Abstract
Colonic epithelial cells are constantly exposed to high levels of bacterial DNA in the intestinal lumen and must recognize and respond appropriately to pathogens, while they maintain a tolerance to nonpathogenic commensal bacterial strains. Bacterial DNA is recognized by Toll-like receptor 9 (TLR9). The aim of this study was to investigate TLR9 expression and localization in colonic epithelial cells under basal conditions and in response to bacterial DNA. HT-29 cells were exposed to DNA from various strains of commensal and pathogenic microbes. TLR9 mRNA expression was determined by real-time reverse transcription-PCR, and interleukin-8 (IL-8) secretion was measured by an enzyme-linked immunosorbent assay. Localization of TLR9 was determined by flow cytometry in HT-29 cells and by immunofluorescence in HT-29 cells and mouse colonic tissue. Immunofluorescence and flow cytometric analyses demonstrated that there was intracellular and surface expression of TLR9 in HT-29 cells under basal conditions. Exposure of cells to DNA from pathogenic strains of Salmonella and Escherichia coli resulted in a significant increase in TLR9 mRNA expression. Salmonella enterica serovar Dublin DNA increased surface TLR9 protein and IL-8 secretion. There was no change in mRNA levels or localization of TLR9 in response to Bifidobacterium breve. Chloroquine did not block IL-8 secretion in response to S. enterica serovar Dublin DNA. TLR9 was expressed on the colonic apical surface in wild-type mice but not in germfree mice. These results demonstrate that intestinal epithelial cells recognize pathogenic bacterial DNA and respond by increasing surface localization and expression of TLR9, suggesting that the epithelial inflammatory response to pathogenic DNA is mediated at least in part by increased TLR9 expression.
Recognition of microbial components and discrimination of harmful pathogens from commensal bacteria and from self are fundamental elements of the innate immune system. Toll-like receptors (TLRs) are responsible for recognizing various pathogen-associated molecular patterns, including lipoproteins (Toll-like receptor 2 [TLR2]), lipopolysaccharides (TLR4), and flagellin (TLR5). Bacterial DNA contains unmethylated 2′-deoxyribo(cytidine-phosphate-guanine) (CpG) dinucleotides flanked by specific sequences that activate the vertebrate innate immune system through TLR9. Bacterial DNA differs from mammalian DNA by its 20-fold-greater frequency of unmethylated CpG sequences (32). These sequences activate a signaling cascade via transcription factors NF-κB and AP-1 and stimulate the proliferation of B cells and the secretion of proinflammatory cytokines (interleukin-6 [IL-6], IL-12, and tumor necrosis factor alpha) required to eliminate an invading pathogen (15, 33). Intestinal epithelial cells are constantly exposed to high levels of bacterial DNA and must recognize and respond appropriately to the presence of pathogenic bacteria. These cells interact with microbes in the lumen, and TLR signaling is a key component of communication between intestinal epithelial cells and underlying immune cells in the lamina propria (31).
There has been variation in the results of studies in which the subcellular location of TLR9 was investigated. In dendritic cells and macrophages, TLR9 is located in the endoplasmic reticulum of resting cells. CpG-containing DNA is endocytosed, moves to the tubular lysosomal compartment, and subsequently binds directly to TLR9 (17). Conversely, surface expression of TLR9 has been reported to occur in HEK293 cells transfected with TLR9-containing expression vectors (6, 7), in gastric epithelium (27), in intestinal epithelial cells (18), and in some peripheral blood mononuclear cells and tonsil cells (9). Recently, a cytosolic innate immune response to DNA which triggers a potent interferon I response has been identified (29). Whether expression or localization of TLR9 changes in response to native bacterial DNA in intestinal epithelial cells which are exposed to high levels of bacterial DNA is not known.
The inflammatory activities of various types of synthetic oligonucleotides have been well studied; however, the activity of native bacterial DNA is understood less well but is known to be more stimulatory to macrophages than synthetic oligonucleotides are (25). Synthetic CpG-containing oligonucleotides have been shown to induce a TLR9-mediated inflammatory cytokine response and to exacerbate dextran sodium sulfate-induced colitis (13, 20). Other investigators have reported conflicting results, and administration of bacterial DNA ameliorated experimental murine colitis and human colitis (11, 22, 23). In an elegant study by Rakhoff-Nahoum et al., TLR ligands were shown to have a substantial role in intestinal homeostasis (24). Nonetheless, the role of bacterial DNA is intestinal immune signaling is incompletely understood.
MATERIALS AND METHODS
Cell culture and treatments.
HT-29 cells were obtained from the American Type Culture Collection (Manassas, VA) and were cultured in RPMI 1640 (Gibco, Burlington, ON, Canada) supplemented with 10% heat-inactivated fetal calf serum (Cansera, Rexdale, ON, Canada). All experiments were conducted in serum-free media. Cells were seeded at a density of 1 × 106 cells/well and were maintained at 37°C in a 5% CO2 atmosphere. Fresh medium was added to cultures daily. Cells were grown in 96-well tissue culture plates (Falcon, New Jersey) for determination of IL-8 secretion, in 6-well plates for the biotinylation assay, in 12-well plates for flow cytometry, and in chamber slides for immunofluorescence analysis.
Bacterial strains and preparation of DNA.
S. enterica serovar Dublin strain Lane (= ATCC 15480) was selected as a source of pathogenic bacterial DNA due to the ability of its DNA to induce IL-8 secretion at relatively low concentrations (10 μg/ml) (11) and was stored in 20% (wt/vol) skim milk at −70°C. Bifidobacterium breve was selected due to the ability of its DNA to reduce basal IL-8 secretion (11) and was stored at 4°C as a lyophilized powder. Bifidobacterium infantis and Lactobacillus acidophilus were used as other representative probiotic organisms, and Enterobacter cloacae was used as an opportunistic pathogen. S. enterica serovar Typhimurium and two virulent Escherichia coli strains were used as representative pathogens. The E. coli strains were selected from strains that were isolated from human biopsies and were fully characterized previously (14). E. coli 146 belonged to the B2 phylogenetic group and contained the Ag43, Sfa, and PAI adhesions. E. coli 147 also belonged to the B2 phylogenetic group and contained the Ag43 and Sfa adhesions (14). Bacteria were inoculated at a concentration of 0.18% (vol/vol) into 25 ml of Mann-Rogosa-Sharpe broth (bifidobacteria and lactobacilli) or tryptone soy broth (Difco catalog no. 0370-17-3) (all other organisms) and grown statically overnight (18 to 20 h) at 37°C. For experiments involving exposure of cells to live bacteria, overnight cultures of S. enterica serovar Dublin and B. breve were centrifuged at 2,000 × g for 10 min and resuspended in phosphate-buffered saline (PBS). Bacteria were diluted in serum-free cell culture media to obtain a concentration of 2 × 106 CFU/ml, and each suspension was applied to the apical surface of cells. This resulted in a multiplicity of infection of 1. After 60 min, the medium was removed, and the cells were washed with PBS. For DNA isolation, cells were centrifuged at 11,700 × g for 10 min, washed with 1× SSC buffer (0.15 M NaCl plus 0.015 M sodium citrate), and resuspended in 0.01 M sodium phosphate buffer with 20% sucrose and 2.5 mg/ml lysozyme for 45 min at 37°C, followed by lysis buffer (10 mM Tris-HCl, 1 mM EDTA, 500 mg pronase B, 1% sodium dodecyl sulfate, pH 8) for 30 min at 37°C. DNA was extracted by adding an equal volume of buffer-saturated phenol-chloroform (1:1) to the bacterial suspension. The mixture was centrifuged for 5 min at 4,000 rpm, and the aqueous layer was removed. The extraction was repeated until no interface was visible. Traces of phenol were removed with chloroform, and the salt concentration was adjusted by addition of 0.1 volume of sodium acetate (pH 5.2). DNA was precipitated with cold 100% ethanol, washed with 70% ethanol, and resuspended in sterile Tris-EDTA buffer. The concentration and purity of DNA preparations were confirmed by determining the optical density at 260 nm (OD260) and the OD260/OD280 ratio and by performing agarose gel electrophoresis. DNA preparations were assayed to determine the presence of endotoxin using the Limulus amebocyte assay (QCL-1000; BioWhittaker, Walkesville, MD). Only preparations with endotoxin levels that were not greater than 0.05 endotoxin unit/ml were used. DNase-treated preparations were used as control preparations in all experiments.
DNase and methylase treatment of DNA preparations.
DNA preparations were incubated overnight at 37°C with 5 mg/ml DNase 1 (Sigma-Aldrich) in the presence of 5 mM MgCl2. DNA depletion was confirmed by agarose gel electrophoresis with ethidium bromide staining (Fig. 1). Prior to cell culture treatment, DNase was heat inactivated. S. enterica serovar Dublin DNA and B. breve DNA were methylated with CpG methylase (3 U SssI/μg DNA; New England Biolabs, Pickering, ON, Canada) in NE buffer 2 supplemented with S-adenosylmethionine (New England Biolabs) at 37°C for 24 h.
FIG. 1.
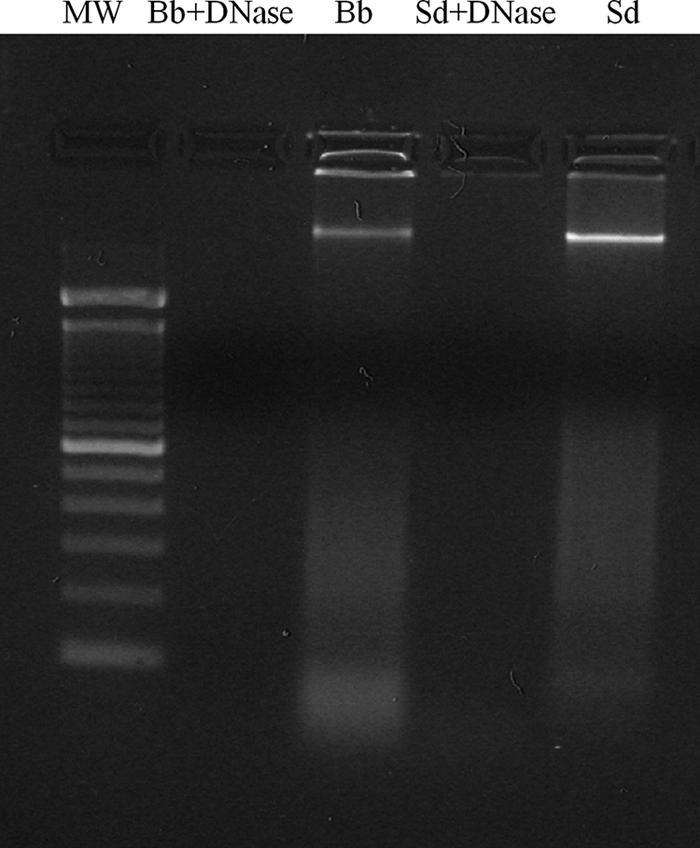
DNase treatment of DNA preparations. Overnight incubation of DNA preparations with 5 μg/ml DNase effectively eliminated DNA. MW, molecular weight markers; Bb, B. breve; Sd, S. enterica serovar Dublin.
Animal studies.
129 Sv/Ev mice were housed behind a barrier under specific-pathogen-free conditions. The mice had access to autoclaved 9% fat rodent blocks and sterile filtered water ad libitum. The facility's sanitation was verified by Health Sciences Lab Animal Services at the University of Alberta. Bacterial culture analyses, parasitological examinations, serological tracking profiles, and histological staining, all of which were negative for known murine viral and bacterial pathogens, indicated that the barrier was intact. Mice were sacrificed by cervical dislocation, and a segment of each colon was removed for immunohistochemical analysis. The mice used in the germfree experiments were derived in an axenic (germfree) fashion in the Health Science Laboratory Animal facility at the University of Alberta and were suckled, weaned, and raised in a sterile portable isolator. All axenic mice were maintained in sterile isolator bubbles and were monitored continually for microbial contamination. All experimental animals were tested at the time of sacrifice for microbial contamination.
Real-time reverse transcription-PCR.
Total RNA was isolated with Trizol (Gibco, Burlington, ON, Canada) by following the manufacturer's instructions. mRNA (1 μg) was reverse transcribed and amplified by PCR using β-actin as an endogenous control. A real-time PCR analysis was performed with an ABI 7700 sequence detector (Applied Biosystems, Branchburg, NJ). The reaction mixture (total volume, 20 μl) consisted of 1 μl cDNA, 7 μl double-distilled H2O, 1 μl target primer (6-carboxyfluorescein labeled; Applied Biosystems), 1 μl control primer (VIC labeled; Applied Biosystems), and 10 μl TaqMan Universal PCR master mixture (Applied Biosystems). The human TLR9 and β-actin sequences were obtained from GenBank and were used to design intron-spanning primers. The primer sequences used for PCR were as follows: TLR9 forward primer, GTGACAGATCCAAGGTGAAGT; TLR9 reverse primer, CTTCCTCTACAAATGCATCACT; actin forward primer, 5′-CGTGGGCCGCCCTAGGCACCA-3′; and actin reverse primer, 5′-TTGGCCTTAGGGTTCAGGGGGG-3′.
Flow cytometry.
Cells were allowed to grow to confluence (approximately 4 × 105 cells) and treated with 50 μg/ml bacterial DNA (S. enterica serovar Dublin or B. breve). After rinsing with PBS, cells were trypsinized with 0.05% trypsin-EDTA for 5 min, rinsed, and blocked with 6% neonatal goat serum for 10 min on ice. Anti-TLR9 (1/100; Imgenex, San Diego, CA) was added to 50 μl of blocked cells and incubated for 45 min on ice. Phycoerythrin-conjugated goat anti-mouse secondary antibody (1/100) was added, and the preparations were incubated for 30 min on ice and fixed in paraformaldehyde or permeabilized in methanol. A flow cytometric analysis was performed using a FACScan (San Jose, CA) equipped with an argon 488-nm laser. For each measurement, 1 × 105 events were collected. Fluorescence emission was collected in the FL2 channel. Files were analyzed using the CellQuest software, version 3.3 (San Jose, CA).
Immunofluorescence.
HT-29 cells were seeded (∼5 × 105 cells/well) into four-chamber slides and incubated overnight, and then they were treated with 50 μg/ml bacterial DNA for 2 h, washed twice with PBS, and incubated in 2% paraformaldehyde for 1 h. After 1 h of blocking with 5% skim milk in PBS, cells were incubated with anti-TLR9 or anti-calnexin antibody (1/100) for 60 min and then with Alexafluor 488-conjugated goat anti-mouse secondary antibody for TLR9 and Cy5-conjugated goat anti-rat secondary antibody for calnexin or Alexafluor 555-conjugated cholera toxin subunit B (CTxB) (Invitrogen, Burlington, ON, Canada). For immunohistochemical analysis, distal colonic tissue samples were harvested and fixed in 10% phosphate-buffered formalin, embedded in paraffin, and sectioned (4-μm sections). The tissue was hydrated by passing slides through xylene, xylene-100% ethanol (1:1), 100% ethanol, 95% ethanol, 70% ethanol, and distilled water washes. The tissue was blocked with 2% goat serum and incubated with anti-TLR9 and then with Alexafluor 488-conjugated goat anti-mouse antibody. The tissue was dehydrated by performing the hydration procedure in reverse order and was mounted with coverslips in Fluorsave. Slides were examined with a Zeiss Axiovert 100 M confocal microscope coupled with a Zeiss LSM510 laser scanning system (Germany). Images were taken with a ×63 Plan-apochromat n.a. 1.4 with a zoom factor of 2. Alexafluor 488 was scanned with an argon laser (excitation wavelength, 488 nm; emission wavelength, 505 nm), and Alexafluor 555-labeled CTxB and Cy5 were scanned with an HeNe laser (excitation wavelength, 543 nm; emission wavelength, 560 nm). For 4′,6′-diamidino-2-phenylindole (DAPI) staining, a Coherent Mira 2-Photon laser tuned at 780 nm was used, and emission was collected with a BP 390-465 IR. Confocal reflectance images were obtained by reflecting excitation wavelengths to a confocal detector.
Surface protein biotinylation.
To label surface proteins, confluent HT-29 cells were incubated with bacterial DNA (50 μg/ml), cooled on ice, washed three times with cold PBS, and incubated with 80 μl of 10 mM sulfosuccinimidyl-2-(biotinamido)-ethyl-1,3-dithiopropionate (Pierce, Rockford, IL) for 30 min on ice. Cells were washed with cold PBS, and free biotin was quenched with 100 mM glycine in PBS. Cell were lysed in 1% Triton X-100-150 mM NaCl-5 mM EDTA-50 mM Tris (pH 7.4) for 1 h at 4°C on an orbital shaker. Samples were electrophoresed on 10% sodium dodecyl sulfate-polyacrylamide gels as previously described and were blotted for TLR9 analysis (12).
IL-8 ELISA.
IL-8 protein levels were determined by an enzyme-linked immunosorbent assay (ELISA) using matched antibodies from R&D Systems as previously described (12).
Statistical analysis.
Data are expressed below as means ± standard deviations for three or more samples unless indicated otherwise. Statistical significance was assessed using analysis of variance or a t test (Sigmastat; Systat Software, Inc., Point Richmond, CA), and differences were considered significant if the P values were <0.05.
RESULTS
TLR9 mRNA expression in response to probiotic and pathogenic bacterial DNA.
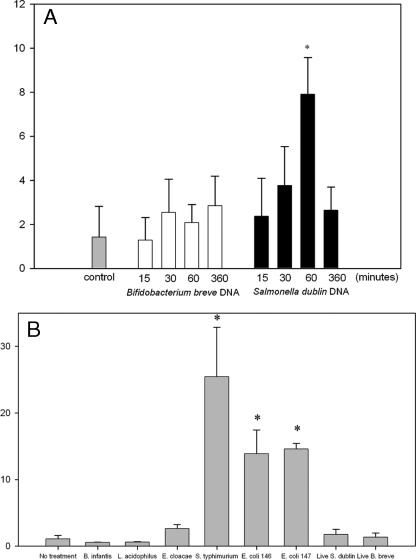
HT-29 cells express TLR9 (2, 21). To determine how TLR9 expression is affected by exposure to bacterial DNA, HT-29 monolayers were treated with DNA from either B. breve, a probiotic organism, or S. enterica serovar Dublin, a pathogenic organism, and the time course of TLR9 mRNA expression was determined using real-time PCR. As shown in Fig. 2A, B. breve DNA had no effect on TLR9 expression. In contrast, treatment with S. enterica serovar Dublin DNA resulted in a significant increase in TLR9 mRNA expression, and the expression was eightfold greater than the expression of resting cells after 60 min of exposure (P < 0.05). To determine if the increase in TLR9 expression was a general response to pathogens or was specific to S. enterica serovar Dublin, cells were exposed to DNA from another strain of Salmonella, an S. enterica serovar Typhimurium strain, and also to pathogenic strains of E. coli and E. cloacae. The results were compared with responses to the probiotic organisms L. acidophilus and B. infantis. As shown in Fig. 2B, TLR9 expression was upregulated after 60 min of exposure to DNA from S. enterica serovar Typhimurium and the pathogenic E. coli strains, marginally upregulated by E. cloacae, and not changed by B. infantis or L. acidophilus (Fig. 2B). This suggests that the response is a generalized response to pathogens and not specific to Salmonella sp. Live B. breve had no effect on TLR expression. However, interestingly, live S. enterica serovar Dublin also did not upregulate TLR9 expression, suggesting that the DNA in viable microbes may not have been accessible to the TLR receptor under these conditions.
FIG. 2.
TLR9 expression (y axes show n-fold expression) was upregulated in response to S. enterica serovar Dublin DNA but not in response to B. breve DNA. (A) Real-time PCR measurements of TLR9 mRNA expression in confluent HT-29 monolayers in response to S. enterica serovar Dublin and B. breve DNA, showing that S. enterica serovar Dublin DNA elicited a substantial increase in TLR9 expression at 60 min. (B) TLR9 expression was upregulated after 60 min of exposure to DNA from S. enterica serovar Typhimurium and the pathogenic E. coli strains, was marginally upregulated by E. cloacae, and was not changed by B. infantis or L. acidophilus. The bars indicate the means and the error bars indicate the standard deviations of triplicate measurements obtained in three separate experiments. An asterisk indicates that the P value is <0.05.
Chloroquine did not inhibit the IL-8 response elicited by S. enterica serovar Dublin DNA.
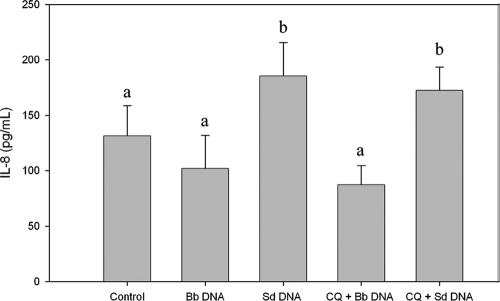
We and other workers have previously shown that bacterial DNA can elicit epithelial cells to secrete IL-8 (2, 11). The location of the signaling event for TLR9 and bacterial DNA in epithelial cells is not known; however, in immune cells, TLR9 is expressed on the endoplasmic reticulum and moves to the lysosome upon cellular uptake of CpG-containing oligonucleotides (17, 19). Chloroquine, an inhibitor of endosomal acidification, has been shown to block CpG-TLR9 binding and thus to eliminate CpG-containing DNA effects on immune cells (26). To determine whether signaling events in epithelial cells are similar, HT-29 cells were treated with chloroquine (10 μg/ml), followed by exposure to S. enterica serovar Dublin and B. breve DNA. IL-8 secretion was measured by ELISA. Exposure to S. enterica serovar Dublin DNA, but not exposure to B. breve DNA, elicited significantly increased secretion of IL-8 compared with the secretion by control cells (P < 0.05) (Fig. 3). Confirming the results obtained with HCA-7 cells (18), chloroquine did not inhibit S. enterica serovar Dublin-induced IL-8 secretion, suggesting that the TLR9 signaling in intestinal epithelial cells is different than the TLR9 signaling in immune cells.
FIG. 3.
Chloroquine treatment did not inhibit S. enterica serovar Dublin DNA-induced IL-8 secretion. HT-29 cells were pretreated with chloroquine (CQ) (10 μg/ml) for 2 h, and then B. breve (Bb) or S. enterica serovar Dublin (Sd) DNA was added for 6 h. The IL-8 concentration was determined by ELISA. Treatment with S. enterica serovar Dublin DNA, but not treatment with B. breve DNA, resulted in an increase in IL-8 secretion, which was not eliminated by chloroquine treatment. All “a” bars and all “b” bars are significantly different at a P value of <0.05. The bars indicate the means and the error bars indicate the standard deviations of triplicate measurements obtained in three separate experiments.
Flow cytometric analysis of TLR9 expression.
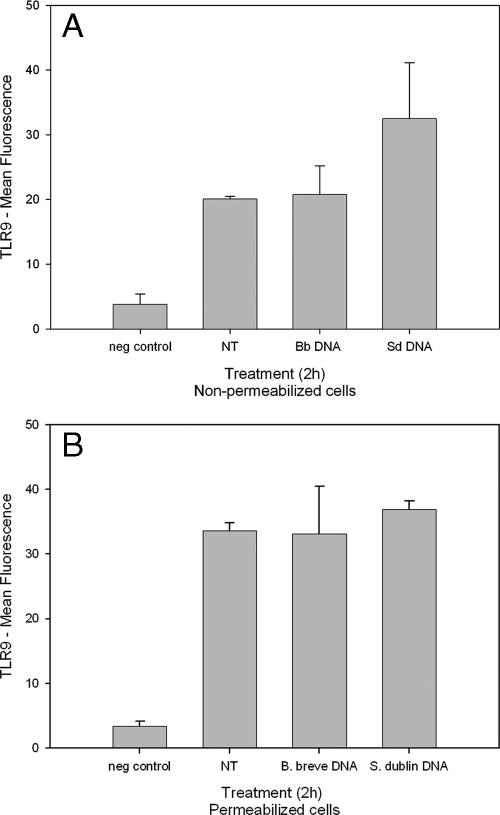
As endosomal uptake of DNA does not appear to be necessary for S. enterica serovar Dublin DNA-induced IL-8 secretion, we investigated localization of TLR9 expression in HT-29 cells. HT-29 cells were fixed in paraformaldehyde or permeabilized with methanol. TLR9 was labeled with phycoerythrin-conjugated secondary antibody, and cells were analyzed by flow cytometry. Untreated and B. breve DNA-treated cells both exhibited limited surface expression of TLR9 (Fig. 4). Cells treated with S. enterica serovar Dublin DNA exhibited increased surface expression of TLR9 compared with B. breve DNA-treated and untreated cells. The mean fluorescence in permeabilized cells was greater than the mean fluorescence in nonpermeabilized cells; however, no increase in expression was observed in response to S. enterica serovar Dublin DNA.
FIG. 4.
Flow cytometric analysis of TLR9. HT-29 cells were treated with B. breve (Bb) or S. enterica serovar Dublin (Sd) DNA for 2 h and preserved in paraformaldehyde (A) or methanol (B), and TLR9 was labeled with phycoerythrin-conjugated goat anti-mouse secondary antibody. Cell suspensions were analyzed by flow cytometry. No antibodies were added for the negative (neg) control. The bars indicate the means and the error bars indicate the standard deviations of triplicate measurements obtained in three separate experiments. NT, no treatment.
Immunofluorescence of TLR9 in HT-29 cell culture.
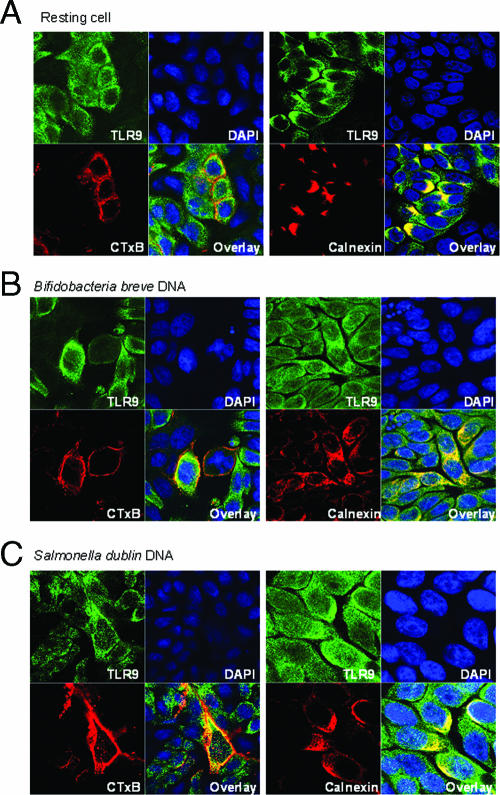
To confirm the results of the flow cytometric analysis, we immunofluorescently labeled TLR9 in HT-29 monolayers containing resting cells and cells after exposure to S. enterica serovar Dublin and B. breve DNA. CTxB (red) was used to localize the cell membrane, and calnexin (red) was used to localize the endoplasmic reticulum. Under resting conditions, TLR9 (green) colocalized with calnexin, and there was little cell surface expression (Fig. 5A). After a 2-h exposure to B. breve DNA (Fig. 5B), some colocalization with CTxB was observed. With S. enterica serovar Dublin DNA (Fig. 5C), colocalization with surface-bound CTxB was evident, and TLR9 was observed beyond the boundary of calnexin staining. No fluorescence was observed in secondary antibody controls (data not shown).
FIG. 5.
Fluorescently labeled TLR9 moves to the surface in response to bacterial DNA: confocal images of HT-29 cells with Alexafluor 488-labeled TLR9 (green) and either Alexafluor 555-labeled CTxB (red) (left panels) or Cy5-labeled calnexin (red) (right panels) in resting cells (A), cells treated with B. breve DNA for 2 h (B), and cells treated with S. enterica serovar Dublin DNA for 2 h (C). Nuclei were stained with DAPI (blue). The stack size is 73 by 73 μm. The images are representative of the results of three separate experiments.
Biotinylation of surface proteins.
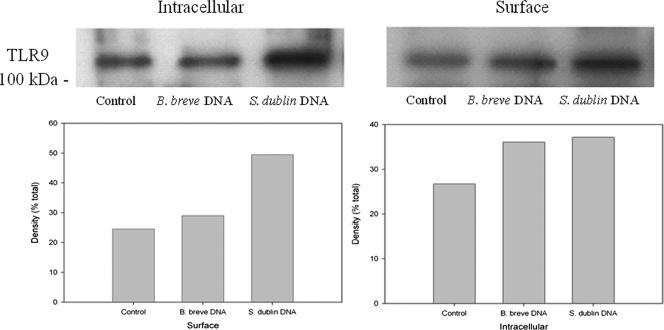
To further substantiate the surface expression of TLR9, biotinylation of surface proteins was carried out using resting HT-29 cells and cells treated with B. breve DNA or S. enterica serovar Dublin DNA. TLR9 was present on the cell surface in the resting cells, and approximately equivalent amounts were present in the cells treated with B. breve DNA (Fig. 6). In the cells treated with S. enterica serovar Dublin DNA, an increase in surface expression of TLR9 was observed. A slight increase in the level of cytoplasmic TLR9 was also observed in the S. enterica serovar Dublin DNA-treated cells, but this increase was not as substantial as the increase in surface expression.
FIG. 6.
Biotinylation of surface proteins. HT-29 cells were treated for 2 h with either B. breve DNA or S. enterica serovar Dublin DNA. Cells were lysed, and surface proteins were separated from cytoplasmic proteins by biotinylation and separation on streptavidin-coated beads. Lysates were electrophoresed on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, transferred to nitrocellulose, and probed with anti-TLR9. Equal protein loading was confirmed by Ponceau S staining. Surface expression of TLR9 occurred in resting cells and was upregulated by S. enterica serovar Dublin DNA treatment. A slight increase in cytoplasmic TLR9 was also observed upon treatment with S. enterica serovar Dublin DNA. The images are representative of the results of three separate experiments.
Localization of TLR9 in mouse colonic tissue.
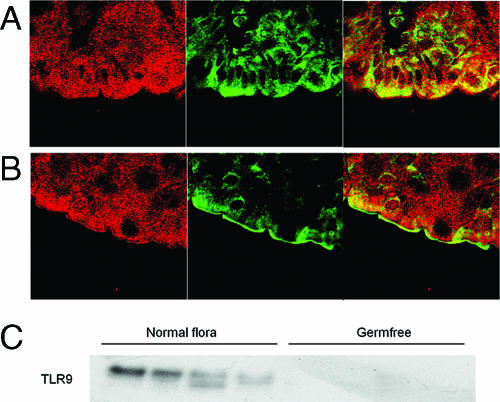
To determine whether surface expression of TLR9 also occurs in vivo, sections of mouse colonic tissue were incubated with anti-TLR9 antibody, followed by Alexafluor 488-conjugated secondary antibody. The brush border membrane was localized by confocal reflectance. In colonic tissue from 129 Sv/Ev germfree mice (Fig. 7A), limited fluorescence was observed, and this fluorescence was markedly different from the fluorescence observed in wild-type 129 Sv/Ev mice that were normally colonized with microflora (Fig. 7B). In these mice, expression clearly occurred on the brush border membrane. Likewise, Western blotting of TLR9 in colonic tissue demonstrated that there was abundant expression in colonized mice and that there was no detectable expression in germfree mice (Fig. 7C).
FIG. 7.
TLR9 was expressed on the apical surface of the intestine of 129 Sv/Ev mice with normal flora but not on the apical surface of the intestine of germfree mice. Colonic tissue was collected from germfree 129 Sv/Ev mice (A) and 129 Sv/Ev mice raised under conventional conditions (B) and was probed to determine the presence of TLR9. Confocal reflectance is indicated by red, and TLR9 labeling is indicated by green. The stack size is 73 by 73 μm. (C) Western blot of colonic TLR9 expression in SvEv/129 mice colonized with the normal flora and in germfree mice. The results are representative of three separate experiments.
Methylation of bacterial DNA.
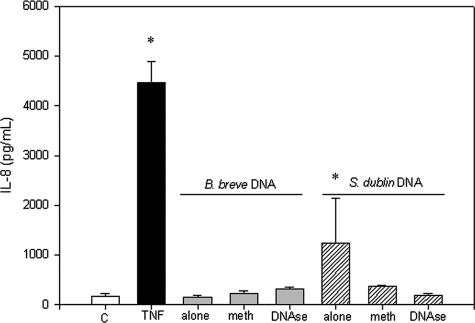
Since it is not known how TLR9 differentiates between different types of bacterial DNA, we sought to determine whether the stimulatory capacities of B. breve and S. enterica serovar Dublin DNA differ due to the methylation status of CpG elements. B. breve and S. enterica serovar Dublin DNA were treated with SssI methylase, which adds a methyl group to CpG motifs, thereby rendering them nonstimulatory. Methylation of B. breve DNA had no effect on IL-8 secretion; however, methylation of S. enterica serovar Dublin DNA significantly reduced the stimulatory effect of S. enterica serovar Dublin DNA (P < 0.05), as determined by IL-8 secretion (Fig. 8). Furthermore, DNase treatment of S. enterica serovar Dublin DNA eliminated its stimulatory effect (P < 0.05).
FIG. 8.
Methylation of bacterial DNA inhibits stimulatory effects. HT-29 monolayers were exposed to methylated (meth) and unmethylated B. breve and S. enterica serovar Dublin DNA, and the cell culture media were analyzed to determine the presence of secreted IL-8 by ELISA. Treatment with S. enterica serovar Dublin DNA, but not treatment with B. breve DNA, resulted in increased IL-8 secretion, which was eliminated when the S. enterica serovar Dublin DNA was methylated. An asterisk indicates that the P value is <0.05 for a comparison with the control values. The bars indicate the means and the error bars indicate the standard deviations of triplicate measurements obtained in three separate experiments. C, control; TNF, tumor necrosis factor.
DISCUSSION
In this study, we demonstrated that TLR9 is expressed on the apical surface of HT-29 cells and in mouse colonic tissue. Furthermore, surface localization increased upon stimulation with proinflammatory DNA from pathogenic bacteria and in response to a normal luminal microflora in vivo. We also confirmed that intestinal epithelial cells do not respond equally to bacterial DNA and are capable of distinguishing between DNA from probiotic strains and DNA from pathogenic strains. Finally, unmethylated CpG appears to involved in epithelial cellular recognition and the differential response to bacterial DNA.
Several investigators have shown that TLR9 is expressed in intracellular compartments in dendritic cells and macrophages and that internalization and endosomal maturation is necessary for CpG-containing DNA to activate TLR9 (1, 17, 19, 30). However, other workers have described surface expression of TLR9 in various types of cells (6, 7, 9, 18, 27). Furthermore, studies using chloroquine to inhibit endosomal acidification have produced different results; some investigators have observed inhibition of TLR9 signaling (1, 10, 30), and other workers have observed no effect, indicating that there may be an alternate pathway (8, 18). In our study, TLR9 was shown by flow cytometry, immunofluorescence, and biotinylation of surface proteins to be expressed on the surface of intestinal epithelial cells. Furthermore, confirming what has recently been shown with HCA-7 cells (18), chloroquine treatment did not eliminate the inflammatory response initiated by bacterial DNA, indicating that the signaling events in intestinal epithelia in response to TLR9 ligands differ from the signaling events in immune cells.
An increase in expression of TLR9 has been shown to occur in response to other ligands, including lipopolysaccharide and gamma interferon, and B-cell receptor triggering (4, 5, 30). We ensured that our DNA preparations were free of lipopolysaccharide contamination in order to eliminate the possibility of cross-stimulation of TLR9. Moreover, the effects of S. enterica serovar Dublin DNA on IL-8 secretion were eliminated by treating the preparations with DNase.
A major question arising from this work and other work is how TLR9 is able to distinguish between different types of bacterial DNA. Unmethylated CpG motifs are present at a 20-fold-higher frequency in bacterial DNA than in mammalian DNA due to CpG suppression and CpG methylation in mammalian DNA (16). Huang et al. (10) showed that, like the effect of E. coli DNA (28), the stimulatory effect of Brucella abortus DNA is related to a high frequency of CpG motifs and that methylation of the DNA eliminated the IL-12p40 response in mouse splenocytes. Likewise, the CpG content of bacterial DNA was related to the capacity of the DNA to activate TLR9. The sequence of the S. enterica serovar Dublin genome is not known yet, although work is in progress. Identifying the frequency of CpG motifs in S. enterica serovar Dublin DNA and B. breve DNA may aid in determining why these bacterial DNA elicit such different effects. The methylation status of the DNA appears to play a role in the stimulatory effect of S. enterica serovar Dublin, as methylated S. enterica serovar Dublin DNA was not able to initiate an IL-8 response in HT-29 cells. This confirms that the stimulatory ability of DNA from S. enterica serovar Dublin resides in unmethylated CpG motifs in the DNA.
The frequency of stimulatory CpG sequences in the DNA obtained from normal, healthy flora and how accessible this DNA is to intestinal epithelial cells in vivo are not known. The fact that the concentrations of microorganisms can reach 1012 CFU/g of tissue in the colon and the fact that surface expression of TLR9 is not observed in germfree mice, although it is clearly present in mice colonized with normal flora, indicate that intestinal epithelial cells respond to normal microflora colonization with surface expression of TLR9.
The biological significance of surface expression versus endoplasmic reticulum expression of TLR9 is not known. It has been shown that a chimeric TLR9 created to localize to the surface of mouse embryo fibroblasts is stimulated by self DNA but not by viral DNA, which led the authors to speculate that the function of intracellular TLR9 is to prevent chronic activation by self DNA, yet allow recognition of invading viral (or bacterial) DNA (3). However, stimulation of TLRs does not always lead to an inflammatory response, and the presence of TLR receptor-ligand interactions is a component of intestinal homeostasis (24). The recent finding that TLR9 activation through apical and basolateral surfaces in polarized epithelial cells activates different intracellular signaling pathways and the finding that apical TLR9 stimulation appears to confer tolerance to subsequent TLR challenge suggest that TLR9 plays an important role in maintaining intestinal homeostasis (18). Recognition of foreign bacterial antigens by the innate immune system is critical to the well-being of the host organism. TLRs are an integral part of this system, as they produce signals that initiate an immune response and lead to the destruction of invading pathogens. Further understanding of the signaling events that occur upon binding of TLR9 to different types of bacterial DNA should aid in determining how bacterial ligands contribute to inflammation and to homeostasis.
Acknowledgments
This study was supported by the Alberta Heritage Foundation for Medical Research, by the Crohn's and Colitis Foundation of Canada, and by the Canadian Institutes for Health Research. Karen Madsen was supported as a senior scholar by the Alberta Heritage Foundation for Medical Research. Julia Ewaschuk was supported by fellowships from the Canadian Association for Gastroenterology and the Alberta Heritage Foundation for Medical Research.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 26 February 2007.
REFERENCES
- 1.Ahmad-Nejad, P., H. Hacker, M. Rutz, S. Bauer, R. M. Vabulas, and H. Wagner. 2002. Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur. J. Immunol. 32:1958-1968. [DOI] [PubMed] [Google Scholar]
- 2.Akhtar, M., J. L. Watson, A. Nazli, and D. M. McKay. 2003. Bacterial DNA evokes epithelial IL-8 production by a MAPK-dependent, NF-kappaB-independent pathway. FASEB J. 17:1319-1321. [DOI] [PubMed] [Google Scholar]
- 3.Barton, G. M., J. C. Kagan, and R. Medzhitov. 2006. Intracellular localization of Toll-like receptor 9 prevents recognition of self DNA but facilitates access to viral DNA. Nat. Immunol. 7:49-56. [DOI] [PubMed] [Google Scholar]
- 4.Bernasconi, N. L., N. Onai, and A. Lanzavecchia. 2003. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood 101:4500-4504. [DOI] [PubMed] [Google Scholar]
- 5.Bourke, E., D. Bosisio, J. Golay, N. Polentarutti, and A. Mantovani. 2003. The Toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood 102:956-963. [DOI] [PubMed] [Google Scholar]
- 6.Chuang, T. H., J. Lee, L. Kline, J. C. Mathison, and R. J. Ulevitch. 2002. Toll-like receptor 9 mediates CpG-DNA signaling. J. Leukoc. Biol. 71:538-544. [PubMed] [Google Scholar]
- 7.Cornelie, S., J. Hoebeke, A. M. Schacht, B. Bertin, J. Vicogne, M. Capron, and G. Riveau. 2004. Direct evidence that Toll-like receptor 9 (TLR9) functionally binds plasmid DNA by specific cytosine-phosphate-guanine motif recognition. J. Biol. Chem. 279:15124-15129. [DOI] [PubMed] [Google Scholar]
- 8.Cortez-Gonzalez, X., I. Pellicciotta, M. Gerloni, M. C. Wheeler, P. Castiglioni, P. Lenert, and M. Zanetti. 2006. TLR9-independent activation of B lymphocytes by bacterial DNA. DNA Cell Biol. 25:253-261. [DOI] [PubMed] [Google Scholar]
- 9.Eaton-Bassiri, A., S. B. Dillon, M. Cunningham, M. A. Rycyzyn, J. Mills, R. T. Sarisky, and M. L. Mbow. 2004. Toll-like receptor 9 can be expressed at the cell surface of distinct populations of tonsils and human peripheral blood mononuclear cells. Infect. Immun. 72:7202-7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang, L. Y., K. J. Ishii, S. Akira, J. Aliberti, and B. Golding. 2005. Th1-like cytokine induction by heat-killed Brucella abortus is dependent on triggering of TLR9. J. Immunol. 175:3964-3970. [DOI] [PubMed] [Google Scholar]
- 11.Jijon, H., J. Backer, H. Diaz, H. Yeung, D. Thiel, C. McKaigney, C. De Simone, and K. Madsen. 2004. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology 126:1358-1373. [DOI] [PubMed] [Google Scholar]
- 12.Jijon, H. B., W. J. Panenka, K. L. Madsen, and H. G. Parsons. 2002. MAP kinases contribute to IL-8 secretion by intestinal epithelial cells via a posttranscriptional mechanism. Am. J. Physiol. Cell Physiol. 283:C31-C41. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi, M., M. N. Kweon, H. Kuwata, R. D. Schreiber, H. Kiyono, K. Takeda, and S. Akira. 2003. Toll-like receptor-dependent production of IL-12p40 causes chronic enterocolitis in myeloid cell-specific Stat3-deficient mice. J. Clin. Investig. 111:1297-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotlowski, R., C. N. Bernstein, S. Sepehri, and D. O. Krause. 6 October 2006, posting date. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut. doi: 10.1136/gut.2006.099796. [DOI] [PMC free article] [PubMed]
- 15.Krieg, A. M. 2002. CpG motifs in bacterial DNA and their immune effects. Annu. Rev. Immunol. 20:709-760. [DOI] [PubMed] [Google Scholar]
- 16.Krieg, A. M., A. K. Yi, S. Matson, T. J. Waldschmidt, G. A. Bishop, R. Teasdale, G. A. Koretzky, and D. M. Klinman. 1995. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature 374:546-549. [DOI] [PubMed] [Google Scholar]
- 17.Latz, E., A. Schoenemeyer, A. Visintin, K. A. Fitzgerald, B. G. Monks, C. F. Knetter, E. Lien, N. J. Nilsen, T. Espevik, and D. T. Golenbock. 2004. TLR9 signals after translocating from the ER to CpG DNA in the lysosome. Nat. Immunol. 5:190-198. [DOI] [PubMed] [Google Scholar]
- 18.Lee, J., D. Rachmilewitz, and E. Raz. 2006. Homeostatic effects of TLR9 signaling in experimental colitis. Ann. N. Y. Acad. Sci. 1072:351-355. [DOI] [PubMed] [Google Scholar]
- 19.Leifer, C. A., M. N. Kennedy, A. Mazzoni, C. Lee, M. J. Kruhlak, and D. M. Segal. 2004. TLR9 is localized in the endoplasmic reticulum prior to stimulation. J. Immunol. 173:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obermeier, F., N. Dunger, L. Deml, H. Herfarth, J. Scholmerich, and W. Falk. 2002. CpG motifs of bacterial DNA exacerbate colitis of dextran sulfate sodium-treated mice. Eur. J. Immunol. 32:2084-2092. [DOI] [PubMed] [Google Scholar]
- 21.Pedersen, G., L. Andresen, M. W. Matthiessen, J. Rask-Madsen, and J. Brynskov. 2005. Expression of Toll-like receptor 9 and response to bacterial CpG oligodeoxynucleotides in human intestinal epithelium. Clin. Exp. Immunol. 141:298-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rachmilewitz, D., F. Karmeli, S. Shteingart, J. Lee, K. Takabayashi, and E. Raz. 2006. Immunostimulatory oligonucleotides inhibit colonic proinflammatory cytokine production in ulcerative colitis. Inflamm. Bowel Dis. 12:339-345. [DOI] [PubMed] [Google Scholar]
- 23.Rachmilewitz, D., F. Karmeli, K. Takabayashi, T. Hayashi, L. Leider-Trejo, J. Lee, L. M. Leoni, and E. Raz. 2002. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology 122:1428-1441. [DOI] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118:229-241. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, T. L., J. A. Dunn, T. D. Terry, M. P. Jennings, D. A. Hume, M. J. Sweet, and K. J. Stacey. 2005. Differences in macrophage activation by bacterial DNA and CpG-containing oligonucleotides. J. Immunol. 175:3569-3576. [DOI] [PubMed] [Google Scholar]
- 26.Rutz, M., J. Metzger, T. Gellert, P. Luppa, G. B. Lipford, H. Wagner, and S. Bauer. 2004. Toll-like receptor 9 binds single-stranded CpG-DNA in a sequence- and pH-dependent manner. Eur. J. Immunol. 34:2541-2550. [DOI] [PubMed] [Google Scholar]
- 27.Schmausser, B., M. Andrulis, S. Endrich, S. K. Lee, C. Josenhans, H. K. Muller-Hermelink, and M. Eck. 2004. Expression and subcellular distribution of Toll-like receptors TLR4, TLR5 and TLR9 on the gastric epithelium in Helicobacter pylori infection. Clin. Exp. Immunol. 136:521-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacey, K. J., G. R. Young, F. Clark, D. P. Sester, T. L. Roberts, S. Naik, M. J. Sweet, and D. A. Hume. 2003. The molecular basis for the lack of immunostimulatory activity of vertebrate DNA. J. Immunol. 170:3614-3620. [DOI] [PubMed] [Google Scholar]
- 29.Stetson, D. B., and R. Medzhitov. 2006. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity 24:93-103. [DOI] [PubMed] [Google Scholar]
- 30.Takeshita, F., C. A. Leifer, I. Gursel, K. J. Ishii, S. Takeshita, M. Gursel, and D. M. Klinman. 2001. Cutting edge: role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J. Immunol. 167:3555-3558. [DOI] [PubMed] [Google Scholar]
- 31.Uematsu, S., and S. Akira. 2006. Toll-like receptors and innate immunity. J. Mol. Med. 84:712-725. [DOI] [PubMed] [Google Scholar]
- 32.Wagner, H. 1999. Bacterial CpG DNA activates immune cells to signal infectious danger. Adv. Immunol. 73:329-368. [DOI] [PubMed] [Google Scholar]
- 33.Zhao, Q., J. Temsamani, R. Z. Zhou, and S. Agrawal. 1997. Pattern and kinetics of cytokine production following administration of phosphorothioate oligonucleotides in mice. Antisense Nucleic Acid Drug Dev. 7:495-502. [DOI] [PubMed] [Google Scholar]