Abstract
Vibrio cholerae NRT36S is a non-cholera toxin-producing, non-O1 strain that causes diarrhea in volunteers. The genome of NRT36S was sequenced to create a draft containing 174 contigs plus the superintegron region. Our analysis of the draft genome revealed several putative toxin genes and colonization factors. Besides confirming the existence of nonagglutinable heat-stable toxin, we also identified the genes for a type three secretion system, a putative exotoxin, two different RTX toxins, and four pilus systems.
Vibrio cholerae is best known as the causative agent of cholera. Cholera epidemics are associated with O1 and O139 serogroup isolates that produce cholera toxin (CT). V. cholerae strains from the other more than 200 serogroups reported for this species may be isolated from sporadic small outbreaks and isolated cases of diarrhea. The mechanisms by which the latter strains cause human disease remain controversial. In this study, we sequenced the genome of a known pathogenic non-O1 V. cholerae clinical isolate and compared it to the fully sequenced genome of V. cholerae O1 El Tor N16961 (6). V. cholerae NRT36S was originally isolated from a Japanese patient with traveler's diarrhea. It is a serogroup O31, CT-negative isolate and produces nonagglutinable heat-stable toxin (NAG-ST) (1). When fed to volunteers, this isolate caused diarrhea, including a 5.3-liter diarrheal purge from one patient (16). Molecular studies have indicated that this isolate is not closely related to epidemic O1 and O139 isolates (18).
The V. cholerae NRT36S genome was sequenced by the company 454 Life Sciences. Genomic DNA was extracted with a QIAamp DNA Mini kit (QIAGEN, Valencia, CA). 454 Life Sciences employed a different sequencing strategy from conventional shotgun sequencing, as described in detail elsewhere (15). Briefly, genomic DNA was randomly sheared to small fragments and ligated to common adaptors. Single fragments were attached to beads in an emulsion. Amplification by PCR was done in the emulsion and produced ∼107 copies of the fragments per bead. After removal of the emulsion, the beads were deposited on a fiber optic slide. The DNAs were sequenced using a pyrosequencing protocol. Sequencing was performed on a Genome Sequencer 20 system. The four sequencing runs generated 1,082,967 reads and output 104,531,256 bp of sequence. The estimated coverage depth was 26×. The draft genome consisted of 174 large contigs plus the superintegron region, with a total length of 4,079,433 bp. The average GC content for the draft genome was 47.5%. The genome was annotated by the National Microbial Pathogen Data Resource and is available online (http://anno-2.nmpdr.org/umd/FIG/index.cgi).
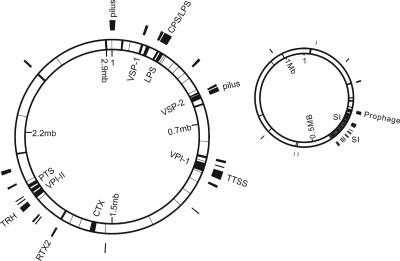
We compared the genome of V. cholerae NRT36S to the published genome of V. cholerae N16961, a serogroup O1 isolate, using the program MUMMER 3.0 (using the default settings of the program) (Fig. 1) (12). The genome sizes were comparable, at approximately 4.1 megabases. About 3.5 megabases (89%) of sequence was common to both genomes. Substantial differences between the genomes of V. cholerae NRT36S and N16961 were noted, especially in the genes for pathogenesis, and surface polysaccharides and in the superintegrons. The sequences identified in only one isolate by MUMMER (cutoff, 70% nucleotide identity) were considered strain specific.
FIG. 1.
Comparison of V. cholerae N16961 and NRT36S genomes. The two closed rings represent two chromosomes of N16961. Chromosome I is on the left, and chromosome II is on the right. The white areas in the rings are sequences conserved between the two genomes. Gray areas are sequences specific to N16961, with labels inside the rings. Black areas outside the rings are sequences specific to NRT36S, with labels outside the rings. VSP-I and VSP-II, Vibrio seventh pandemic islands I and II; LPS, lipopolysaccharide; VPI-1 and VPI-2, Vibrio pathogenic islands 1 and 2; CTX, cholera toxin genes; PTS, phosphoenolpyruvate phosphotransferase system; CPS, capsule polysaccharide; TTSS, type III secretion system; TRH, thermostable direct hemolysin; SI, superintegron.
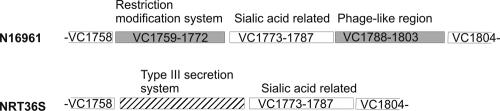
We confirmed that the genes related to pathogenesis in V. cholerae N16961 were strain specific. These genes made up 30% of the N16961-specific sequences. The CTXφ prophage (VC1452 to VC1478), which encodes CT in V. cholerae O1 (20), was absent from NRT36S. The genome of CTXφ also carries genes responsible for phage morphogenesis and its insertion into the host genome. None of these genes was present in NRT36S. The toxin-coregulated pilus (TCP), the major colonization factor for V. cholerae O1 (7, 19), was also missing from NRT36S. TCP is encoded in a 39.5-kb region of the V. cholerae genome designated Vibrio pathogenic island 1 (VC0819 to VC0845) (10). Together with the TCP gene, the genes carried in Vibrio pathogenic island 1 were absent from NRT36S. Another genomic island, described as Vibrio pathogenic island 2 for V. cholerae O1 (VC1758 to -1803), was altered in NRT36S (Fig. 2). VC1759 to -1772 encode a restriction modification system, and VC1773 to -1787 encode enzymes, including a sialidase (neuraminidase; EC 3.2.1.18), involved in the transport and utilization of sialic acid. VC1788 to -1803 encode phage proteins (8). Neuraminidase was found to be an important virulence factor that can increase the amount of the human receptor for CT (5a). Most of Vibrio pathogenic island 2 (VC1759 to -1772 and VC1788 to -1803) was absent from NRT36S, but an internal section (VC1773 to -1787), the sialic acid-related region, was present. This interesting mosaic structure suggests that this region of the genome is a hot spot for lateral gene transfer.
FIG. 2.
Mosaic pattern of Vibrio pathogenic island 2. The three regions in the Vibrio pathogenic island were described previously (8). White boxes are conserved regions between N16961 and NRT36S. Shaded boxes indicate genes specific to N16961. The hatched box indicates genes specific to NRT36S.
Two other genomic islands, named Vibrio seventh pandemic islands I and II (VC0175 to -0185 and VC0490 to -0497), have been reported to be specific to V. cholerae O1 El Tor and related O139 strains (8). The functions of these two islands are not clear. Neither of these islands was present in NRT36S.
The genes related to pathogenesis in NRT36S were different from those in N16961. In a previous study (16), Morris et al. suggested that the heat-stable enterotoxin NAG-ST and the ability to colonize were the major factors contributing to the occurrence of disease in human volunteers ingesting NRT36S. NAG-ST causes fluid accumulation in a suckling mouse model (1, 16, 17). NAG-ST was not present in N16961. Interestingly, the NAG-ST gene was located in the superintegron region of the NRT36S genome (described below).
The colonization factor(s) in NRT36S was not identified in previous studies. We found four pilus systems in NRT36S. First, a type 1 pilus assembly system was identified to be encoded by an 8-kb sequence specific to the genome of NRT36S. Second, a pilus system in NRT36S related to a mating pilus system in Salmonella enterica serovar Typhi (2) was identified. Third, a type IV pilus system, which was a mannose-sensitive hemagglutinin described by Jonson et al. (9), was present in both NRT36S and N16961. The protein sequences of the mannose-sensitive hemagglutinin systems shared 60 to 99% identity between the two isolates. Fourth, another type IV pilus system, which was described previously (5), was also conserved in NRT36S and N16961. None of the pilus systems identified in NRT36S was similar to TCP, which is a type IV pilus.
We also identified a type III secretion system in a 48-kb gene cluster specific to NRT36S. It is highly similar to the one described for another non-O1/non-O139 V. cholerae isolate, AM-19226 (4), sharing 99% sequence similarity. The location of the type III secretion system in NRT36S was next to the homolog of VC1758, while in N16961, the genes next to VC1758 were designated as Vibrio pathogenic island 2. In NRT36S, we also found two additional potential toxin gene clusters that were not found in N16961. The first was an exotoxin A precursor gene. This gene had homologs in the non-O1/non-O139 V. cholerae isolates AM-19226 (4) and V51 (http://www.ncbi.nlm.nih.gov/). The predicted protein product is related to an NAD-dependent ADP-ribosyltransferase of Pseudomonas aeruginosa. Second, there were two RTX toxin genes present in NRT36S. The first one was similar to rtxA of N16961, sharing 99% amino acid identity (AAI). The second RTX toxin gene was similar to the RTX toxin gene in Aeromonas salmonicida (E value = 0; score = 640) and was very divergent from the rtxA gene found in N16961.
We identified two additional phage-like gene clusters in NRT36S. One prophage-like gene cluster was 33 kb long and was located adjacent to genes which are on chromosome II in N16961. The other cluster was 6 kb long and showed similarity to the filamentous bacteriophages KSF-1φ and VGJφ. Whether these two prophages play a role in virulence remains unknown. Altogether, the putative virulence genes and phage sequences made up 33% of the strain-specific sequences in NRT36S.
Besides the genes related to toxin and colonization, V. cholerae N16961 and NRT36S differed in the genes associated with the synthesis of surface polysaccharide, which might also contribute to their virulence and survival. These genes made up 9% and 13% of the strain-specific sequences in N16961 and NRT36S, respectively. The genes for O and K antigens in the two isolates were entirely different. N16961 is a serogroup O1 isolate and has no capsule; its O antigen biogenesis region has been identified (14), occurring between gmhD and rjg in chromosome II. NRT36S is a serogroup O31 isolate and has a capsule in addition to the O31 antigen. The O antigen and capsule shared the same genetic locus in NRT36S, also located between gmhD and rjg (3a). Despite the similar locations, the gene contents of this locus in N16961 and NRT36S were very different.
A superintegron exists in all Vibrio genomes examined so far (3, 6, 13), and NRT36S was no exception. The superintegron is a highly variable region. Our sequencing strategy generated only short reads (an average length of 96.5 bp) that could not read through the 128-bp Vibrio cholerae repeat and caused problems when assembling the superintegron of NRT36S. Therefore, our knowledge of the superintegron region is not complete. Among the genes we could confirm to be in the superintegron region, most of them encoded hypothetical proteins, as expected. Several genes were homologous to the recognized genes in the N16961 superintegron: genes for a killer protein, four putative acetyltransferases, and some hypothetical proteins were conserved in the superintegrons of both NRT36S and N16961. There were also several other genes recognized to be specific to the NRT36S superintegron. As mentioned above, the superintegron encoded NAG-ST, which may be the major toxin of NRT36S. We also identified a quinone oxidoreductase gene and a hydrolase gene in the NRT36S superintegron.
In addition to the genes for toxins, pili, and surface polysaccharide and those in the superintegron, the differences between N16961 and NRT36S extended to other genes in other functional categories, such as chemotaxis, transportation and metabolism, and transcriptional regulation. These genes made up 32% and 24% of the strain-specific sequences in N16961 and NRT36S, respectively.
To gain insight into the variations between homologous genes in V. cholerae, we also compared the proteomes of the two isolates by BLASTP (settings for BLASTP, no filter and E values of <1e−10). All best-hit pairs were identified for the two genomes. “Best-hit pair” was defined as a pair in which one gene in one genome found the other in the other genome as its best match and vice versa. Genes with 85% or more AAI that covered at least 70% of the full length were considered conserved. Eighty-four percent of the genes were conserved between the two genomes, <1% of the genes had low AAI (between 21% and 84%), another 1% of the genes represented paralogs in the genome, and the other 14% of genes from NRT36S did not match those in N16961. Genes that had <85% AAI were considered strain specific.
Our genome analysis revealed extensive variation among pathogenic strains of V. cholerae. Sixteen percent of genes were not conserved between the two genomes, suggesting the occurrence of lateral gene transfer (with the presence of prophages raising the possibility of phage-mediated gene transfer). NRT36S and the O1 strain N169061 clearly have entirely different sets of virulence-associated genes. The observed variations in their surface polysaccharides and membrane transportation systems may reflect adaptation to different niches during their life cycles. The function of the superintegron remains cryptic. The NAG-ST gene is the first functional virulence gene found in the superintegron, which strongly suggests that the superintegron is a mechanism by which members of this species can import exogenous genes and convert them for their own use.
Acknowledgments
We thank Ross Overbeek and Michael Fonstein of the National Microbial Pathogen Data Resource for their kindly help in the annotation of the V. cholerae NRT36S genome and Lutz Krause of the University of Bielefeld, Germany, for allowing us to use the program GISMO (11) in the initial gene calling. We also thank Shiladitya DasSarma and Beenish Bhatia (Center of Marine Biotechnology, Baltimore, MD) for their help in the initial analysis of the genome.
Editor: A. Camilli
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Arita, M., T. Takeda, T. Honda, and T. Miwatani. 1986. Purification and characterization of Vibrio cholerae non-O1 heat-stable enterotoxin. Infect. Immun. 52:45-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Chen, Y., P. Bystricky, J. Adeyeye, P. Panigrahi, A. Ali, J. A. Johnson, C. A. Bush, J. G. Morris, Jr., and O. C. Stine. 2007. The capsule biogenesis genes are embedded in the LPS region in non-O1 Vibrio cholerae NRT36S. BMC Microbiol. 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dziejman, M., D. Serruto, V. C. Tam, D. Sturtevant, P. Diraphat, S. M. Faruque, M. H. Rahman, J. F. Heidelberg, J. Decker, L. Li, K. T. Montgomery, G. Grills, R. Kucherlapati, and J. J. Mekalanos. 2005. Genomic characterization of non-O1, non-O139 Vibrio cholerae reveals genes for a type III secretion system. Proc. Natl. Acad. Sci. USA 102:3465-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fullner, K. J., and J. J. Mekalanos. 1999. Genetic characterization of a new type IV-A pilus gene cluster found in both classical and El Tor biotypes of Vibrio cholerae. Infect. Immun. 67:1393-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5a.Galen, J. E., J. M. Ketley, A. Fasano, S. H. Richardson, S. S. Wasserman, and J. B. Kaper. 1992. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect. Immun. 60:406-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herrington, D. A., R. H. Hall, G. Losonsky, J. J. Mekalanos, R. K. Taylor, and M. M. Levine. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J. Exp. Med. 168:1487-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jermyn, W. S., and E. F. Boyd. 2002. Characterization of a novel Vibrio pathogenicity island (VPI-2) encoding neuraminidase (nanH) among toxigenic Vibrio cholerae isolates. Microbiology 148:3681-3693. [DOI] [PubMed] [Google Scholar]
- 9.Jonson, G., J. Holmgren, and A. M. Svennerholm. 1991. Identification of a mannose-binding pilus on Vibrio cholerae El Tor. Microb. Pathog. 11:433-441. [DOI] [PubMed] [Google Scholar]
- 10.Karaolis, D. K., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause, L., A. C. McHardy, A. Puhler, J. Stoye, and F. Meyer. 2007. GISMO—gene identification using a support vector machine for ORF classification. Nucleic Acids Res. 35:540-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurtz, S., A. Phillippy, A. L. Delcher, M. Smoot, M. Shumway, C. Antonescu, and S. L. Salzberg. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5:R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V. cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 14.Manning, P. A., M. W. Heuzenroeder, J. Yeadon, D. I. Leavesley, P. R. Reeves, and D. Rowley. 1986. Molecular cloning and expression in Escherichia coli K-12 of the O antigens of the Inaba and Ogawa serotypes of the Vibrio cholerae O1 lipopolysaccharides and their potential for vaccine development. Infect. Immun. 53:272-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris, J. G., Jr., T. Takeda, B. D. Tall, G. A. Losonsky, S. K. Bhattacharya, B. D. Forrest, B. A. Kay, and M. Nishibuchi. 1990. Experimental non-O group 1 Vibrio cholerae gastroenteritis in humans. J. Clin. Investig. 85:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogawa, A., J. Kato, H. Watanabe, B. G. Nair, and T. Takeda. 1990. Cloning and nucleotide sequence of a heat-stable enterotoxin gene from Vibrio cholerae non-O1 isolated from a patient with traveler's diarrhea. Infect. Immun. 58:3325-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stine, O. C., S. Sozhamannan, Q. Gou, S. Zheng, J. G. Morris, Jr., and J. A. Johnson. 2000. Phylogeny of Vibrio cholerae based on recA sequence. Infect. Immun. 68:7180-7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]