial periplasm independent of the Ptl transporter (41). Thus, PT is believed to interact initially with the Ptl transporter in the periplasm rather than the cytoplasm and therefore must interact with the Ptl transporter in a manner distinctly different from the manner in which the substrates of the VirB/D4 transporter initially interact with that assembly. Currently, little is known about how and when PT initially interacts with its transporter.
FIG. 1.
The ptx-ptl region and conserved motifs in transfer ATPases. (A) Schematic representation of the structure and nucleotide numbering of the ptx-ptl operon. Pr represents the promoter region of ptx-ptl operon. (B) Schematic representation of the conserved motifs of transfer ATPases that have been previously described (28). The consensus sequence of each motif is shown along with the corresponding sequence in PtlH.
In our previous studies, we examined mutants of B. pertussis that lack specific Ptl proteins or the toxin itself in order to gain a better understanding of the transporter and its interaction with PT. In the course of these studies, we noted that subcellular localization of PtlH, a critical component of the Ptl transporter, is altered in certain of these mutants. PtlH is a member of a family of type IV transporter proteins that possess ATPase activity and which have been demonstrated to be peripheral inner membrane proteins (9, 27). Because tight association of peripheral membrane proteins with the membrane can depend on their interaction with other integral membrane proteins (19, 29), we monitored the subcellular localization of PtlH in specific mutant strains of B. pertussis in order to gain insight into the requirements for assembly of PtlH with the remainder of the membrane-embedded Ptl transporter. In this study, we examined the subcellular localization of PtlH in mutants of B. pertussis lacking PT and/or specific Ptl proteins. We also examined localization of PtlH in the absence of ATP binding. We found that tight association of PtlH with the membrane is dependent on the toxin substrate, certain other Ptl proteins, and ATP binding and/or hydrolysis, information that sheds light on the requirements for transporter assembly, interactions of PT with the transporter, and the sequence of events that take place during secretion of PT from B. pertussis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains of B. pertussis and Escherichia coli as well as the plasmids used in this study are listed in Table 1. B. pertussis strains were grown at 37°C on Bordet-Gengou (BG) agar.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− ϕ80dlacZΔM15 Δ(lacZYA-argF)U169 deoR recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | Gibco-BRL |
| SM10λpir | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu λpirR6K | 31 |
| BL-21(DE3) | F−ompT gal[dcm][lon] hsdSB (rB− mB−); an E. coli B strain with DE3, a λ prophage carrying the T7 RNA polymerase gene | Stratagene |
| B. pertussis strains | ||
| BP536 | Wild-type, nalidixic acid-resistant, streptomycin-resistant derivative of Tohama I | 36 |
| BP536Δptx(507-3683) | BP536 with an in-frame deletion in the ptx region, from nucleotide 507 to nucleotide 3683 | This study |
| BP536Δptl | BP536 with a 7.4-kb in-frame deletion in the ptx-ptl region, from nucleotide 3626 to nucleotide 11039 | 5 |
| BP536Δptxptl | BP536 with an 11.5-kb in-frame deletion in ptx-ptl genes, from nucleotide 425 to nucleotide 11971 | 15 |
| BP536ΔptlH | BP536 with a deletion in the ptx-ptl region from nucleotide 10973 to nucleotide 11992 | This study |
| BP536ΔptlD | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 6804 to nucleotide 8191 | This study |
| BP536ΔptlI | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 8201 to nucleotide 8323 | 14 |
| BP536ΔptlF | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 9078 to nucleotide 9845 | This study |
| BP536ΔptlA | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 3687-3989 | This study |
| BP536ΔptlE | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 8378 to nucleotide 9053 | This study |
| BP536ΔptlG | BP536 with an in-frame deletion in the ptx-ptl region, from nucleotide 9974 to nucleotide 10972 | This study |
| Plasmids | ||
| pUFR047 | Broad-host-range (IncW) vector, Mob+, lacZα+, gentamicin resistant | 11 |
| pGEM-T Easy | Ampicillin-resistant cloning vector | Promega |
| pSS1129 | Genr, Ampr, Sms allelic exchange vector | 35 |
| pQE80 | Ampr, cis-repressed, high-level expression vector of N-terminally His6-tagged proteins | QIAGEN |
| pTH18 | pUFR047 containing ptxS2, ptxS4, ptxS5, and ptxS3 under lacZ promoter control | 15 |
| pTH19 | pRK415 containing the ptx-ptl promoter region and ptxS1 | 15 |
| pAV9 | pGEX6P-1 containing ptlH (nucleotides 10973 to 11992) at the 3′ end of the gene encoding GST | This study |
| pAV21 | pGEX6P-1 containing ptlH(K176A) at the 3′ end of the gene encoding GST | This study |
| pAV31 | pUFR047 containing ptlH under ptx-ptl promoter control | This study |
| pAV32 | pUFR047 containing ptlH(K176A) under ptx-ptl promoter control | This study |
| pAV69 | PQE80 containing ptlH with a His6 tag at the 5′ end of the gene | This study |
| pTH22 | pUFR047 containing the phoA gene of E. coli under lacZ promoter control | 15 |
Construction of B. pertussis in-frame deletion mutants.
B. pertussis in-frame chromosomal deletion mutants were constructed by homologous recombination as previously described (14, 35). Briefly, approximately 900-bp regions flanking upstream and downstream of the desired gene to be deleted were amplified using primers mapping in upstream and downstream regions, respectively. Amplified products were combined either by overlapping PCR or by ligation and cloned in vector pSS1129 (a suicide vector that can replicate in E. coli but not in B. pertussis). The resulting constructs were transformed into E. coli SM10λpir and finally transferred to B. pertussis BP536 by biparental conjugation. The BP536 cells in which the required construct had integrated into the chromosome by homologous recombination were selected on BG plates containing gentamicin (10 μg/ml) and nalidixic acid (50 μg/ml). A second homologous recombination was achieved by selection on BG plates containing streptomycin (100 μg/ml), resulting in the loss of the plasmid. Streptomycin-resistant exconjugants with the desired gene deletion were identified by PCR.
Production of polyclonal antibodies against recombinant PtlH.
pAV69, a construct containing the full-length wild-type ptlH gene in pQE80 expression vector (QIAGEN), was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) to express the His6-PtlH fusion protein in E. coli BL-21. Fusion protein was purified using Ni-nitrilotriacetic acid resin (QIAGEN) and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE gel slice containing the purified His6-PtlH fusion protein was used to raise antibodies in mice at Alpha Diagnostic Inc. (TX).
Introduction of genes in trans into BP536.
Genes were introduced in trans into B. pertussis by cloning them into a pUFR047 vector. The resulting plasmids were then introduced into BP536 by triparental conjugation as described previously (3). Exconjugants were selected on BG agar containing gentamicin (10 μg/ml) and streptomycin (100 μg/ml).
Preparation of cell extracts and fractionation. (i) Separation of soluble and total membrane fractions.
B. pertussis strains were grown on BG agar plates and suspended in phosphate-buffered saline (PBS) to an A550 of 2.0. Cells (6 ml of suspended culture) were collected by centrifugation at 10,000 × g, suspended in 1.25 ml lysis buffer (10 mM Tris-HCl, pH 8.0, 20% sucrose, 1 mM EDTA, 0.1 μg/ml lysozyme, protease inhibitor cocktail, and 1 mM phenylmethylsulfonyl fluoride) and incubated on ice for 10 min. Lysates were subjected to a freeze-thaw cycle. Cells were broken by sonication. Samples were cleared of unbroken cells by centrifugation at 2,000 × g for 10 min. Cleared lysates were subjected to ultracentrifugation at 144,000 × g for 1 h. After ultracentrifugation, the pellet (total membrane fraction) and the supernatant (periplasmic/cytoplasmic fraction) were treated with 1.5% Sarkosyl overnight at 4°C on a rotator. Detergent solubilized fractions were subjected to ultracentrifugation at 144,000 × g for 1 h. Proteins were precipitated from detergent-solubilized fractions (100 μl) by ethanol, suspended in SDS-PAGE loading buffer, and subjected to SDS-PAGE according to procedure of Laemmli (21), followed by transfer to nitrocellulose and immunoblot analysis using antiserum raised against His6-PtlH at dilution of 1:2,000. Blots were developed using the ECL system (Amersham). Densitometry was performed using AlphaEaseFC software (Alpha Innotech Corp. USA).
(ii) Triton X-100 solubilization of the total membrane fraction.
The membrane fraction, prepared as described above, was suspended in 10 mM Tris-HCl (pH 8.0) containing 2% Triton X-100. The preparation was incubated overnight at 4°C and then centrifuged at 144,000 × g for 1 h. The pellet (Triton X-100-insoluble fraction) was suspended in 10 mM Tris-HCl (pH 8.0). Equivalent portions of the Triton X-100 fractions were precipitated by ethanol and then subjected to immunoblot analysis.
(iii) Isolation of periplasmic and cytoplasmic fractions.
BP536Δptx(507-3683)(pTH22), which contains a plasmid capable of expressing the phoA gene of E. coli, was grown on BG agar plates. Cells were suspended in PBS to an A550 of 2.0, collected by centrifugation at 10,000 × g, and resuspended in 1 ml of 0.2 M Tris-HCl, pH 8.0. An equal volume of 0.2 M Tris-HCl, pH 8.0, containing 1 M sucrose, 0.5% Zwittergent-316, (Calbiochem, La Jolla, CA), and 0.1 mg of lysozyme per ml was added. Cells were then exposed to mild osmotic shock by the addition of 4 ml of water to release the periplasmic contents of the cell as previously described (32). After a shaking for 2 h at room temperature, the cell suspension was centrifuged for 30 min at 8,000 × g and the supernatant (periplasmic fraction) was collected. Spheroplasts were resuspended in 6 ml of 50 mM Tris-HCl, pH 8.0, and subjected to sonication. Samples were cleared of unbroken spheroplasts by centrifugation at 2,000 × g for 10 min, and the supernatant (cytoplasmic fraction) was collected. Cytoplasmic and periplasmic supernatants were subjected to ultracentrifugation at 144,000 × g for 1 h to remove any membrane debris. After ultracentrifugation, aliquots of the supernatants were used to measure alkaline phosphatase activity as described by Brickman and Beckwith (4). For immunoblot analysis, periplasmic and cytoplasmic fractions were treated with 1.5% Sarkosyl overnight at 4°C on a rotator. Detergent solubilized fractions were again subjected to ultracentrifugation at 144,000 × g for 1 h. Proteins were precipitated from detergent-solubilized fractions (100 μl) by ethanol and subjected to immunoblot analysis as described above.
Depletion of ATP from B. pertussis.
BP536 strain was inoculated in Stainer-Scholte medium at an initial A550 of 0.25 to 0.30 and grown to an A550 of 1.1 to 1.5 for approximately 40 h. After 40 h of culture growth, cells were treated with 50 μM of carbonyl cyanide m-chlorophenylhydrazone (CCCP; Sigma) or 50 mM of sodium arsenate for 30 min with shaking. After treatment, cells were harvested, centrifuged, and suspended in an amount of PBS equal to that of the harvested culture volume. Cellular ATP levels were measured in untreated or CCCP- or sodium arsenate-treated cells by using an ATP bioluminescence assay kit (Promega) as per the manufacturer's instructions. Untreated and treated cells were also subjected to ultracentrifugation for separation of soluble and membrane fractions, treated with 1.5% Sarkosyl, and subjected to immunoblot analysis as described above.
Cloning, expression, and purification of GST-PtlH in E. coli.
A 1,019-bp fragment of the ptx-ptl operon extending from nucleotide 10973 through nucleotide 11992, corresponding to ptlH, was amplified using primers (5′ GGAATTCATGAATGATGCCGCGCCGGATCG 3′ [with its EcoRI site underlined]) and (5′ CCGCTCGAGTCATGGCGCCGGGAGGCCATCC 3′ [with its XhoI site underlined]) and genomic DNA isolated from the B. pertussis Tohama I strain. The nucleotide numbering system used for the ptx-ptl operon has been previously described (25, 41). The amplified product was cloned into the pGEM-T Easy plasmid (Promega) and sequenced. For expression, the ptlH gene was further subcloned in frame into the EcoRI and XhoI sites at the 3′ end of the gene encoding glutathione S-transferase (GST) of pGEX-6P-1 (Amersham). The resulting construct, pAV9, was transferred to E. coli BL-21 for expression and purification. The following procedure was used to purify the GST fusion protein from pAV9. Briefly, cells were inoculated into LB medium containing 100 μg/ml ampicillin and allowed to grow to an A600 of 0.6 at 30°C. Protein expression was induced with 1 mM IPTG for 4 h. Cells were harvested and resuspended in lysis buffer (1 × PBS, 25% sucrose, 1% Triton X-100, protease inhibitor cocktail [Calbiochem], 1 mM phenylmethylsulfonyl fluoride). Cells were lysed by sonication and bacterial debris was pelleted at 10,000 × g for 20 min. The clear supernatant was added to washed glutathione Sepharose 4B beads (Amersham) and mixed gently at 4°C overnight. The beads were centrifuged at 500 × g for 5 min and washed several times with PBS containing 0.1% Triton X-100. GST fusion proteins were eluted from glutathione Sepharose beads using 10 mM of reduced glutathione. Similar methods were used to purify a mutant form of GST-PtlH expressed in pAV21.
ATPase activity assays.
ATPase activity of GST-PtlH and GST-PtlH(K176A) was assayed on native gels using a method previously described (22). Briefly, 20 μg of eluted GST-PtlH or GST-PtlH(K176A) fusion protein was subjected to electrophoresis on native 4 to 20% acrylamide gels in 25 mM Tris-190 mM glycine buffer. The gels were incubated in 50 mM Tris-HCl (pH 7.8)-10 mM MgCl2-1 mM dithiothreitol-2 mM ATP-10% glycerol for 60 min at 37°C followed by staining with 0.034% malachite green, 0.1% Triton X-100, and 10.5 g of ammonium molybdate per liter in 1 M HCl, which results in formation of a precipitate in the presence of inorganic phosphate ion (Pi). Parallel gels were stained with Coomassie brilliant blue to determine the position of the respective GST fusion proteins.
PCR mutagenesis in Walker A box motif of PtlH.
A point mutation was made in the coding sequence of ptlH to change the conserved lysine at amino acid position 176 of PtlH to alanine by using the QuikChange II site-directed mutagenesis kit (Stratagene). Primers mapping at the mutagenesis site (5′ CCGGCCAGACCGGTTCGGGCGCGACCACATTGATGAACGCGTTGAGCGC 3′ and 5′ CCGCTCAACGCGTTCATCAATGTGGTCGCGCCCGAACCGGTCTGGCCGG 3′ [underlining indicates the mutagenesis site]) were used for the amplification. After mutagenesis, plasmids were sequenced for the desired mutations and to ensure that no other mutation was introduced in the ptlH gene. Mutated ptlH, designated ptlH(K176A), was further subcloned in frame into the EcoRI and XhoI sites at the 3′ end of the gene encoding GST of pGEX-6P-1, resulting in pAV21.
Statistical analysis.
Statistical comparisons between ptl mutant and parent strains or between ptx mutant and parent strains were performed by analysis of variance (ANOVA) and tested with Tukey's HSD test for multiple comparisons.
RESULTS
Subcellular localization of PtlH in a wild-type strain of B. pertussis.
In order to localize PtlH in B. pertussis, we fractionated bacterial cells into soluble (cytoplasmic and periplasmic) and total (inner and outer) membrane fractions. Visualization of PtlH on immunoblots by using polyclonal anti-PtlH antiserum required treatment of the fractions with 1.5% Sarkosyl. Sarkosyl treatment most likely disrupts higher-molecular-weight complexes of PtlH, such as oligomeric forms of PtlH or PtlH complexed with other proteins, thus allowing PtlH to enter the gel and migrate as a monomeric species.
When we examined the wild-type strain of B. pertussis, BP536, we found that PtlH migrated as a protein of approximately 37 kDa that was detected exclusively in the membrane fraction of the cells (Fig. 2A, lanes 1 and 2). Immunoblot analysis of a strain of B. pertussis containing an in-frame deletion of PtlH, BP536ΔptlH, showed no reactivity in either the membrane or soluble fraction (Fig. 2A, lanes 3 and 4), thereby confirming the identity of the 37-kDa band as PtlH.
FIG. 2.
Localization of PtlH in BP536. (A) Soluble (S) and total membrane (M) fractions of different B. pertussis strains were prepared as described in Materials and Methods and subjected to immunoblot analysis using polyclonal antibodies specific for PtlH. Lanes 1 and 2, BP536; lanes 3 and 4, BP536ΔptlH. (B) Total membrane fraction of BP536 was solubilized with 2% Triton X-100 and separated as soluble (S) and insoluble (I) fractions by centrifugation. Equivalent amounts of the soluble and insoluble fractions were then subjected to immunoblot analysis with PtlH antibodies. Lane 1, Triton X-100-soluble fraction; lane 2, Triton X-100-insoluble fraction. Positions of molecular size markers (in kilodaltons) are indicated at the left. The arrow indicates the protein band corresponding to PtlH.
In order to further localize the membrane-bound form of PtlH, we treated the membrane fraction with Triton X-100 to separate the inner and outer membranes, since this detergent is thought to preferentially solubilize inner membrane proteins of B. pertussis (1, 13, 17). We used this technique, since isopycnic centrifugation cannot separate the membranes of B. pertussis into discrete fractions corresponding to the inner and outer membranes (13). As shown in Fig. 2B, PtlH fractionated almost exclusively to the Triton X-100-soluble fraction, suggesting that the membrane-bound form of PtlH is associated with the inner membrane. Association of PtlH with the inner membrane as opposed to the outer membrane of the bacterium is consistent with the subcellular localization of other members of this family of transfer ATPases (27).
Subcellular localization of PtlH in strains lacking PT and PT subunits.
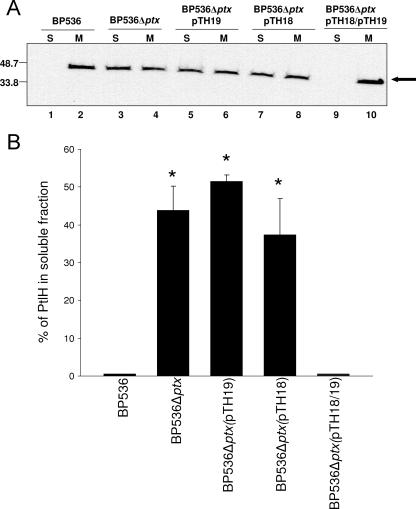
We next determined whether the presence of PT influenced the subcellular localization of PtlH. We examined a strain of B. pertussis, BP536Δptx(507-3683), in which there is an in-frame deletion in the ptx region and which does not produce any of the PT subunits but which does produce a functional Ptl apparatus. Immunoblot analysis of this strain showed distribution of PtlH in both the soluble and membrane fractions (Fig. 3A, lanes 3 and 4) in contrast to the exclusive localization of PtlH to the membrane fraction seen with the wild-type strain (Fig. 3A, lanes 1 and 2).
FIG. 3.
Localization of PtlH in BP536 strains lacking PT or its subunits. Soluble (S; 100 μl) and membrane (M; 100 μl) fractions of BP536 strains lacking PT or PT subunits and complemented strains were prepared as described in Materials and Methods and subjected to immunoblot analysis using polyclonal antibodies specific for PtlH. (A) Lanes 1 and 2, BP536; lanes 3 and 4, BP536Δptx(507-3683); lanes 5 and 6, BP536Δptx(507-3683)(pTH19), which produces only the S1 subunit; lanes 7 and 8, BP536Δptx(507-3683)(pTH18), which produces only the B oligomer; lanes 9 and 10, BP536Δptx(507-3683)(pTH18)(pTH19), which produces both the S1 subunit and the B oligomer. Positions of molecular size markers (in kilodaltons) are indicated at the left. The arrow indicates the protein band corresponding to PtlH. (B) Percentages of PtlH found in soluble fractions. Immunoblots from three independent experiments were analyzed by densitometry to determine the percentage of PtlH found in the soluble fraction. Results are reported as means ± standard deviations (error bars). Asterisks indicate significant (P < 0.05) differences in the percentage of PtlH found in the soluble fraction of BP536Δptx(507-3683), BP536Δptx(507-3683)(pTH19), or BP536Δptx(507-3683)(pTH18) compared to either BP536 or BP536Δptx(507-3683)(pTH18)(pTH19) as determined by Tukey's HSD test following ANOVA. The percentages of PtlH found in the soluble fractions of BP536Δptx(507-3683), BP536Δptx(507-3683)(pTH19), and BP536Δptx(507-3683)(pTH18) were not found to be significantly different from each other.
Because our results indicated that PT is required for localization of PtlH exclusively to the membrane, we next determined whether either the enzymatically active S1 subunit or the B oligomer (subunits S2, S3, S4, and S5) might be sufficient to direct localization of PtlH exclusively to the membrane. We introduced plasmids expressing ptxS1 (pTH19) or ptxS2-ptxS5 (pTH18) into BP536Δptx(507-3683). In both cases, we observed that approximately 35 to 50% of the cellular PtlH partitioned to the soluble fraction (Fig. 3A, lanes 5 to 8) and that the localization of PtlH in these strains did not differ significantly from that of BP536Δptx(507-3683) (Fig. 3B). However, when the enzymatically active S1 subunit and B oligomer were produced simultaneously in BP536Δptx(507-3683) by introducing both pTH19 and pTH18, we observed that PtlH localized exclusively to the membrane fraction (Fig. 3A, lanes 9 and 10), indicating that an intact PT molecule is necessary for tight association of PtlH with the membrane.
In the experiment whose data are shown in Fig. 3, the soluble fraction consisted of both the cytoplasmic and periplasmic contents of the cell. In order to further localize the soluble form of PtlH, we separated the cytoplasmic and periplasmic contents of BP536Δptx(507-3683)(pTH22). This strain expresses a gene encoding alkaline phosphatase, which can be used as a marker for the periplasmic contents of the cell (12). After separating cytoplasmic and periplasmic fractions of the cell by using the Zwittergent-lysozyme procedure described in Materials and Methods, we found that approximately 90% of the alkaline phosphatase was found in the periplasmic fraction, with only about 10% contaminating the cytoplasmic fraction (Fig. 4A), indicating that the method satisfactorily removes the periplasmic contents of the cell from its cytoplasmic contents. We then examined both the periplasmic and cytoplasmic fractions for PtlH content. As shown in Fig. 4B, PtlH partitioned exclusively to the cytoplasmic fraction, indicating that the soluble form of PtlH is located in the cytoplasm.
FIG. 4.
Localization of PtlH in BP536Δptx(507-3683)(pTH22), a strain that produces the periplasmic marker enzyme, alkaline phosphatase. Cytoplasmic (C) and periplasmic (P) fractions were separated by the procedure described in Materials and Methods. (A) Alkaline phosphatase activities of the fractions were assessed to determine the efficiency of separation of the periplasm from the cytoplasm. Error bars indicate standard deviations of triplicate measurements. (B) Fractions were also subjected to immunoblot analysis with anti-PtlH antibodies. Lanes 1 and 2 show cytoplasmic and periplasmic fractions, respectively. Positions of molecular size markers (in kilodaltons) are indicated at the left. The arrow indicates the protein band corresponding to PtlH.
Localization of PtlH in strains lacking other Ptl proteins.
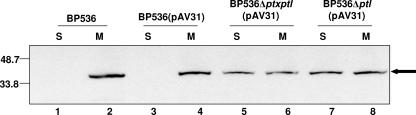
We next asked whether the presence of the other Ptl proteins influences localization of PtlH. BP536Δptl, which does not produce the Ptl proteins but does produce PT, was complemented in trans with a plasmid that expresses wild-type ptlH (pAV31). As a control, the wild-type strain was also complemented with this plasmid. Immunoblot analysis revealed that PtlH partitioned exclusively to the membrane fraction of the wild-type strain into which pAV31 had been introduced (Fig. 5, lanes 3 and 4). Approximately 50% of the total cellular PtlH could be seen in the soluble fraction of BP536Δptl(pAV31), a strain of B. pertussis lacking all Ptl proteins except PtlH, which was provided in trans (Fig. 5, lanes 7 and 8). These observations suggest that the presence of other Ptl proteins is critical for tight association of PtlH with the membrane.
FIG. 5.
Localization of PtlH in strains lacking other Ptl proteins. A plasmid containing ptlH (pAV31) was introduced into BP536 (wild-type B. pertussis), BP536Δptxptl (lacking PT and Ptl proteins), or BP536Δptl (lacking Ptl proteins). Soluble (S; 100 μl) and membrane (M; 100 μl) fractions of these strains were prepared as described in Materials and Methods and subjected to immunoblot analysis using polyclonal antibodies specific for PtlH. Lanes 1 and 2, BP536; lanes 3 and 4, BP536(pAV31); lanes 5 and 6, BP536Δptxptl(pAV31); lanes 7 and 8, BP536Δptl(pAV31). Positions of molecular size markers (in kilodaltons) are indicated at the left. The arrow indicates the protein band corresponding to PtlH. Results are representative of two independent experiments.
Because both PT and the Ptl proteins are required for wild-type localization of PtlH, we asked whether the subcellular location of PtlH would be more perturbed in a strain lacking both of these components than in a strain lacking only the Ptl proteins. When we examined the cellular localization of PtlH in BP536Δptxptl(pAV31), a strain lacking all ptx and ptl genes except ptlH, which was provided in trans, we found that PtlH partitioned in a manner similar to that of BP536Δptl(pAV31) (Fig. 5, compare lanes 5 and 6 to lanes 7 and 8). The finding that about half of the PtlH of the cell associates with the membrane even in the absence of other members of the Ptl transporter-PT complex may be due to an intrinsic affinity of PtlH for the membrane. Other homologs of PtlH, in particular, VirB11, are found to be associated with the inner membrane of the bacterium, even in the absence of other transporter proteins (27).
In order to gain insight into which Ptl protein(s) might be necessary for tight association of PtlH with the membrane, we examined the localization of PtlH in strains of B. pertussis which contained an in-frame deletion of ptlA, ptlD, ptlE, ptlF, ptlG, or ptlI. The in-frame deletion of ptlA, ptlD, ptlE, ptlF, ptlG, or ptlI resulted in deletion of almost the entire coding region for the respective protein. Unfortunately, repeated attempts to generate mutants lacking the entire coding region of ptlB or ptlC were unsuccessful.
When we examined the localization of PtlH in these mutants, we found that localization was dependent on the particular deletion. PtlH partitioned exclusively to the membrane in the ptlA mutant. A small, but detectable, amount of PtlH was found in the soluble fraction of the ptlI mutant, and approximately 30 to 50% of total cellular PtlH was found in the soluble fraction of the ptlD, ptlE, ptlF, and ptlG mutants (Fig. 6). Statistical analysis of the data indicates that the localization of PtlH in the ptlD, ptlE, ptlF, and ptlG mutants is significantly different from that in either the wild-type strain or the ptlA mutant (P < 0.05). Moreover, the localization of PtlH in the ptlI mutant was significantly different from its localization in the ptlD, ptlE, and ptlF mutants (P < 0.05). We note that the amount of PtlH observed in each of the mutant strains, with the exception of the ptlD mutant, was similar to that of the wild-type strain. Thus, as expected, the in-frame ptlA, ptlE, ptlF, ptlG, and ptlI mutations do not appear to affect expression of the downstream ptlH gene. The decrease in the total amount of PtlH observed in the strain lacking PtlD is likely due to stabilization of PtlH by PtlD since complementation of the mutant with ptlD can restore PtlH levels to those of the wild-type strain (A. Cheung and D. Burns, unpublished data).
FIG. 6.
Localization of PtlH in BP536 strains lacking different Ptl proteins. Soluble (S; 100 μl) and membrane (M; 100 μl) fractions of BP536 strains lacking specific Ptl proteins were prepared as described in Materials and Methods and subjected to immunoblot analysis using polyclonal antibodies specific for PtlH. (A) Lanes 1 and 2, BP536; lanes 3 and 4, BP536ΔptlA; lanes 5 and 6, BP536ΔptlD. (B) Lanes 1 and 2, BP536; lanes 3 and 4, BP536ΔptlE; lanes 5 and 6, BP536ΔptlF; lanes 7 and 8, BP536ΔptlG; lanes 9 and 10, BP536ΔptlI. Positions of molecular size markers (in kilodaltons) are indicated at the left. The arrow indicates the protein band corresponding to PtlH. (C) Percentage of PtlH found in soluble fraction. Immunoblots from three independent experiments were analyzed by densitometry to determine the percentage of PtlH found in the soluble fraction. Results are reported as means ± standard deviations (error bars). An asterisk indicates significant (P < 0.05) differences between the percentages of PtlH found in the soluble fraction of the strain and that of either BP536 or BP536ΔptlA. A plus sign indicates significant (P < 0.05) differences between the percentages of PtlH found in the soluble fraction of the strain and that of BP536ΔptlI. Significant differences were determined by Tukey's HSD test following ANOVA.
Localization of PtlH in cells from which ATP has been depleted and localization of altered forms of PtlH lacking ATPase activity.
PtlH is homologous to transfer ATPases that can exhibit ATP-dependent alterations in conformation (30). Others have suggested that these conformational changes might affect membrane binding and interaction with other components of the transport apparatus (9, 30). We therefore examined the localization of PtlH in bacteria from which ATP has been depleted or of mutant forms of PtlH that exhibit altered ATPase activity in order to assess the dependence of PtlH localization on ATP.
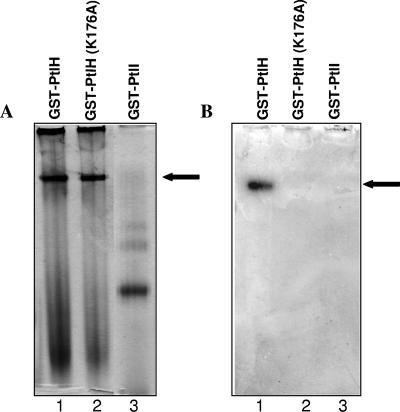
Previously, PtlH was classified as an ATPase, based on homology with other members of this family of transfer ATPases. PtlH contains the amino acid sequence motifs characteristic of this family of proteins that are thought to be critical for ATPase activity (Fig. 1B). In this study, we confirmed the ATPase activity of PtlH by examining the ability of a GST-PtlH fusion that had been produced in E. coli to hydrolyze ATP. Affinity-purified GST-PtlH fusion protein, along with a negative control protein, affinity-purified GST-PtlI, was subjected to nondenaturing PAGE. The gel was then stained to assess the formation of Pi formed after incubation with ATP. The purified protein band corresponding to GST-PtlH exhibited ATPase activity (Fig. 7B, lane 1), whereas the negative control protein did not (Fig. 7B, lane 3). Mutation of the highly conserved alanine residue (mutated from lysine) at amino acid position 176 located in the Walker A box motif of PtlH (Fig. 1B) resulted in loss of this ATPase activity (Fig. 7B, lane 2) as expected since the Walker A box motif of nucleotide-binding proteins has been shown to interact directly with nucleotides and to be essential for the NTP-dependent functions of these proteins (27, 34, 38).
FIG. 7.
ATPase activity associated with GST-PtlH. Proteins were separated on native polyacrylamide gels and either stained with Coomassie brilliant blue (A) or incubated with ATP (B). (B) Pi released due to hydrolysis of ATP formed a dark green precipitate in the gel upon reaction with malachite green. (A and B) Lane 1, GST-PtlH; lane 2, GST-PtlH(K176A); lane 3, GST-PtlI fusion protein. Arrows indicate purified GST-PtlH and the corresponding Pi precipitate formed after malachite green staining in panels A and B, respectively.
In order to assess whether ATP affects the localization of PtlH, we treated B. pertussis cells with either CCCP, which collapses the transmembrane potential and dissipates cellular ATP, or sodium arsenate, which lowers ATP levels without affecting the transmembrane potential (6, 23). After treatment of the bacteria with either CCCP or sodium arsenate, ATP levels were measured to be 2% of the level in untreated cells. Treatment of cells with either energy poison perturbed the localization of PtlH. As can be seen in Fig. 8A, when cells were exposed to either CCCP or sodium arsenate, the portion of PtlH that partitioned to the soluble fraction was larger than that for untreated cells.
FIG. 8.
Localization of PtlH in BP536 in which ATP has been depleted or in a mutant strain of BP536 that lacks the ATPase activity of PtlH. (A) BP536 was treated with either CCCP or sodium arsenate as described in Materials and Methods. Soluble (S) and membrane (M) fractions were prepared and subjected to immunoblot analysis using polyclonal antibodies specific for PtlH. Lanes 1 and 2, untreated; lanes 3 and 4, treated with CCCP; lanes 5 and 6, treated with sodium arsenate. (B) Soluble (S) and membrane (M) fractions of BP536ΔptlH(pAV31), which produces native PtlH, and BP536ΔptlH(pAV32), which produces PtlH(K176A), were prepared and subjected to immunoblot analysis using polyclonal antibodies specific for PtlH. Lanes 1 and 2, BP536ΔptlH(pAV31); lanes 3 and 4, BP536ΔptlH(pAV32). The percentages of PtlH found in the soluble fraction of BP536ΔptlH(pAV32) were quantified to be 28% in one experiment and 35% in another independent experiment.
We also examined whether PtlH(K176A), which lacks ATPase activity, localized in a manner distinct from that of the native form of PtlH. In order to accomplish this, we introduced a plasmid (pAV32) capable of expressing ptlH(K176A) into BP536ΔptlH and examined localization of PtlH(K176A) in this strain. For a control, we also introduced a plasmid (pAV31) capable of expressing wild-type ptlH into BP536ΔptlH. BP536ΔptlH(pAV32) exhibited partial distribution of PtlH(K176A) in the soluble fraction as well as the membrane fraction (Fig. 8B, lanes 3 and 4), whereas native PtlH localized strictly to the membrane fraction in BP536ΔptlH(pAV31) (Fig. 8, lanes 1 and 2). We have previously shown that production of PtlH(K176A) in a wild-type background of B. pertussis results in a dominant negative phenotype (20) suggesting that this form of the protein can properly fold and interact with other proteins.
DISCUSSION
In this study, we found that localization of PtlH in the B. pertussis bacterium is dependent both on Ptl proteins and on the toxin substrate. Our finding that both the toxin and other Ptl proteins are required for proper localization of PtlH suggests that PtlH may interact either directly or indirectly with one or more of these proteins. Given the homologies that exist between the Ptl system and other type IV transporters, it is expected that PtlH would interact directly with other Ptl transporter proteins (9), possibly PtlF, since data from yeast two-hybrid studies of the VirB proteins have demonstrated a direct interaction between VirB11 (PtlH homolog) and VirB9 (PtlF homolog) (39). While direct interactions of PtlH with other Ptl proteins would be predicted, our finding that PT is required for proper localization of PtlH was unexpected and provides important clues as to when and how PT initially interacts with its transporter.
The finding that PT is required for proper localization of PtlH suggests that PtlH tightly engages with the transport apparatus only when PT is present. We found that neither the S1 subunit nor the B oligomer, composed of subunits S2, S3, S4, and S5, can substitute for the holotoxin. These results are consistent with and extend our previous finding, that secretion of PT from the bacterium requires assembly of the S1 subunit with subunits of the B oligomer (15). Importantly, these findings suggest that final assembly of the transporter does not occur in the absence of the holotoxin substrate.
Localization of PtlH in specific ptl mutants provides further clues as to the events that may occur during Ptl transporter morphogenesis and the interaction of the transporter with PT. Mutants of B. pertussis lacking PtlD, PtlE, PtlF, and PtlG displayed the greatest abnormalities in PtlH localization, suggesting that these proteins may interact either directly with PtlH or indirectly, perhaps by stabilizing the protein(s) that interact with PtlH. PtlD, PtlE, PtlF, and PtlG proteins are homologs of VirB6, VirB8, VirB9, and VirB10, respectively, which are part of the membrane-associated core complex of VirB/D4 transporter proteins, the formation of which is believed to represent the first step in the assembly of that transporter (9). Our results suggest that, likewise, association of PtlD, PtlE, PtlF, and PtlG may represent an initial step in Ptl transporter assembly and that assembly of this complex would occur before PtlH tightly engages with the transporter assembly. Subcellular localization of PtlH in the ptlI mutant did not differ significantly from that of the wild-type strain, suggesting that PtlI, which is homologous to another VirB/D4 core-complex protein (VirB7), has only minimal effects on the interaction of PtlH with other proteins of the transporter complex. PtlA, which is homologous to the pilin subunit VirB2, has essentially no effect on PtlH localization. The finding that PtlA is not required for correct localization of PtlH indicates that the full complement of Ptl transporter proteins is not required for PtlH to tightly engage with the transporter. Importantly, the finding that PT but not PtlA is required for correct localization of PtlH suggests that PT can interact with an incomplete form of the transporter that lacks PtlA.
In this study, we also examined the requirement for ATP binding to PtlH and/or hydrolysis of ATP by PtlH for wild-type localization of PtlH. We believe that our result demonstrating a dependence of PtlH localization on ATP helps to shed light on the dynamics of the interaction of PtlH with the transporter-PT complex. In cells from which ATP has been deleted by the energy poisons CCCP or sodium arsenate, PtlH associates only loosely with the membrane, suggesting that normal interactions of PtlH with the membrane likely occur only if ATP is bound to the protein. These results are consistent with our finding that a mutant form of PtlH with alterations in its Walker A box also showed aberrant localization. Since the Walker A box of PtlH is essential for the ATPase activity of the protein and it is thought that this region is involved in nucleotide binding (38), this form of PtlH is predicted to be defective in ATP binding.
Savvides et al. (30) have demonstrated that HP0525, the Helicobacter pylori type IV transporter homolog of PtlH, can exist in three forms, an ATP-bound form, an ADP-bound form, and an apo form devoid of any nucleotides. These three forms differ in conformation with the largest differences observed between the apo form and the two nucleotide-bound forms. These workers suggest that, because of the similarity in conformation between the ADP- and ATP-bound forms, nucleotide binding is likely the critical step for biological activity and that ATP hydrolysis would serve only to accelerate nucleotide release, regenerating the apo form for another cycle of activity. Our results are consistent with a model in which binding of ATP to PtlH results in tighter association of PtlH with the membrane, likely through interactions with other protein members of the transport apparatus and/or PT. Hydrolysis of ATP to ADP and subsequent release of the nucleotide would convert PtlH back to the form that is only loosely associated with the membrane because it exhibits a lower affinity for the transport apparatus.
When taken together, our results on the effects of PT, Ptl proteins, and ATP on the localization of PtlH allow us to significantly extend our model of PT secretion (Fig. 9). In this model, toxin subunits are first individually secreted across the inner membrane into the periplasm. The periplasmic space contains the signal peptidase that would then cleave the signal sequences of each of the toxin subunits (26), followed by assembly of the toxin. At the same time, portions of the Ptl apparatus would assemble, in particular, a core complex consisting, at least in part, of PtlD, PtlE, PtlF, and PtlG. PT would then interact with the Ptl subassembly followed by tight association of the ATP-bound form of PtlH with the complex. The pilus-like protein PtlA, which has been shown to be essential for toxin secretion (10), may not be recruited to the transporter complex until the final stage of transporter assembly. Only after assembly of the transporter-PT complex is completed would secretion of the toxin proceed. Because we were unable to obtain mutants of B. pertussis from which ptlB and ptlC were completely deleted, we cannot, as yet, say when these proteins might interact with the transporter apparatus during the assembly process.
FIG. 9.
Model for secretion of PT. After synthesis, pertussis toxin subunits are individually transported across the inner membrane, likely by a Sec-like system. The signal sequences of the individual polypeptide chains are then cleaved, followed by assembly of the holotoxin. Simultaneously, the core of the Ptl transporter, which consists of PtlD, PtlE, PtlF, and PtlG, assembles. This complex may also contain PtlB, PtlC, and PtlI. The toxin is predicted to then interact with the Ptl transporter subassembly followed by tight association of the ATP-bound form of PtlH with the complex. PtlA may not associate with the complex until the final stage of transporter assembly which would then be followed by secretion of the toxin across the outer membrane.
The mechanism by which PT, which is located in the periplasm, interacts with its secretion apparatus has remained an enigma. Because of its homology with other type IV transporters, the Ptl apparatus is believed to span both the inner and outer membranes of the bacterium, forming a translocation channel through which the substrate would pass (9). The substrates for other type IV transporters are cytoplasmic and are thought to interact with the transporter at the cytoplasmic face of the inner membrane through initial interactions with VirD4 (9), a protein that is absent in the Ptl system. Thus, current models of type IV transport do not easily accommodate a periplasmic substrate such as PT. The model shown in Fig. 9, which is based on the data presented in this study, depicts PT interacting with a partially assembled Ptl apparatus. Only after PT interacts with this subassembly would complete assembly of the apparatus occur. Such a model would explain how the periplasmic toxin is secreted by a transport apparatus that is believed to span the inner and outer membranes.
Acknowledgments
We thank Anissa Cheung and Karen Farizo for the construction of BP536Δptx(507-3683), BP536ΔptlD, and BP536ΔptlF.
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 5 March 2007.
REFERENCES
- 1.Armstrong, S. K., and C. D. Parker. 1986. Heat-modifiable envelope proteins of Bordetella pertussis. Infect. Immun. 54:109-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmakuri, K., E. Cascales, and P. J. Christie. 2004. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol. Microbiol. 54:1199-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, E. M., A. A. Weiss, I. E. Ehrmann, M. C. Gray, E. L. Hewlett, and M. S. M. Goodwin. 1991. Bordetella pertussis adenylate cyclase toxin and hemolytic activities require a second gene, cyaC, for activation. J. Bacteriol. 173:720-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brickman, E., and J. Beckwith. 1975. Analysis of the regulation of Escherichia coli alkaline phosphatase using deletions and 80 transducing phages. J. Mol. Biol. 96:307-316. [DOI] [PubMed] [Google Scholar]
- 5.Burns, D. L., S. Fiddner, A. M. Cheung, and A. Verma. 2004. Analysis of subassemblies of pertussis toxin in vivo and their interaction with the Ptl transport apparatus. Infect. Immun. 72:5365-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascales, E., and P. J. Christie. 2004. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc. Natl. Acad. Sci. USA 101:17228-17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cascales, E., and P. J. Christie. 2004. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science 304:1170-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 179:3085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craig-Mylius, K. A., and A. A. Weiss. 1999. Mutants in the ptlA-H genes of Bordetella pertussis are deficient for pertussis toxin secretion. FEMS Microbiol. Lett. 179:479-484. [DOI] [PubMed] [Google Scholar]
- 11.De Feyter, R., Y. Yang, and D. W. Gabriel. 1993. Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant-Microbe Interact. 6:225-237. [DOI] [PubMed] [Google Scholar]
- 12.Ehrmann, M., D. Boyd, and J. Beckwith. 1990. Genetic analysis of membrane protein topology by a sandwich gene fusion approach. Proc. Natl. Acad. Sci. USA 87:7574-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ezzell, J. W., W. J. Dobrogosz, W. E. Kloos, and C. R. Manclark. 1981. Phase shift markers in Bordetella: alterations in envelope proteins. J. Infect. Dis. 143:562-569. [DOI] [PubMed] [Google Scholar]
- 14.Farizo, K. M., T. G. Cafarella, and D. L. Burns. 1996. Evidence for a ninth gene, ptlI, in the locus encoding the pertussis toxin secretion system of Bordetella pertussis and formation of a PtlI-PtlF complex. J. Biol. Chem. 271:31643-31649. [DOI] [PubMed] [Google Scholar]
- 15.Farizo, K. M., T. Huang, and D. L. Burns. 2000. Importance of holotoxin assembly in Ptl-mediated secretion of pertussis toxin from Bordetella pertussis. Infect. Immun. 68:4049-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilman, A. G. 1987. G proteins: transducers of receptor generated signals. Annu. Rev. Biochem. 56:615-649. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, F. D., and D. L. Burns. 1994. Detection and subcellular localization of three Ptl proteins involved in the secretion of pertussis toxin from Bordetella pertussis. J. Bacteriol. 176:5350-5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katada, T., and M. Ui. 1982. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc. Natl. Acad. Sci. USA 79:3129-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerppola, R. E., V. K. Shyamala, P. Klebba, and G. F. Ames. 1991. The membrane-bound proteins of periplasmic permeases form a complex. Identification of the histidine permease HisQMP complex. J. Biol. Chem. 266:9857-9865. [PubMed] [Google Scholar]
- 20.Kotob, S. I., and D. L. Burns. 1997. Essential role of the consensus nucleotide-binding site of PtlH in secretion of pertussis toxin from Bordetella pertussis. J. Bacteriol. 179:7577-7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lanzetta, P. A., L. J. Alvarez, P. S. Reinach, and O. A. Candia. 1979. An improved assay for nanomole amounts of inorganic phosphate. Anal. Biochem. 100:95-97. [DOI] [PubMed] [Google Scholar]
- 23.Letellier, L., S. P. Howard, and J. T. Buckley. 1997. Studies on the energetics of proaerolysin secretion across the outer membrane of Aeromonas species. J. Biol. Chem. 272:11109-11113. [DOI] [PubMed] [Google Scholar]
- 24.Locht, C., and J. M. Keith. 1986. Pertussis toxin gene: nucleotide sequence and genetic organization. Science 232:1258-1264. [DOI] [PubMed] [Google Scholar]
- 25.Nicosia, A., M. Perugini, C. Franzini, M. C. Casagli, M. G. Borri, G. Antoni, M. Almoni, P. Neri, G. Ratti, and R. Rappuoli. 1986. Cloning and sequencing of the pertussis toxin genes: operon structure and gene duplication. Proc. Natl. Acad. Sci. USA 83:4631-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Randall, L. L., S. J. S. Hardy, and J. R. Thom. 1987. Export of protein: a biochemical view. Annu. Rev. Biochem. 41:507-541. [DOI] [PubMed] [Google Scholar]
- 27.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivas, S., S. Bolland, E. Cabezon, F. M. Goni, and F. de la Cruz. 1997. TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J. Biol. Chem. 272:25583-25590. [DOI] [PubMed] [Google Scholar]
- 29.Sandkvist, M., M. Bagdasarian, S. P. Howard, and V. J. DiRita. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14:1664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savvides, S. N., H.-J. Yeo, M. R. Beck, F. Blaesing, R. Lurz, E. Lanka, R. Buhrdorf, W. Fischer, R. Haas, and G. Waksman. 2003. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 22:1969-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 32.Stabel, T. J., Z. Sha, and J. E. Mayfield. 1994. Periplasmic location of Brucella abortus Cu/Zn superoxide dismutase. Vet. Microbiol. 38:307-314. [DOI] [PubMed] [Google Scholar]
- 33.Stein, P. E., A. Boodhoo, G. D. Armstrong, S. A. Cockle, M. H. Klein, and R. J. Read. 1994. The crystal structure of pertussis toxin. Structure 2:45-57. [DOI] [PubMed] [Google Scholar]
- 34.Stephens, K. M., C. Roush, and E. Nester. 1995. Agrobacterium tumefaciens VirB11 protein requires a consensus nucleotide-binding site for function in virulence. J. Bacteriol. 177:27-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stibitz, S. 1994. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 235:458-465. [DOI] [PubMed] [Google Scholar]
- 36.Stibitz, S., and M.-S. Yang. 1991. Subcellular localization and immunochemical detection of proteins encoded by the vir locus of Bordetella pertussis. J. Bacteriol. 173:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura, M., K. Nogimori, S. Murai, M. Yajima, K. Ito, T. Katada, M. Ui, and S. Ishii. 1982. Subunit structure of islet-activating protein, pertussis toxin, in conformity with the A-B model. Biochemistry 21:5516-5522. [DOI] [PubMed] [Google Scholar]
- 38.Walker, M. E., M. Saraste, M. J. Runswick, and N. J. Gay. 1982. Distantly related sequences in the alpha and beta subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1:945-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward, D. V., O. Draper, J. R. Zupan, and P. C. Zambryski. 2002. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc. Natl. Acad. Sci. USA 99:11493-11500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weiss, A. A., E. L. Hewlett, G. A. Myers, and S. Falkow. 1984. Pertussis toxin and extracytoplasmic adenylate cyclase as virulence factors in Bordetella pertussis. J. Infect. Dis. 150:219-222. [DOI] [PubMed] [Google Scholar]
- 41.Weiss, A. A., F. D. Johnson, and D. L. Burns. 1993. Molecular characterization of an operon required for pertussis toxin secretion. Proc. Natl. Acad. Sci. USA 90:2970-2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winans, S. C., D. L. Burns, and P. J. Christie. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]