Abstract
We used a whole-genome microarray screening system (Affymetrix human GeneChips covering 47,000 different transcripts) to examine the gene expression in duodenal mucosa during acute cholera. Biopsies were taken from the duodenal mucosa of seven cholera patients 2 and 30 days after the onset of diarrhea, and the gene expression patterns in the acute- and convalescent-phase samples were compared pairwise. Of about 21,000 transcripts expressed in the intestinal epithelium, 29 were defined as transcripts that were up-regulated and 33 were defined as transcripts that were down-regulated during acute cholera. The majority of the up-regulated genes characterized were found to have an established or possible role in the innate defense against infections; these genes included the LPLUNC1, LF, VCC1, TCN1, CD55, SERPINA3, MMP1, MMP3, IL1B, LCN2, SOCS3, GDF15, SLPI, CXCL13, and MUC1 genes. The results of confirmative PCR correlated well with the microarray data. An immunohistochemical analysis revealed increased expression of lactoferrin in lamina propria cells and increased expression of CD55 in epithelial cells, whereas increased expression of the SERPINA3 protein (α1-antichymotrypsin) was detected in both lamina propria and epithelial cells during acute cholera. The expression pattern of CD55 and SERPINA3 in cholera toxin (CT)-stimulated Caco-2 cells was the same as the pattern found in the intestinal mucosa during acute cholera, indicating that the activation of the CD55 and SERPINA3 genes in intestinal epithelium was induced by CT. In conclusion, during acute cholera infection, innate defense mechanisms are switched on to an extent not described previously. Both direct effects of CT on the epithelial cells and changes in the lamina propria cells contribute to this up-regulation.
Enteric infections caused by bacteria, viruses, and parasites cause 2 to 3 billion diarrheal disease episodes and at least 3 million deaths each year. Approximately one-half of these infections are caused by bacteria that produce one or more enterotoxins (34, 35). Cholera is the archetype for these diseases and has been endemic for at least 500 years in the Ganges River delta of Bangladesh and India. From this source, it has spread to other countries in epidemic waves. The causative agent, Vibrio cholerae, is transferred via contaminated water and food to the small intestine, where bacterial colonization occurs. After uptake of the lysogenic phage CTXΦ, V. cholerae can produce cholera toxin (CT), the main cause of cholera symptoms (40). CT is composed of a homopentamer of B-subunits and a single toxic active A-subunit, and its structure and function have been well defined (35). After attachment via its B-subunits, CT invades cells in the intestinal epithelium, where it exerts its toxic effects, leading to massive secretion of electrolytes and fluid from the serosal side to the mucosal side of the epithelium. Within 1 or 2 days the infected persons have massive diarrhea, which often leads to dehydration and in up to 30% of the cases to death without proper treatment (34, 35).
In the search for effective vaccines, adaptive immunity against V. cholerae and CT has been investigated intensely (19). Much less is known about the innate defense mechanisms during cholera that may be involved in the early defense against the infection and also in initiation of the adaptive immune response. Although there is no pronounced inflammation during cholera, in several studies workers have noted increased infiltration of neutrophils, degranulation of mast cells and eosinophils, and production of some innate defense molecules during acute cholera (24, 31, 32). We previously reported that CT can induce the migration of CD8+ intraepithelial lymphocytes (IEL) from the epithelium to the lamina propria region in the small intestine of rats (12).
In the present investigation we studied the response of genes expressed in the intestinal mucosa in Bangladeshi cholera patients. By using wide screening microarray chips, covering the whole human genome, we compared intestinal gene expression during acute-phase and convalescent-phase disease. The results showed that many genes coding for secreted proteins implicated as proteins that are involved in the innate defense against pathogens were up-regulated during the acute phase of cholera.
MATERIALS AND METHODS
Study group.
In this study we evaluated seven adult male patients who were 18 to 49 years old and had cholera caused by V. cholerae O1 El Tor. All of the patients were treated at the International Centre for Diarrhoeal Disease Research, Bangladesh (ICDDR, B) hospital in Dhaka, Bangladesh. Physician assessment using the Denver system showed that the patients were severely dehydrated (42). For microbiological diagnosis, stool samples were analyzed by dark-field microscopy and for reactivity with antibodies specific for the V. cholerae O1 serogroup. To confirm the causative agent of the disease, stool samples were plated on taurocholate-tellurite-gelatin agar and gelatin agar. Suspected vibrio colonies were identified by slide agglutination. Stool samples were also screened for other common enteric pathogens, including both bacteria and parasites (32), but none were detected. All patients received intravenous rehydration, oral rehydration solution, and antibiotics (five patients were given doxycycline, and two patients were given ciprofloxacin) on the day of admission, which was 1 day before biopsies were first collected. The study was approved by the ethics review committee of ICDDR, B and by the human subjects research board in Göteburg.
Sample collection.
Mucosal punch biopsies were collected from the second part of the duodenum, as described previously (31), on the second day of hospitalization (about 2 days after the onset of diarrhea), which we termed the acute phase. Samples were also collected on day 30 (convalescence), at least 3 weeks after the intestine had recovered from the cholera episode (34, 42). Biopsy specimens were immediately put into RNAlater solution (Ambion, Huntingdon, United Kingdom) and kept at −70°C until they were used for isolation of RNA, or they were fixed in buffered formaldehyde and stored at 4°C until they were used for immunohistochemical analysis.
Cell culture and CT challenge.
We used the human intestinal epithelial cell line Caco-2 for stimulation experiments with CT (List Biological Laboratories, Inc., Campbell, CA). Cells were grown in Dulbecco's modified Eagle's medium with nonessential amino acids (Gibco, Invitrogen, Stockholm, Sweden) supplemented with 10% fetal calf serum, glutamine, β-mercaptoethanol, and gentamicin under a 5% CO2 atmosphere at 37°C. The culture medium was changed every other day. For the experiments, the cells were seeded into 35-mm wells at a density of 0.6 × 105 cells/cm2. The cells were grown for 9 days after confluence before stimulation with CT (1 μg/ml). Fetal calf serum was not present in the culture medium during CT challenge or during the 12-h period preceding stimulation. For control experiments, we used serum-free medium without CT. The cells were challenged for 18 h before RNA was isolated.
RNA isolation.
Total RNA was isolated from RNAlater-preserved biopsies and Caco-2 cells using a GenElute mammalian total RNA kit (Sigma, St. Louis, MO) according to the manufacturer's instructions. The RNA concentration was determined spectrophotometrically, and the quality was checked with an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Palo Alto, CA). Total RNA was used for real-time PCR (RT-PCR) and microarray analyses.
Microarray experiments.
RNA isolated from the cholera patients was converted into labeled target cRNA by in vitro transcription, performed as described in the GeneChip expression analysis manual (Affymetrix Inc., Santa Clara, CA), and was hybridized to an Affymetrix human GeneChip U133 plus 2.0 containing 54,000 probe sets representing 47,000 different transcripts. The hybridization procedure was followed by washing, staining, and scanning, as described in the Affymetrix manual. The resulting images were analyzed with the Affymetrix GeneChip operating software. All probe sets were used for scaling and normalization. Log2 ratios for all transcripts for the acute- and convalescent-phase samples were calculated for each patient. Two predetermined criteria had to be fulfilled for characterizing a transcript as up- or down-regulated. First, the mean log2 ratio of the acute phase to the convalescent phase for all patients examined had to be more than 1.0 for up-regulation or less than −1.0 for down-regulation. Second, to minimize the effect of individual outliers, a log2 ratio greater than 0.5 for up-regulation or less than −0.5 for down-regulation had to be observed for at least five of the seven patients.
Relative quantification by RT-PCR.
For each sample, we converted 2 μg of total RNA into cDNA as described previously (11). Oligonucleotide primers (Table 1) purchased from TAG Copenhagen A/S (Copenhagen, Denmark) were used for relative quantification with the Applied Biosystems 7500 RT-PCR system used according to the manufacturer's protocol. The glyceraldehyde-3-phosphate dehydrogenase gene was used as a reference gene in all experiments. The relative levels of transcripts expressed as log2 ratios of acute-phase values to convalescent-phase values for samples from each patient were calculated by using the formula described by Pfaffl (30). For the Caco-2 experiments a log2 ratio of the target gene value to the reference gene value was derived for each sample using the same formula. The relative expression values were adjusted so that the mean in the control group was 0.
TABLE 1.
Primers used for RT-PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| LPLUNC1 | 5′-TGGGATTGGCTGGTTCCA-3′ | 5′-CCCCAGATCTTAATTTGCCATTCT-3′ |
| LF | 5′-GCCATGGCCCCGAATC-3′ | 5′-CAGATCCATTTCTCCCAAATTTAGC-3′ |
| TCN1 | 5′-ACCTGCCCTGATGGGAAAGA-3′ | 5′-CAGTTATAGGCTCATCAGCGGAGAT-3′ |
| CD55 | 5′-CAACCATCTCCTTCTCATGTAACACA-3′ | 5′-GTTCACGGAGGATGCCAAGA-3′ |
| SERPINA3 | 5′-GTTCACGGAGGATGCCAAGA-3′ | 5′-TCTTCACGTAGTCGTTGATGAGCTT-3′ |
| LCN2 | 5′-GCAGCAGAACTTCCAGGACAAC-3′ | 5′-ACATCTTTTGCGGGTCTTTGTCT-3′ |
| GAPDHa | 5′-GAGCACCAGGTGGTCTCCTCTGACTTC-3′ | 5′-GCCAAATTCGTTGTCATACCAGGAAATG-3′ |
GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Immunohistochemistry.
Paraffin-embedded duodenal sections from the cholera patients were subjected to immunohistochemical analyses (31). Before the immunohistochemical analyses, the gross histology of the sections was evaluated by periodic acid-Schiff and hematoxylin staining, which did not reveal any consistent differences between the acute- and convalescent-phase samples. Commercially available monoclonal or polyclonal primary antibodies were used to detect human neutrophil defensins (Novocastra Laboratories Ltd., Newcastle, United Kingdom), CD8 (DAKO, Glostrup, Denmark), CD55 (Santa Cruz Biotechnology Inc., Santa Cruz, CA), lactoferrin, and α1-antichymotrypsin (Abcam Inc., Cambridge, United Kingdom). Alkaline phosphatase-conjugated antibodies (Jackson ImmunoResearch Europe Ltd., Soham, United Kingdom) were used as secondary reagents with nitroblue tetrazolium/5-bromo-4-chloro-3-indolylphosphate as the substrate for detection of CD55, lactoferrin, and α1-antichymotrypsin. For detection of human neutrophil defensins and CD8, the PAP procedure (DAKO) was used, and the sections were developed using diaminobenzidine as the substrate before they were lightly counterstained with hematoxylin.
Statistical analyses.
We used the Student t test to evaluate differences between gene expression in CT-stimulated Caco-2 cells and gene expression in nonstimulated Caco-2 cells. Correlations between gene expression data generated by the microarray technique and RT-PCR were determined with Pearson's test.
RESULTS AND DISCUSSION
All cholera patients studied by whole-genome microarray screening had signs of severe dehydration during the acute phase of illness. Of the 47,000 transcripts covered on the microarray chip, about 21,000 (corresponding to approximately 17,000 genes) were expressed in the intestinal mucosa of the cholera patients. Of these transcripts, 29 were defined as transcripts that were up-regulated and 33 were defined as transcripts that were down-regulated during the acute phase (Tables 2 and 3), as calculated using a threshold value of a mean twofold change and differential expression found in five or more of the seven patients.
TABLE 2.
Genes up-regulated during acute cholera
| Affymetrix ID | Mean log2 ratioa | No. of patientsb | Gene | Gene product | Subcellular localizationc |
|---|---|---|---|---|---|
| 226067_at | 2.81 | 6 | LPLUNC1 | von Ebner minor salivary gland protein | S |
| 202018_s_at | 2.24 | 5 | LF | Lactoferrin | S |
| 226960_at | 1.89 | 6 | VCC1 | VEGFd coregulated chemokine 1 | S |
| 205886_at | 1.77 | 5 | REG1B | Regenerating protein 1β | C |
| 205513_at | 1.69 | 5 | TCN1 | Transcobalamin 1 | S |
| 201925_s_at | 1.64 | 6 | CD55 | CD55 (complement decay accelerating factor) | M |
| 202376_at | 1.64 | 5 | SERPINA3 | α1-Antichymotrypsin | S |
| 1562283_at | 1.56 | 5 | EST (GenBank accession no. BQ889971) | ||
| 204475_at | 1.49 | 5 | MMP1 | Matrix metalloproteinase 1 | S |
| 209519_at | 1.49 | 5 | ESTe (GenBank accession no. BG108193) | ||
| 39402_at | 1.46 | 5 | IL1B | IL-1β | S |
| 205067_at | |||||
| 242592_at | 1.43 | 5 | Hypothetical protein DKFZp762F0713 | ||
| 212531_at | 1.39 | 5 | LCN2 | Lipocalin 2 | S |
| 217678_at | 1.36 | 6 | SLC7A11 | Solute carrier family 7A11 | M |
| 209921_at | |||||
| 242685_at | 1.33 | 5 | Hypothetical protein HSPC135 | ||
| 1557459_at | 1.29 | 5 | cDNA clone (GenBank accession no. AL831884) | ||
| 208473_s_at | 1.27 | 6 | GP2 | Glycoprotein 2 (zymogen granule protein) | S/M |
| 206681_x_at | |||||
| 219795_at | 1.24 | 5 | SLC6A14 | Solute carrier 6A14 | M |
| 206359_at | 1.24 | 5 | SOCS3 | Suppressor of cytokine signaling 3 | C |
| 228058_at | 1.20 | 5 | EST (GenBank accession no. AI559190) | ||
| 221577_x_at | 1.13 | 5 | GDF15 | Growth differentiation factor 15 | S |
| 203021_at | 1.11 | 7 | SLPI | Secretory leukocyte protease inhibitor | S |
| 205242_at | 1.11 | 5 | CXCL13 | Cytokine B13 (B lymphocyte chemoattractant) | S |
| 1559051_s_at | 1.11 | 5 | Hypothetical protein LOC115004 | ||
| 211695_x_at | 1.10 | 6 | MUC1 | Mucin 1 | M |
| 213693_s_at | |||||
| 207847_s_at | |||||
| 205828_at | 1.07 | 7 | MMP3 | Matrix metalloproteinase 3 | S |
| 214135_at | 1.06 | 5 | CLDN18 | Claudin 18 | M |
| 210282_at | 1.04 | 5 | ZNF198 | Zinc finger protein 198 | N |
| 243003_at | 1.04 | 5 | EST (GenBank accession no. AV702197) |
The log2 ratios are the acute-phase/convalescent-phase ratios.
Number of patients that showed differential expression resulting in a log2 ratio of >0.5.
S, secreted; M, membrane associated; C, cytosolic; N, nuclear.
VEGF, vascular endothelial growth factor.
EST, expressed sequence tag.
TABLE 3.
Genes down-regulated during acute cholera
| Affymetrix ID | Mean log2 ratioa | No. of patientsb | Gene | Gene product | Subcellular localizationc |
|---|---|---|---|---|---|
| 205749_at | −2.73 | 5 | CYP1A1 | Cytochrome P-450, 1A1 | M, Li |
| 238245_at | −1.77 | 6 | ENPP7 | Ectonucleotide pyrophosphatase/phosphodiesterase 7 | M, Li |
| 222938_x_at | −1.74 | 6 | ENPP3 | Ectonucleotide pyrophosphatase/phosphodiesterase 3 | M |
| 244044_at | |||||
| 232737_s_at | |||||
| 204450_x_at | −1.70 | 6 | APOA1 | Apolipoprotein A-I | S, Li |
| 217073_x_at | |||||
| 229725_at | −1.46 | 6 | ACSL6 | Long-chain acyl-coenzyme A synthetase 6 | M, Li |
| 231416_at | −1.46 | 5 | DHDH | Dimeric dihydrodiol dehydrogenase | C |
| 241811_x_at | −1.46 | 6 | ESTd (GenBank accession no. BE645279) | ||
| 205820_s_at | −1.43 | 6 | APOC3 | Apolipoprotein C-III | S, Li |
| 244014_x_at | −1.41 | 5 | EST (GenBank accession no. BF433962) | ||
| 239229_at | −1.41 | 6 | EST (GenBank accession no. AI342246) | ||
| 1555338_s_at | −1.40 | 6 | AQP10 | Aquaporin 10 | M |
| 242009_at | −1.36 | 6 | EST (GenBank accession no. AI082692) | ||
| 204561_x_at | −1.34 | 7 | APOC2 | Apolipoprotein C-II | S, Li |
| 1552279_a_at | −1.30 | 6 | Hypothetical protein FLJ00234 | ||
| 206292_s_at | −1.23 | 6 | SULT2A1 | Alcohol sulfotransferase | C, Li |
| 206293_at | |||||
| 223579_s_at | −1.21 | 6 | APOB | Apolipoprotein B | S, Li |
| 224412_s_at | −1.21 | 5 | TRPM6 | Transient receptor potential cation channel M6 | M |
| 228377_at | −1.20 | 5 | KLHL14 | Kelch-like 14 | |
| 210096_at | −1.17 | 5 | CYP4B1 | Cytochrome P-450, 4B1 | M, Li |
| 227480_at | −1.16 | 6 | SUSD2 | Sushi domain containing 2 | M |
| 207519_at | −1.16 | 7 | SLC6A4 | Solute carrier family 6A4 | M |
| 1553486_a_at | −1.16 | 6 | Hypothetical protein FLJ39647 | ||
| 1556250_at | −1.16 | 5 | ST18 | Suppression of tumorigenicity 18 | N |
| 223732_at | −1.13 | 6 | SLC23A1 | Solute carrier 23A1 | M |
| 207885_at | −1.09 | 5 | S100G | Calbindin D9k | C |
| 220851_at | −1.09 | 5 | Hypothetical protein PRO1600 | ||
| 207378_at | −1.06 | 5 | TREH | Trehalase (brush border membrane glycoprotein) | M |
| 229793_at | −1.06 | 6 | EST (GenBank accession no. AI656964) | ||
| 1552362_a_at | −1.06 | 5 | LEAP-2 | Liver-expressed antimicrobial peptide 2 | S |
| 205939_at | −1.04 | 5 | CYP3A7 | Cytochrome P-450, 3A7 | M, Li |
| 207362_at | −1.04 | 5 | SLC30A4 | Solute carrier family 30A4 | M |
| 207125_at | −1.01 | 5 | ZNF225 | Zinc finger protein 225 | N |
| 1558703_at | −1.00 | 6 | Hypothetical protein DKFZp451K1917 |
The log2 ratios are the acute-phase/convalescent-phase ratios.
Number of patients that showed differential expression resulting in a log2 ratio of <0.5.
S, secreted; M, membrane associated; C, cytosolic; N, nuclear; Li, involved in the metabolism or transport of lipids.
EST, expressed sequence tag.
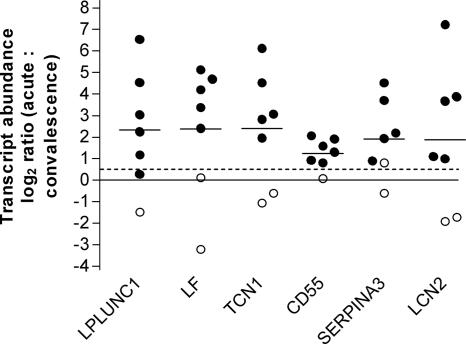
In the present study, we found a clear distinction between the highly up-regulated and down-regulated genes. Of the 29 up-regulated transcripts, 21 code for proteins with a known function (Table 2). The majority of these proteins, many of which are secreted, may be functionally grouped as proteins that are part of the innate defense response. Of the 33 down-regulated transcripts, 22 code for proteins whose function is known to some extent (Table 3). Of these, 9 proteins are enzymes and 10 are transporting proteins. In contrast to the up-regulated genes, most of the down-regulated genes encode membrane proteins, and many of these proteins are involved in the metabolism or transport of lipids. For six of the genes, the LPLUNC1 (von Ebner minor salivary gland protein), LF (lactoferrin), TCN1 (transcobalamin 1), CD55, SERPINA3 (α1-antichymotrypsin), and LCN2 (lipocalin 2) genes, the up-regulation indicated by the microarray analysis was examined and confirmed by RT-PCR (Fig. 1). The correlation between the results generated by the microarray technique and the results generated by the RT-PCR assays was very high (P < 0.0001; r = 0.93), as determined by Pearson's test, indicating that the microarray data are reliable.
FIG. 1.
Differentially expressed genes during acute cholera compared to convalescence, expressed as log2 ratios determined by RT-PCR. The horizontal lines indicate means. The circles represent measurements for patients with (filled circles) and without (open circles) up-regulation, as determined by the microarray technique using an acute-phase/convalescent-phase log2 ratio of >0.5 to define individual up-regulation. The dashed line indicates the individual threshold value used during the microarray screening.
Antibacterial response is induced during acute cholera.
In order to colonize the small intestine, which is relatively nonpermissive for bacterial growth, V. cholerae must adapt to and counteract many different enteric defense mechanisms. Thus, the V. cholerae ToxR protein, which regulates important virulence factors, including CT production in response to environmental changes, through its regulation of the OmpU porin, also seems to counteract the action of the bactericidal BPI protein, which is distributed on the surface of intestinal epithelial cells (25). Our finding that the gene encoding the BPI-like von Ebner minor salivary gland protein was highly up-regulated during acute cholera might suggest an alternative way that the host defense attacks the outer cell wall of the bacteria (2, 21). Furthermore, the up-regulation of lactoferrin, transcobalamin 1, and lipocalin 2 detected could also increase the protection against V. cholerae. All of these proteins are antibacterial proteins that are known to be secreted from granulated epithelial cells and neutrophils (4, 10, 15, 22, 23). Lactoferrin has been shown previously to have a strong bactericidal effect on V. cholerae (1), and this was confirmed in this study (data not shown). In addition, the lactoferrin level is elevated in the mucosa, blood, and stools of cholera patients (31, 32).
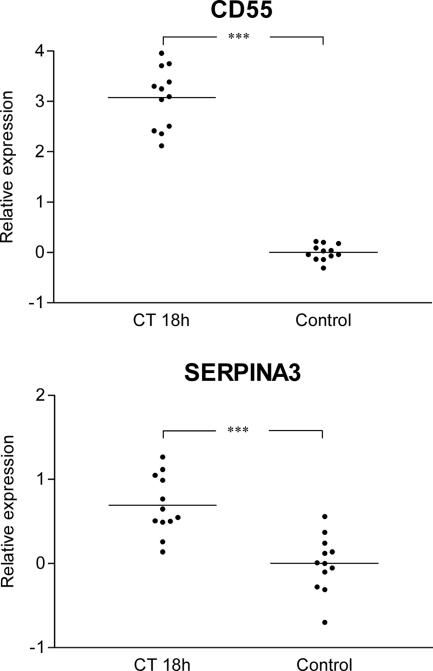
In order to investigate if the observed changes in gene expression during acute cholera were due to direct effects of the secreted CT on the epithelial cells, we tried to extend our array findings to see if we could mimic the effects in tissue-cultured human intestinal epithelial cells exposed to CT. Cultured Caco-2 cells were stimulated with CT (1 μg/ml for 18 h), and after this RNA was isolated from CT-treated cells and untreated control cells and expression of the six RT-PCR-confirmed up-regulated genes was analyzed (Fig. 1) by quantitative PCR. The changes in the expression of the CD55 and SERPINA3 genes were also observed in Caco-2 cells exposed to CT, indicating that these effects are triggered at least in part by the action of CT on the epithelial cells during acute cholera (Fig. 2). In contrast, CT did not induce the expression of any of the four other genes encoding the antibacterial proteins examined in the Caco-2 cell model (data not shown), possibly because the induction effects are restricted to specific epithelial cells, such as paneth cells, or because the expression is stimulated by bacterial components other than CT. Alternatively, the up-regulation of these genes observed in the cholera patients may be ascribed to nonepithelial cells. Neutrophils have been described as cells that infiltrate the mucosa during acute cholera (24, 31, 32), and increased numbers were also found in the acute-phase biopsy samples in this study (data not shown). However, most of the granular products of neutrophils are preformed before they enter the target tissue, and thus their contribution to the transcriptional profile should be limited (13, 28). In contrast, the degranulation of mast cells and eosinophils, which has also been observed during the acute stage of cholera (31), may affect the outcome of a microarray analysis, since massive degranulation is likely to be followed by resynthesis of granule products in these cells.
FIG. 2.
Expression of the CD55 and SERPINA3 genes in Caco-2 cells stimulated with CT for 18 h. The relative expression values are adjusted log2 transcript abundance ratios of the target gene to the glyceraldehyde-3-phosphate dehydrogenase gene, as determined by RT-PCR. Means are indicated by horizontal lines. Three asterisks indicate that there is a significant difference between the CT-challenged group and the control (P < 0.001).
Both epithelial and lamina propria cells contribute to the up-regulation of genes involved in the innate defense during acute cholera.
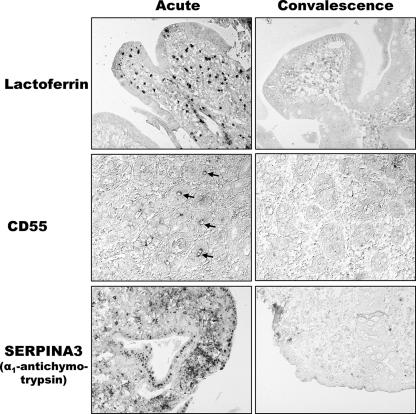
The proteins corresponding to three of the up-regulated genes, LF, CD55, and SERPINA3, for which we had access to specific antibodies, were studied by performing an immunohistochemical analysis (Fig. 3). We confirmed previous data showing that the level of lactoferrin was increased in the subepithelial mucosa during acute cholera (31, 32). Our results also indicate that cells other than neutrophils contribute to the increased expression of lactoferrin, since the number of mucosal cells positive for lactoferrin clearly exceeded the number of mucosal neutrophils detected in this study. In addition, a different localization pattern was observed for the cells expressing lactoferrin compared to the pattern for the neutrophils. The immunohistochemical analysis also revealed increased levels of CD55 and α1-antichymotrypsin (Fig. 3). Both of these proteins seem to be important proteins that protect the mucosa during an immune response by inhibiting the lysis of host cells by the complement system and by inhibiting neutrophil and mast cell proteases, respectively (20, 26). In contrast to lactoferrin, but consistent with the findings with Caco-2 cells exposed to CT (see above), up-regulation of CD55 and up-regulation of α1-antichymotrypsin (SERPINA3) were detected in epithelial cells of the intestinal mucosa, although increased expression of α1-antichymotrypsin was also evident in many lamina propria cells, which again outnumbered the mucosal neutrophils detected. Thus, changes in both epithelial cells and various lamina propria cell populations seem to contribute to the altered gene expression pattern during acute cholera.
FIG. 3.
Immunolocalization of lactoferrin, CD55, and α1-antichymotrypsin in duodenal biopsies during acute- and convalescent-phase cholera. The arrows indicate CD55 expression in the crypt epithelium.
Up-regulation of many innate defense factors during acute cholera is mediated via IL-1β.
Two other genes, the interleukin-1β (IL-1β) and SOCS3 genes, which were detected as up-regulated genes during acute cholera, are known to be induced by CT. CT is known to activate IL-1β in both macrophages and epithelial cells (5, 6). In addition, in a recent microarray study, in which a mixture of human monocytes and lymphocytes was stimulated with CT, the workers also detected up-regulation of IL-1β (33). The activation and increased production of IL-1β seem to trigger the immediate innate response and to have a potent adjuvant function in promoting a specific immune response (39). Induction of the SOCS3 gene, which encodes an intracellular protein that is believed to prevent excessive responses to cytokines, is up-regulated in cultivated leukocytes by CT (14).
As mentioned above, IL-1β is a pivotal cytokine and may mediate many of the immunomodulatory effects of CT (39). Although several previously reported transcriptional effects of IL-1β (9) were not observed to be significant up-regulation effects in this study, most likely due to the limited sensitivity of the microarray technique, a number of the observed up-regulation effects during acute cholera are probably IL-1β-induced effects. The up-regulated GDF15, MMP1, MMP3, and SERPINA3 genes are all known to be induced by IL-1β (3, 7, 16). The GDF15 gene codes for growth differentiation factor 15, a member of the transforming growth factor β family that is expressed at high levels in epithelial cells, and, like SOCS3, seems to play a role in limiting the host response (36). The MMP1 and MMP3 genes encode two metalloproteinases that are secreted in the gut (29). Thus, it appears that CT may affect innate immunity via IL-1β by stimulating many genes that trigger the immune response, as well as genes that protect against immune stimulation that is too vigorous and potentially tissue damaging.
Additional up-regulated genes during acute cholera also contribute the innate defense and protection of the mucosa.
Some other up-regulated genes implicated in the innate defense systems are the VCC1, MUC1, SLPI, and CXCL13 genes. The VCC1 gene codes for a chemokine which so far has not been well characterized. The MUC1 gene encodes mucin 1, which contributes to the buildup of the protective glycocalyx layer. The SLPI gene codes for a leukocyte protease inhibitor which, like α1-antichymotrypsin, may prevent damage of the mucosa caused by neutrophil proteases (37). In addition, the SLPI product exerts its protective effect not only by inhibiting various proteinases secreted by the host but also by displaying antimicrobial activity and by being involved in tissue repair reactions (18). The CXCL13 gene codes for cytokine B13, which is secreted in the mucosa and is chemotactic for B lymphocytes (17). The up-regulation of this gene, like the up-regulation of the IL-1β gene, may be important for the initiation of an adaptive immune response against V. cholerae.
Furthermore, the up-regulation of REG1B and the up-regulation of CLD18 may enhance the general protection of the mucosal surface. The REG1B gene codes for the regenerating protein 1β, a C-type lectin that is induced during inflammation and seems to play a role in mucosal regeneration (27), whereas the CLD18 gene codes for claudin 18, an integral membrane protein involved in the tight junction-specific obliteration of the intercellular space. Thus, claudin 18 may be important for maintaining the primary cellular barrier in the gut. Only four of the up-regulated genes with known functions, the GP2 and ZNF198 genes and the genes encoding two transmembrane solute carriers, have no obvious role in mucosal protection.
Genes involved in lipid metabolism are down-regulated during acute cholera.
The massive inhibition of genes involved in lipid metabolism, including genes that encode enzymes, as well as genes that encode major apolipoproteins, which are constituents of the lipid-transporting plasma lipoproteins, during acute cholera has to our knowledge not been reported previously. However, Wang et al. found that apolipoprotein A-I can bind and inhibit an antibacterial protein/peptide in humans (41). Down-regulation of the apolipoprotein genes may be a way to potentiate the antibacterial response further. It should also be noted that all apolipoproteins that were determined to be down-regulated proteins during acute cholera are major components of the chylomicrons, which are a type of lipoproteins essential for transport of dietary fats to appropriate peripheral tissues.
Three different cytochrome P-450 genes were determined to be genes that were down-regulated during the acute phase of cholera. This down-regulation may be mediated via bacterial components, since it has been shown that P-450 genes are down-regulated in hepatocytes in response to lipopolysaccharide (8). It has also been proposed that down-regulation of cytochrome P-450 genes is permissive for elevated levels of leukotriens and prostaglandins, which are inflammatory mediators that have been suggested to play a role during cholera (8).
Mucosal CD8+ cells have an altered localization pattern during acute cholera.
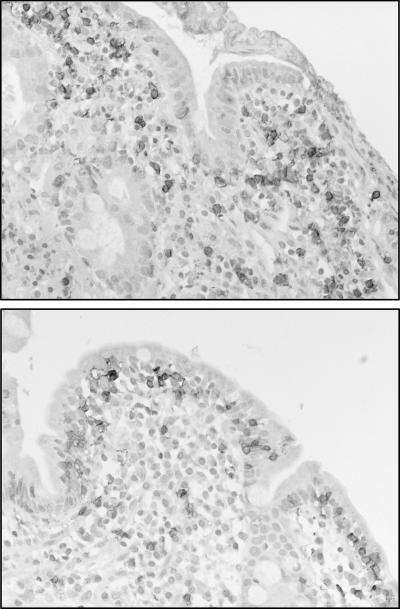
We previously reported that CT can induce the migration of CD8+ IEL from the epithelium to the lamina propria region in the small intestine of rats (12). In this study a different localization pattern for mucosal CD8+ cells, consistent with the findings in rats, was evident in three of the seven patients. In these patients, numerous CD8+ cells were found in the lamina propria region during the acute stage; this was in contrast to the location in convalescent-phase samples, in which the majority of these cells were found in the epithelium (Fig. 4). The change in the localization pattern of the mucosal CD8+ cells is not, however, likely to affect the transcriptional profile of the samples, even though this cellular event in rats was originally detected as a reduction in various IEL-associated transcripts by using the microarray technique. A major difference between the study in rats and the present study was that in the former, samples comprising only the upper parts of the villi were analyzed by the microarray technique. This made it possible to detect, by the microarray technique, the migration of CD8+ IEL from the epithelium to the lamina propria region, since the cells predominantly migrated to lower parts of the villi. In the present study, because biopsies comprising the whole mucosa were analyzed, the microarray technique should not have been able to detect a change in the cellular localization pattern within the mucosa. Accordingly, no differences between the levels of IEL-associated transcripts in acute-phase samples and the levels of IEL-associated transcripts in convalescent-phase samples were detected in the present study.
FIG. 4.
Differential localization of CD8+ cells in duodenal biopsies during acute-phase (upper panel) and convalescent-phase (lower panel) cholera as revealed by immunohistochemical analysis.
Study limitations and perspectives.
This study had several limitations that warrant comment. First, it is obvious that the gene expression in the acute-phase biopsies represented only a snapshot of the dynamic transcriptional response in the intestine to cholera infection. However, even so, it is striking how many of the up-regulated genes could be associated with an innate immune defense function and how relatively consistent their up-regulation was in the cholera patients. The argument could be made that since the up-regulation of innate defense genes was observed in cholera patients who were already ill, this response may have little significance in protecting against cholera disease. However, it should be recalled that there are at least 10 cholera-infected persons to each clinically ill patient (34), and therefore the early activation of the observed (and additional) innate defense genes could play an important role in preventing many infections from causing illness. In addition, the early activation of innate immune defense genes, such as the IL-1β gene, is probably very important for initiating the adaptive mucosal immune response that is needed both for terminating the cholera illness and for protecting against new infection and disease (35).
Since all patients received rehydration therapy and antibiotics before the biopsies were taken, we cannot eliminate the possibility that this treatment was responsible for some of the effects detected in the microarray screening. However, as mentioned above, this study and previous studies have shown that many of the changes detected can be mediated, directly or indirectly, via CT and other bacterial components and are thus probably not due to an effect caused by medical treatment. Nevertheless, it should be noted that antibiotic treatment can cause a massive release of bacterial components from killed bacteria and that this might induce a response that may not be fully characteristic of the natural disease.
On the technical side, it could be argued that in human gene expression studies using the microarray technique workers have often observed great variability and sometimes reported false-positive results due to biological variation and possibly also technical imprecision. However, by comparing material from the acute and convalescent phases (i.e., by using each patient as his own control), we decreased the biological variation significantly. In addition, an excellent correlation was found between data generated by the microarray technique and data generated by the RT-PCR assays, indicating that the data are reliable. A recent joint effort of the FDA and several microarray manufacturers, the Microarray Quality Control study, also showed that there was both a high degree of intraplatform consistency and interplatform concordance when the same RNA samples were analyzed on different microarray platforms at different test sites in parallel (38). Furthermore, of the seven different microarray platforms tested in the Microarray Quality Control study, the Affymetrix system that was used in the present study generated the most reproducible data with the smallest within-site and between-site variability.
In conclusion, our findings show that during acute cholera infection, a broader range of innate immune mechanisms than previously recognized are activated in the intestinal mucosa, both in the epithelium and in lamina propria cells. Some of the effects are evidently induced by CT, but it seems likely that other bacterial factors also contribute.
Acknowledgments
This work was supported by grants from the Swedish Research Council (Medicine), the Knut and Alice Wallenberg Foundation, the Marianne and Marcus Wallenberg Foundation, the Sahlgrenska University Hospital (LUA/ALF project support), and the Wilhelm and Martina Lundgren Foundation, by project support to ICDDR, B provided by the Swedish International Cooperation and Development Agency (SIDA), and by general support from the MIVAC network through the Swedish Strategic Foundation.
We gratefully acknowledge help with the microarray analyses from the SweGene microarray unit at Lund University established with support from the Knut and Alice Wallenberg Foundation.
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Arnold, R. R., M. F. Cole, and J. R. McGhee. 1977. A bactericidal effect for human lactoferrin. Science 197:263-265. [DOI] [PubMed] [Google Scholar]
- 2.Bingle, C. D., and C. J. Craven. 2002. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum. Mol. Genet. 11:937-943. [DOI] [PubMed] [Google Scholar]
- 3.Bootcov, M. R., A. R. Bauskin, S. M. Valenzuela, A. G. Moore, M. Bansal, X. Y. He, H. P. Zhang, M. Donnellan, S. Mahler, K. Pryor, B. J. Walsh, R. C. Nicholson, W. D. Fairlie, S. B. Por, J. M. Robbins, and S. N. Breit. 1997. MIC-1, a novel macrophage inhibitory cytokine, is a divergent member of the TGF-beta superfamily. Proc. Natl. Acad. Sci. USA 94:11514-11519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borregaard, N., and J. B. Cowland. 1997. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood 89:3503-3521. [PubMed] [Google Scholar]
- 5.Bromander, A., J. Holmgren, and N. Lycke. 1991. Cholera toxin stimulates IL-1 production and enhances antigen presentation by macrophages in vitro. J. Immunol. 146:2908-2914. [PubMed] [Google Scholar]
- 6.Bromander, A. K., M. Kjerrulf, J. Holmgren, and N. Lycke. 1993. Cholera toxin enhances alloantigen presentation by cultured intestinal epithelial cells. Scand. J. Immunol. 37:452-458. [DOI] [PubMed] [Google Scholar]
- 7.Cichy, J., J. Potempa, R. K. Chawla, and J. Travis. 1995. Regulation of alpha 1-antichymotrypsin synthesis in cells of epithelial origin. FEBS Lett. 359:262-266. [DOI] [PubMed] [Google Scholar]
- 8.Cui, X., A. Kalsotra, A. M. Robida, D. Matzilevich, A. N. Moore, C. L. Boehme, E. T. Morgan, P. K. Dash, and H. W. Strobel. 2003. Expression of cytochromes P450 4F4 and 4F5 in infection and injury models of inflammation. Biochim. Biophys. Acta 1619:325-331. [DOI] [PubMed] [Google Scholar]
- 9.Dinarello, C. A. 1996. Biologic basis for interleukin-1 in disease. Blood 87:2095-2147. [PubMed] [Google Scholar]
- 10.Farnaud, S., and R. W. Evans. 2003. Lactoferrin—a multifunctional protein with antimicrobial properties. Mol. Immunol. 40:395-405. [DOI] [PubMed] [Google Scholar]
- 11.Flach, C. F., S. Lange, E. Jennische, and I. Lonnroth. 2004. Cholera toxin induces expression of ion channels and carriers in rat small intestinal mucosa. FEBS Lett. 561:122-126. [DOI] [PubMed] [Google Scholar]
- 12.Flach, C. F., S. Lange, E. Jennische, I. Lonnroth, and J. Holmgren. 2005. Cholera toxin induces a transient depletion of CD8+ intraepithelial lymphocytes in the rat small intestine as detected by microarray and immunohistochemistry. Infect. Immun. 73:5595-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouret, P., R. M. du Bois, J. F. Bernaudin, H. Takahashi, V. J. Ferrans, and R. G. Crystal. 1989. Expression of the neutrophil elastase gene during human bone marrow cell differentiation. J. Exp. Med. 169:833-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gasperini, S., L. Crepaldi, F. Calzetti, L. Gatto, C. Berlato, F. Bazzoni, A. Yoshimura, and M. A. Cassatella. 2002. Interleukin-10 and cAMP-elevating agents cooperate to induce suppressor of cytokine signaling-3 via a protein kinase A-independent signal. Eur. Cytokine Netw. 13:47-53. [PubMed] [Google Scholar]
- 15.Goetz, D. H., M. A. Holmes, N. Borregaard, M. E. Bluhm, K. N. Raymond, and R. K. Strong. 2002. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol. Cell 10:1033-1043. [DOI] [PubMed] [Google Scholar]
- 16.Gooz, M., M. Shaker, P. Gooz, and A. J. Smolka. 2003. Interleukin 1beta induces gastric epithelial cell matrix metalloproteinase secretion and activation during Helicobacter pylori infection. Gut 52:1250-1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn, M. D., V. N. Ngo, K. M. Ansel, E. H. Ekland, J. G. Cyster, and L. T. Williams. 1998. A B-cell-homing chemokine made in lymphoid follicles activates Burkitt's lymphoma receptor-1. Nature 391:799-803. [DOI] [PubMed] [Google Scholar]
- 18.Hiemstra, P. S. 2002. Novel roles of protease inhibitors in infection and inflammation. Biochem. Soc. Trans. 30:116-120. [DOI] [PubMed] [Google Scholar]
- 19.Holmgren, J., J. Adamsson, F. Anjuere, J. Clemens, C. Czerkinsky, K. Eriksson, C. F. Flach, A. George-Chandy, A. M. Harandi, M. Lebens, T. Lehner, M. Lindblad, E. Nygren, S. Raghavan, J. Sanchez, M. Stanford, J. B. Sun, A. M. Svennerholm, and S. Tengvall. 2005. Mucosal adjuvants and anti-infection and anti-immunopathology vaccines based on cholera toxin, cholera toxin B subunit and CpG DNA. Immunol. Lett. 97:181-188. [DOI] [PubMed] [Google Scholar]
- 20.Kalsheker, N. A. 1996. Alpha 1-antichymotrypsin. Int. J. Biochem. Cell Biol. 28:961-964. [DOI] [PubMed] [Google Scholar]
- 21.Leclair, E. E. 2003. Four BPI (bactericidal/permeability-increasing protein)-like genes expressed in the mouse nasal, oral, airway and digestive epithelia. Biochem. Soc. Trans. 31:801-805. [DOI] [PubMed] [Google Scholar]
- 22.Lin, J. C., N. Borregaard, H. A. Liebman, and R. Carmel. 2001. Deficiency of the specific granule proteins, R-binder/transcobalamin I and lactoferrin, in plasma and saliva: a new disorder. Am. J. Med. Genet. 100:145-151. [DOI] [PubMed] [Google Scholar]
- 23.Marcoullis, G., J. P. Nicolas, Y. Parmentier, M. Jimenez, and P. Gerard. 1980. A derivative of R-type cyanocobalamin binding proteins in the human intestine. A candidate antibacterial molecule. Biochim. Biophys. Acta 633:289-294. [DOI] [PubMed] [Google Scholar]
- 24.Mathan, M. M., G. Chandy, and V. I. Mathan. 1995. Ultrastructural changes in the upper small intestinal mucosa in patients with cholera. Gastroenterology 109:422-430. [DOI] [PubMed] [Google Scholar]
- 25.Mathur, J., and M. K. Waldor. 2004. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect. Immun. 72:3577-3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miwa, T., and W. C. Song. 2001. Membrane complement regulatory proteins: insight from animal studies and relevance to human diseases. Int. Immunopharmacol. 1:445-459. [DOI] [PubMed] [Google Scholar]
- 27.Miyaoka, Y., Y. Kadowaki, S. Ishihara, T. Ose, H. Fukuhara, H. Kazumori, S. Takasawa, H. Okamoto, T. Chiba, and Y. Kinoshita. 2004. Transgenic overexpression of Reg protein caused gastric cell proliferation and differentiation along parietal cell and chief cell lineages. Oncogene 23:3572-3579. [DOI] [PubMed] [Google Scholar]
- 28.Nagaoka, I., M. Hirata, K. Sugimoto, Y. Tsutsumi-Ishii, A. Someya, K. Saionji, and J. Igari. 1998. Evaluation of the expression of human CAP18 gene during neutrophil maturation in the bone marrow. J. Leukoc. Biol. 64:845-852. [DOI] [PubMed] [Google Scholar]
- 29.Pender, S. L., C. McKenzie, A. Shaida, and T. T. MacDonald. 1999. Regulation of matrix metalloproteinases in human intestinal mucosa. Ann. N. Y. Acad. Sci. 878:581-582. [DOI] [PubMed] [Google Scholar]
- 30.Pfaffl, M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:E45-E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qadri, F., T. R. Bhuiyan, K. K. Dutta, R. Raqib, M. S. Alam, N. H. Alam, A. M. Svennerholm, and M. M. Mathan. 2004. Acute dehydrating disease caused by Vibrio cholerae serogroups O1 and O139 induce increases in innate cells and inflammatory mediators at the mucosal surface of the gut. Gut 53:62-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qadri, F., R. Raqib, F. Ahmed, T. Rahman, C. Wenneras, S. K. Das, N. H. Alam, M. M. Mathan, and A. M. Svennerholm. 2002. Increased levels of inflammatory mediators in children and adults infected with Vibrio cholerae O1 and O139. Clin. Diagn. Lab. Immunol. 9:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Royaee, A. R., C. Mendis, R. Das, M. Jett, and D. C. Yang. 2006. Cholera toxin induced gene expression alterations. Mol. Immunol. 43:702-709. [DOI] [PubMed] [Google Scholar]
- 34.Sack, D. A., R. B. Sack, G. B. Nair, and A. K. Siddique. 2004. Cholera. Lancet 363:223-233. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez, J., and J. Holmgren. 2005. Virulence factors, pathogenesis and vaccine protection in cholera and ETEC diarrhea. Curr. Opin. Immunol. 17:388-398. [DOI] [PubMed] [Google Scholar]
- 36.Schlittenhardt, D., A. Schober, J. Strelau, G. A. Bonaterra, W. Schmiedt, K. Unsicker, J. Metz, and R. Kinscherf. 2004. Involvement of growth differentiation factor-15/macrophage inhibitory cytokine-1 (GDF-15/MIC-1) in oxLDL-induced apoptosis of human macrophages in vitro and in arteriosclerotic lesions. Cell Tissue Res. 318:325-333. [DOI] [PubMed] [Google Scholar]
- 37.Sehnert, B., A. Cavcic, B. Bohm, J. R. Kalden, K. S. Nandakumar, R. Holmdahl, and H. Burkhardt. 2004. Antileukoproteinase: modulation of neutrophil function and therapeutic effects on anti-type II collagen antibody-induced arthritis. Arthritis Rheum. 50:2347-2359. [DOI] [PubMed] [Google Scholar]
- 38.Shi, L., L. H. Reid, W. D. Jones, R. Shippy, J. A. Warrington, S. C. Baker, P. J. Collins, F. de Longueville, E. S. Kawasaki, K. Y. Lee, Y. Luo, Y. A. Sun, J. C. Willey, R. A. Setterquist, G. M. Fischer, W. Tong, Y. P. Dragan, D. J. Dix, F. W. Frueh, F. M. Goodsaid, D. Herman, R. V. Jensen, C. D. Johnson, E. K. Lobenhofer, R. K. Puri, U. Scherf, J. Thierry-Mieg, C. Wang, M. Wilson, P. K. Wolber, L. Zhang, S. Amur, W. Bao, C. C. Barbacioru, A. B. Lucas, V. Bertholet, C. Boysen, B. Bromley, D. Brown, A. Brunner, R. Canales, X. M. Cao, T. A. Cebula, J. J. Chen, J. Cheng, T. M. Chu, E. Chudin, J. Corson, J. C. Corton, L. J. Croner, C. Davies, T. S. Davison, G. Delenstarr, X. Deng, D. Dorris, A. C. Eklund, X. H. Fan, H. Fang, S. Fulmer-Smentek, J. C. Fuscoe, K. Gallagher, W. Ge, L. Guo, X. Guo, J. Hager, P. K. Haje, J. Han, T. Han, H. C. Harbottle, S. C. Harris, E. Hatchwell, C. A. Hauser, S. Hester, H. Hong, P. Hurban, S. A. Jackson, H. Ji, C. R. Knight, W. P. Kuo, J. E. Leclerc, S. Levy, Q. Z. Li, C. Liu, Y. Liu, M. J. Lombardi, Y. Ma, S. R. Magnuson, B. Maqsodi, T. McDaniel, N. Mei, O. Myklebost, B. Ning, N. Novoradovskaya, M. S. Orr, T. W. Osborn, A. Papallo, T. A. Patterson, R. G. Perkins, E. H. Peters, R. Peterson, et al. 2006. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24:1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staats, H. F., and F. A. Ennis, Jr. 1999. IL-1 is an effective adjuvant for mucosal and systemic immune responses when coadministered with protein immunogens. J. Immunol. 162:6141-6147. [PubMed] [Google Scholar]
- 40.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Y., B. Agerberth, A. Lothgren, A. Almstedt, and J. Johansson. 1998. Apolipoprotein A-I binds and inhibits the human antibacterial/cytotoxic peptide LL-37. J. Biol. Chem. 273:33115-33118. [DOI] [PubMed] [Google Scholar]
- 42.World Health Organization. 1990. Manual for the treatment of diarrhoea: for use by physicians and other healthworkers. World Health Organization, Geneva, Switzerland.