Abstract
Only a minority of the genes, identified in the Caenorhabditis elegans genome sequence data by computer analysis, have been characterized experimentally. We attempted to determine the expression patterns for a random sample of the annotated genes using reporter gene fusions. A low success rate was obtained for evolutionarily recently duplicated genes. Analysis of the data suggests that this is not due to conditional or low-level expression. The remaining explanation is that most of the annotated genes in the recently duplicated category are pseudogenes, a proportion corresponding to 20% of all of the annotated C. elegans genes. Further support for this surprisingly high figure was sought by comparing sequences for families of recently duplicated C. elegans genes. Although only a preliminary analysis, clear evidence for a gene having been recently inactivated by genetic drift was found for many genes in the recently duplicated category. At least 4% of the annotated C. elegans genes can be recognized as pseudogenes simply from closer inspection of the sequence data. Lessons learned in identifying pseudogenes in C. elegans could be of value in the annotation of the genomes of other species where, although there may be fewer pseudogenes, they may be harder to detect.
[Online supplementary material available at www.genome.org.]
The complete genome sequence with its annotation, for the nematode worm Caenorhabditis elegans (C. elegans Consortium 1998), is a considerable resource with which to investigate biology. The most recent estimates predict, on the basis of sequence data, that this worm's genome contains 18,959 protein-coding genes (http://www.wormbase.org), although only ∼4000 of these have been genetically or biochemically characterized, despite the intense study of this experimentally highly tractable system. A primary aim of C. elegans research is to understand how the genome, via the developmental program, generates the animal, but, as yet, there is little functional knowledge for the vast majority of genes predicted in the genome.
This laboratory has been determining gene expression patterns for the annotated C. elegans genes using reporter gene fusion technology as one approach through which to explore genome function (Hope 1991; Young and Hope 1993; Lynch et al. 1995; Hope et al. 1998). Although there are caveats in using this technology (e.g., the need for caution in assuming that an expression pattern observed is an accurate reflection of the expression of the endogenous gene), this approach has the advantage that an expression pattern is linked absolutely to an annotated gene in the genome. In our current strategy, the annotated C. elegans genes are effectively assayed at random, sampling the genome annotation. While analyzing our data, we have noticed that genes duplicated relatively recently in C. elegans' evolution are much less likely to drive reporter gene expression. One interpretation implies that many of the predicted genes, possibly a fifth of the genes in the annotated genome, are nonfunctional pseudogenes.
RESULTS
We have examined the expression of 364 of the annotated C. elegans genes using our current reporter gene fusion approach. After shotgun cloning of 5–7-kb genomic DNA restriction fragments into lacZ or GFP reporter gene expression vectors (Fire et al. 1990), the fusion junction was sequenced for randomly selected clones. Plasmids with a C. elegans gene to reporter gene translational fusion that would be appropriate for expression analysis, according to the genome annotation in ACeDB/WormBase, were thereby identified. The point of fusion for any particular gene was random and could therefore be at any position within the predicted protein-coding region. The translational reading frame was corrected when fusions were to an exon, in the appropriate orientation, but in the wrong reading frame. Expression of the reporter was examined in situ in worm strains generated by transformation with the identified plasmids. The expression pattern data generated are presented on our laboratory web site (http://bgypc086.leeds.ac.uk) and in the C. elegans database WormBase/ACeDB (http://www.wormbase.org).
Of the 364 effectively randomly selected annotated genes examined, 186 (51%) failed to drive reporter gene expression to observable levels. There are a number of potential reasons for lack of reporter gene expression. The endogenous gene may be expressed at very low levels or only under specific environmental conditions. Transgene expression in the germ line can be suppressed (Kelly et al. 1997). Approximately one quarter of C. elegans genes are organized into polycistronic units (Zorio et al. 1994), and no attempt was made to avoid such operons in this study, because operons are difficult to predict on the basis of sequence data alone. A reporter gene fusion to a gene that is downstream in an operon may not contain distant upstream promoter elements, and may therefore fail to show expression. Finally, the structure of many of the annotated genes is based primarily on predictions by the computer program Genefinder. It is thought that 5.45% of all exons may be mispredicted in a way that a correct translational fusion would not be formed within the reporter gene fusions assayed (Reboul et al. 2001), and splitting of one gene into two predicted genes can mean an assayed fragment would not contain the necessary promoter elements.
While analyzing our data, we noted a remarkable correlation that addresses this issue of why a C. elegans gene's promoter region may fail to drive reporter expression. The proteins encoded by the 364 genes analyzed for reporter gene expression were classified as unique, duplicated, or conserved on the basis of BLAST comparisons with the C. elegans, Drosophila melanogaster, Saccharomyces cerevisiae, and human genomes. Either BLAST scores were extracted from the Proteome database (Costanzo et al. 2001) or generated using the BLASTP algorithm applied to the NCBI nonredundant database (http://www.ncbi.nlm.nih.gov). Genes were considered homologous if a BLAST score E value was less than or equal to 10−6. By this criterion, genes that have no homolog were classified as unique. For the genes that do have homologs, those for which there is only a C. elegans homolog or the C. elegans homolog is a markedly better match than the best non-C. elegans homolog were classified as duplicated. [The criterion used was that the −log10 (E value of the C. elegans homolog) was more than twice the −log10 (E value of the best other organism homolog)]. The remaining genes, which have close homologs in other species, were simply classified as conserved.
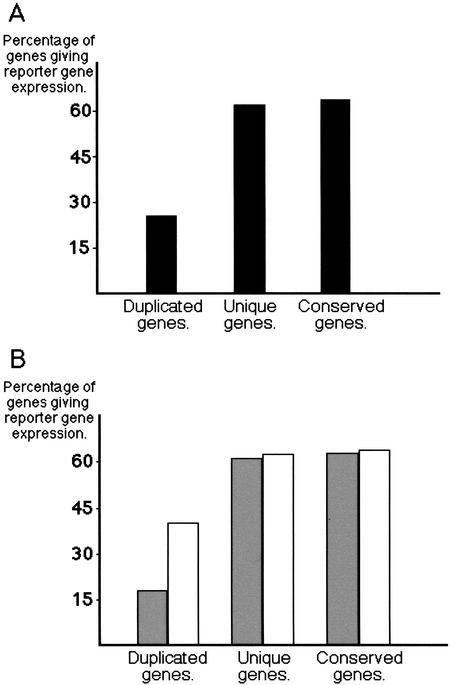
Duplicated genes were far less likely to drive reporter gene expression than genes in the unique or conserved categories. Whereas 62% (36 of 58) of the unique genes, and 64% (104 of 162) of the conserved genes gave reporter gene expression, only 26% (38 of 144) of the duplicated genes did so (Fig. 1A).
Figure 1.
Percentage of genes that gave reporter gene expression. Genes are classified as duplicated, unique, or conserved as explained in the text. (A) All genes examined. (B) Genes examined divided between those with ESTs (open bars), and those without (shaded bars). A list of the genes examined, divided into these various categories, is available as supplementary data.
A number of possible explanations can be identified for why a smaller proportion of the genes, which have undergone relatively recent duplication within C. elegans evolution, give reporter gene expression as compared with nonduplicated genes. Duplicated genes may be more likely to be expressed to lower levels or only upon environmental induction. Alternatively, the annotation for a large proportion of the duplicated genes may be incorrect, either with regard to intron/exon structure or because they are, in fact, pseudogenes.
Low or conditional expression could, at best, only partially explain our observations, according to the following considerations. Genes expressed to very low levels or only under specific environmental conditions are less likely to have associated ESTs/cDNAs. According to ACeDB, 207 of the 364 genes examined have identifiable ESTs/cDNAs (with at least 95% nucleotide identity between the cDNA and gDNA sequence), and the proportion is lower for the duplicated genes: 60% (35 of 58) of the unique genes, 37% (53 of 144) of the duplicated genes, and 73% (119 of 162) of the conserved genes have ESTs. Nevertheless, possession of an EST/cDNA only makes it very slightly more likely that a unique or conserved gene will give reporter gene expression (Fig. 1B). Therefore, level of expression does not appear to correlate with ability to drive reporter gene expression for genes in these categories. Furthermore, whereas possession of an EST/cDNA increases the likelihood of obtaining reporter gene expression for a duplicated gene, the probability of obtaining expression still does not reach that for the other gene categories. The proportion of genes with ESTs that are able to drive reporter gene expression is 63% for unique genes (22 of 35), 40% for duplicated genes (21 of 53), and 65% for conserved genes (77 of 119) (Fig. 1B). An explanation for why duplicated genes with ESTs are less likely to give reporter gene expression than unique or conserved genes with or without ESTs and why duplicated genes without an EST are even less likely to give reporter gene expression is still wanting.
The remaining explanation for our observations is that a significant proportion of the duplicated genes are really pseudogenes or have an incorrect intron/exon structure prediction, both being errors in the C. elegans genome annotation. Gene structure predictions might be expected to be least reliable for the unique genes, rather than the duplicated genes, because they lack homology that can be used to guide exon identification. The key difference between the duplicated genes and the unique or conserved genes is that duplicated genes have an identifiable paralog that must have arisen through a genetic duplication that occurred since the evolutionary split with D. melanogaster. Only rarely will a duplicated gene acquire a new essential function and become fixed within a population. For most duplicated genes, one copy is expected to accumulate mutations and become a nonfunctional pseudogene (Darnell et al. 1990). Depending upon the order in which mutations accumulate over evolutionary time, a pseudogene may still be transcribed and, therefore, even annotated genes with ESTs may be pseudogenes, as implied in our data. Pseudogenes will continue to drift until they are either deleted or become unrecognizable as a genetic copy. The number of pseudogenes in a genome will depend on the relative rates of gene duplication and pseudogene loss.
If the low frequency of reporter gene expression for the duplicated genes was mainly due to the presence of pseudogenes, then 20% of the annotated genes in the C. elegans genome would be pseudogenes. The basis of this claim is as follows. Given that 64% of the unique and conserved genes gave reporter gene expression, then, for 38 genes in the duplicated category to give reporter gene expression as observed, 59 of the 144 duplicated genes would need to be real genes, leaving 85 as pseudogenes. No conclusions can be drawn as to the number of pseudogenes in the unique or conserved categories, but for the purpose of this calculation, there are assumed to be none. If 85 of the 364 genes examined are pseudogenes, then >4000 of the 18,959 annotated genes in the C. elegans genome would be predicted to be pseudogenes. A similar, but less reliable extrapolation, based only on the results for genes with ESTs, suggests that approximately one-quarter of these pseudogenes are transcribed. Of course, some predicted genes that have given reporter gene expression could also be pseudogenes.
Such a large number of pseudogenes in the C. elegans genome is not inconsistent with the observations of other investigators. Biochemical or genetic evidence concerning function has been generated for only a minority of the 18,959 predicted C. elegans genes (http://www.wormbase.org). Extensive analysis of clones from cDNA libraries have identified ESTs for just 10,000 genes (Maeda et al. 2001), and microarray analysis has been able to detect transcripts for only 56%–59% of C. elegans genes (Hill et al. 2000; Reinke et al. 2000). Transcripts could be specifically amplified by PCR, from a cDNA library, for 84% of predicted C. elegans genes (Reboul et al. 2001), a percentage consistent with our estimates of the numbers of transcribed and nontranscribed pseudogenes in the C. elegans genome. There are already 543 predicted pseudogenes identified in ACeDB/Wormbase. In bioinformatic analysis of specific large gene families in C. elegans, such as chemoreceptor genes (Robertson. 2000), an even higher percentage of pseudogenes has been identified. Bioinformatic analysis found 2168 genomic sequences, which do not overlap with annotated pseudogenes or genes, but nevertheless have homology to known or predicted C. elegans exons, and the presence of stop codons or frameshift mutations suggest that these are pseudogenic (Harrison et al. 2001). Finally, on the basis of a number of close paralogs, it has been proposed that C. elegans has a very high rate of gene duplication, generating 383 duplicated genes every million years, as compared with 31 and 52 for D. melanogaster and S. cerevisiae, respectively (Lynch and Conery 2000). However, we cannot totally rule out the possibility that another peculiar and unrecognized property of the recently duplicated genes, such as use of more distant promoter elements or distinct splicing mechanisms, is causing the differential rates of success in our reporter gene fusion experiments.
Proving that a gene unit is totally nonfunctional, and is therefore definitely a pseudogene, is impossible. Nevertheless, a search of the sequence data was undertaken for obvious evidence that might suggest that some of the annotated genes we had assayed were likely to be nonfunctional. It was anticipated that stop codons, translational frameshifts, or deletions in otherwise conserved protein-coding regions may have been avoided in the gene structure predictions.
The sequences of the 74 annotated genes in the duplicated category that had no EST, and which failed to give reporter gene expression, were examined. Three-quarters of these annotated genes would need to be pseudogenes if our interpretations are correct. The predicted amino acid sequence was used in a BLAST search of C. elegans WormPep, and the closest homologs were aligned using CLUSTALX (Jeanmougin et al. 1998). These alignments and the alignments presented within ACeDB by BLIXEM (Sonnhammer and Durbin 1996) were simply visually inspected, because only clear examples of pseudogenes were sought. Annotated genes would need to show extensive sequence identity if faults were to be apparent in this preliminary analysis, and for 26, the homology wasn't good enough to draw any firm conclusions. For two annotated genes, the point of reporter gene fusion lay in potentially mispredicted coding regions, nonhomologous regions that are probably in introns. For 33 annotated genes, the coding region did appear to be intact. However, the integrity of the promoter region would not be assessed in this analysis and for three of these (F14H8.4, K02E2.3, F14H8.4), either another gene or repetitive DNA was located very close upstream of the initiation codon, suggesting that the promoter region may not be intact. Despite the cursory nature of this analysis, potential faults were identified for 13 of these 74 annotated genes (Table 1).
Table 1.
Annotated Genes that Appear to be Pseudogenes From Inspection of the Sequence, Listed According to the Fault Found
| Annotated genes with deletions | Annotated genes with frameshifts | Annotated genes with stop codons |
|---|---|---|
| F10D2.8 | B0281.4 | E02C12.7 |
| F19G12.2 | F56D6.1 | F09D12.2 |
| F22E5.13 | T05A1.4 | F56D6.1 |
| F46A8.9 | Y73C8C.10 | |
| F56D5.9 | ||
| F56D6.1 | ||
| T07D3.1 | ||
| Y73C8C.10 | ||
| ZK666.10 |
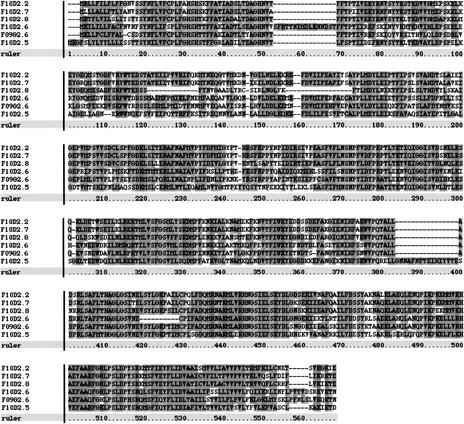
For nine of the annotated genes examined (e.g., F10D2.8; Fig. 2), coding region for well-conserved amino acid residues appears to have been deleted. Gene structure predictions across these incongruities often either incorporate nonhomologous, presumably intronic DNA in the coding region, and/or designate homologous, presumably coding DNA as intron to maintain the integrity of the coding region. One annotated gene, F19G12.2, appears to consist of two pseudogenes joined together. The downstream pseudogene is homologous to the ribonuclease-diphosphate reductase encoding gene, C03C10.3, and has a coding-region deletion. The remaining upstream unit is then a single predicted exon, a rare structure for a real C. elegans gene, with multiple deleted homologs scattered around the genome.
Figure 2.
CLUSTALX alignment of the predicted amino acid sequences for F10D2.2, F10D2.5, F10D2.6, F10D2.7, F10D2.8, and F09G2.6. Each of these genes appears to encode UDP-glucuronosyl transferase. The amino acids in the gap in the alignment for F10D2.8 (positions 120–174) are encoded by an extension of the predicted exon 2 into nonhomologous, presumably intronic DNA. No vestiges of the missing coding region can be seen in this 175-bp second intron. The deletion of this conserved region suggests that F10D2.8 is a pseudogene. Apparent deletions or insertions for the other sequences in the alignment can be accounted for by errors in gene structure prediction, suggesting that the other annotated genes in this cluster have intact coding regions. Extra, nonhomologous amino acids predicted at the amino terminus for F10D2.2 and F09G2.6 have been removed for this alignment.
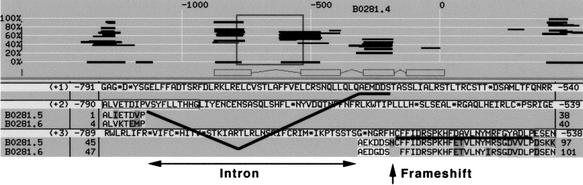
Whereas coding region deletions seem unlikely to be a consequence of simple errors in the sequence data, apparent frameshifting alterations or stop codons could be. Apparent frameshifting alterations were found in four of the annotated genes examined (e.g., B0281.4; Fig. 3), two being associated with coding-region deletions. Three of the annotated genes examined had stop codons within well-conserved protein-coding regions with one of these, F56D6.1, also showing a coding-region deletion and a frameshifting alteration.
Figure 3.
An apparent frameshift in the annotated gene B0281.4. This image was generated from a BLIXEM window of ACeDB. The top of the figure shows the predicted gene structure for B0281.4, and each horizontal line represents a homology. The ruler is marked in base pairs. The region within the box is expanded to give the lower part of the figure. The theoretical translation across this window is given in the three reading frames, (+1), (+2), and (+3). The amino acid sequence encoded at the end of the predicted first exon, ALVETDIPVSYFLLTHHG, lies in the second reading frame, whereas the amino acid sequence encoded at the start of the predicted second exon, ESEN, lies in the third reading frame. The homologous sequence for the other two genes in this tandem cluster, B0281.5 and B0281.6, is retained from the BLIXEM window, whereas additional homologous sequences have been removed for clarity. The homology extends from the second exon into the predicted first intron in the third reading frame (underlined). The homology continues with the sequences AEKDDS for B0281.5 and AEDGDS for B0281.6 (these sequences have been added to the BLIXEM window), but this homology has shifted to the first reading frame (underlined). A splice to a position inside the predicted first exon would then remove nonhomologous residues of the predicted B0281.4 protein, yielding a full coding-region match between B0281.4, B0281.5, and B0281.6, and other homologs.
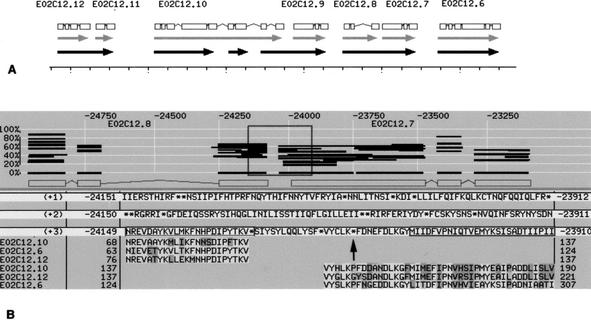
The stop codon found for E02C12.7 is an interesting example. E02C12.7 is one predicted gene in a cluster of tandem duplications. E02C12.6, E02C12.7, E02C12.8, E02C12.9, E02C12.10, E02C12.11, and E02C12.12 show strong homology with each other and with an unlinked predicted gene, F56A4.5. Realigning the predicted protein sequences (Fig. 4) revealed that E02C12.8 forms a gene unit with E02C12.7, as does E02C12.12 with E02C12.11, and the end of E02C12.10 with E02C12.9. The protein-coding regions are interrupted by stop codons at a different position in each case, and these separate the previously predicted gene units. Whereas the start of E02C12.10 appears to be an intact gene unit, like E02C12.6, the middle of E02C12.10 is another truncated copy.
Figure 4.
A stop codon in the annotated gene E02C12.7. (A) A representation, derived from ACeDB, of the tandem gene cluster containing E02C12.6, E02C12.7, E02C12.8, E02C12.9, E02C12.10, E02C12.11, and E02C12.12, which shows homology to a putative choline kinase. The grey arrows indicate the extent of the annotated genes, with the gene structure predictions depicted at top. The black arrows represent the gene units after realignment of the coding sequences as described in the text. The ruler is marked in 500 base pair units. (B) A BLIXEM window from ACeDB with E02C12.8 and E02C12.7 depicted in the top half of the window. Each horizontal line is a homology and the region within the box, covering the gap between the two annotated genes, is expanded to give the lower part of the figure. The theoretical translation across this window is given in the three reading frames, (+1), (+2), and (+3), although only the third reading frame is relevant here. The homologous sequence for three other genes in this tandem cluster, E02C12.6, E02C12.10, and E02C12.12 is retained from the BLIXEM window, whereas other homologous sequences have been removed for clarity. The sequence homology extends upstream from the predicted start of E02C12.7 (MIIDFVPNIQ…) into the predicted intergenic region. A small intron would then link from the homology of E02C12.8 to that of E02C12.7, matching the coding regions of E02C12.6, E02C12.10, E02C12.12, and others, seamlessly. This perfect alignment only fails because of the stop codon (arrow) in the sequence VYCLK*FDNE, which led to the prediction of two gene units. In fact, E02C12.8 and E02C12.7 appear to form a single pseudogene.
These apparently faulty genes could still be functional units having acquired novel function or mode of expression since their duplication, they could appear faulty because of errors in the sequence data, or they are fully defective genes, that is, pseudogenes. The final explanation seems the most likely. Although apparent faults were found in only 13 of the 74 annotated genes examined, a smaller proportion than expected to be pseudogenes from our interpretation of the reporter gene fusion results, the mode of analysis means that this must be an underestimate. Effectively, only annotated genes with close homologs in the C. elegans genome were assessed, and it may be easier to generate potential gene structures for more diverged gene units with lower sequence homology (26 of the 74). Furthermore, the examination was only cursory. Nevertheless, there is obvious sequence evidence suggesting that at least 13 of the 364 annotated genes randomly assayed using reporter gene technology are faulty and, by implication, that at least 4% of the annotated genes in the C. elegans genome are pseudogenes.
DISCUSSION
Pseudogenes may be difficult to distinguish from functional genes by sequence analysis alone or even when combined with experimental analysis. The predominant fate of duplicated genes will be to accumulate mutations that render them nonfunctional pseudogenes. Premature stop codons and frameshifting mutations are the most obvious defining characteristics of a pseudogene, but gene structure prediction programs may find alternative splicing patterns around such obstacles, particularly if there is no good homology with a functionally well-characterized gene or EST data to act as a guide. Genes that have been disabled by damaged splice sites or promoters will be even harder to recognize as pseudogenes, and such genes may linger before genetic drift makes them clearly pseudogenes from inspection of the sequence alone. Although it might have been anticipated that the integrity of the protein-coding region of a recently duplicated gene may be more sensitive to genetic drift than the promoter, our results suggest that this may not be the case. The conservation of protein-coding regions beyond that of introns for these recently duplicated genes suggests that these genes were initially functional and subject to evolutionary selection before they became inactivated by genetic drift. These findings raise many questions about the evolution of the C. elegans genome and, more generally, molecular evolution.
We suggest that many of the considerable number of recently duplicated genes in the C. elegans genome, being in fact pseudogenes, explains the low rate of reporter gene expression among recently duplicated genes. This implies that the C. elegans genome contains substantially fewer real genes than current annotation suggests, and that as many as a fifth of the predicted genes are pseudogenes. Other sequenced animal genomes may contain fewer pseudogenes, which could have made this problem easier to detect in C. elegans. Nevertheless, this problem may be present (Schmid and Aquadro 2001), but harder to deal with in other species in which gene structure is even more difficult to predict, and experience gained with C. elegans may guide this aspect of genome annotation.
METHODS
Reporter Gene Fusion Construction and Analysis
Generation of the reporter gene fusions involved standard molecular biology procedures as described previously (Hope 1991; Lynch et al. 1995). Genomic DNA fragments were derived from the standard wild-type strain, Bristol N2. The vectors were modified from pPD21.28 (lacZ), pPD95.67 (gfp), or pPD95.70 (gfp) (Fire et al. 1990; Miller et al. 1999) (http://www.ciwemb.edu/) by insertion of a 31-bp frameshifting cassette between the multiple cloning site and the reporter gene. This cassette allows the reading frame to be corrected simply by digestion with either AscI or NotI, depending on the shift needed and recircularization. Expression of the reporter gene was examined in wild-type N2 C. elegans that had been transformed by microinjection, using the dominant marker gene rol-6(su1006) (Mello et al. 1991) to identify the transgenics. Details of the genomic DNA fragments assayed and expression patterns obtained are presented on our web site (http://bgypc086.leeds.ac.uk) and have been submitted to ACeDB/WormBase. All assayed plasmids are available on request.
Bioinformatic Analysis
All 74 annotated genes in the duplicated category, which had failed to drive reporter gene expression, were analyzed. The predicted protein sequence was extracted from ACeDB/WormBase (http://www.sanger.ac.uk/Projects/C_elegans/) and used in a BLASTP search of WormPep. The 5 to 10 closest homologs of each, the precise number depending on the distribution of E values obtained, were aligned using CLUSTALX (version 1.81; ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/). Obvious defects in what were otherwise excellent alignments were sought by direct inspection. A gene would be investigated further if the predicted protein product lacked several consecutive amino acid residues that were highly conserved across the protein family. The gene structure prediction in the vicinity of the potential defect was examined using BLIXEM (Sonnhammer and Durbin 1996) in a local version of ACeDB. Translations in all three reading frames and homologys identified in ACeDB, both presented in the BLIXEM window, were searched for the missing protein-coding region. Frequently, part of the missing coding region could be found, but had been omitted from the gene structure prediction because a smaller coding-region deletion, a translational reading frame-shift, or a stop codon prevented their inclusion in any potentially functional gene structure.
WEB SITE REFERENCES
http://bgypc086.leeds.ac.uk; The Hope laboratory web pages with descriptions of reporter gene expression patterns.
http://www.ciwemb.edu/; Access to the Fire laboratory web pages with descriptions of the reporter gene plasmid vectors.
http://www.ncbi.nlm.nih.gov; the NCBI (National Centre for Biotechnology Information) home page.
http://www.sanger.ac.uk/Projects/C_elegans/; The C. elegans BLAST server at The Sanger Institute.
http://www.wormbase.org; The C. elegans database WormBase.
Acknowledgments
We thank Dave McCarroll and Pam Gaunt for technical support, The Sanger Centre for nucleic acid sequencing, and David Coates, Alan Coulson, Jonathan Hodgkin, and John Sulston for reading the manuscript. This work depended crucially upon free access to the C. elegans sequence data and the C. elegans database ACeDB/WormBase. This work was supported by a grant from the Medical Research Council and the Leverhulme Trust/Royal Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
NOTE ADDED IN PROOF
Information recently added to WormBase (http://www.wormbase.org), from the transcriptome project and the Caenorhabditis briggsae genomic sequence, appears consistent with the interpretations presented here for F10D2.8 and E02C12.7.
Footnotes
E-MAIL i.a.hope@leeds.ac.uk; FAX (44) 113 343 2835.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr208802. Article published online before print in April 2002.
REFERENCES
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Costanzo MC, Crawford ME, Hirschman JE, Kranz JE, Olsen P, Robertson LS, Skrzypek MS, Braun BR, Hopkins KL, Kondu P, et al. YPD (TM), PombePD (TM) and WormPD (TM): Model organism volumes of the BioKnowledge (TM) Library, an integrated resource for protein information. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J, Lodish H, Baltimore D. Molecular cell biology. New York: Scientific American Books; 1990. [Google Scholar]
- Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–98. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- Harrison PM, Echols N, Gerstein MB. Digging for dead genes: An analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res. 2001;29:818–830. doi: 10.1093/nar/29.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill AA, Hunter CP, Tsung BT, Tucker-Kellog G, Brown EL. Genomic analysis of gene expression in C. elegans. Science. 2000;290:809–813. doi: 10.1126/science.290.5492.809. [DOI] [PubMed] [Google Scholar]
- Hope IA. Promoter trapping in Caenorhabditis elegans. Development. 1991;113:399–408. doi: 10.1242/dev.113.2.399. [DOI] [PubMed] [Google Scholar]
- Hope IA, Arnold JM, McCarroll D, Jun G, Krupa AP, Herbert R. Promoter trapping identifies real genes in C. elegans. Mol Gen Genet. 1998;260:300–308. doi: 10.1007/s004380050899. [DOI] [PubMed] [Google Scholar]
- Jeanmougin F, Thompson JD, Gouy M, Higgins DG, Gibson TJ. Multiple sequence alignment with Clustal X. Trends Biochem Sci. 1998;23:403–405. doi: 10.1016/s0968-0004(98)01285-7. [DOI] [PubMed] [Google Scholar]
- Kelly WG, Xu SQ, Montgomery MK, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch AS, Briggs D, Hope IA. Developmental expression pattern screen for genes predicted in the C. elegans genome sequencing project. Nature Genet. 1995;11:309–313. doi: 10.1038/ng1195-309. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Maeda I, Kohara Y, Yamamoto M, Sugimoto A. Large-scale analysis of gene function in Caenorhabditis elegans by high-throughput RNAi. Curr Biol. 2001;11:171–176. doi: 10.1016/s0960-9822(01)00052-5. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Desai NS, Hardin DC, Piston DW, Patterson GH, Fleenor J, Xu S, Fire A. Two-color GFP expression system for C. elegans. BioTechniques. 1999;26:914–921. doi: 10.2144/99265rr01. [DOI] [PubMed] [Google Scholar]
- Reboul J, Vaglio P, Tzellas N, Thierry-Mieg N, Moore T, Jackson C, Shin-I T, Kohara Y, Thierry-Mieg D, Thierry-Mieg J, et al. Open-reading-frame sequence tags (OSTs) support the existence of at least 17,300 genes in C. elegans. Nat Genet. 2001;27:332–336. doi: 10.1038/85913. [DOI] [PubMed] [Google Scholar]
- Reinke V, Smith HE, Nance J, Wang J, Van Doren C, Begley R, Jones SJM, Davis EB, Scherer S, Ward S, et al. A global profile of germline gene expression in C. elegans. Mol Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Robertson HM. The large srh family of chemoreceptor genes in Caenorhabditis nematodes reveals processes of genome evolution involving large duplications and deletions and intron gains and losses. Genome Res. 2000;10:192–203. doi: 10.1101/gr.10.2.192. [DOI] [PubMed] [Google Scholar]
- Schmid KJ, Aquadro CF. The evolutionary analysis of “orphans”. from the Drosophila genome identifies rapidly diverging and incorrectly annotated genes. Genetics. 2001;159:589–598. doi: 10.1093/genetics/159.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer ELL, Durbin R. A dot-matrix program with dynamic threshold control suited for genomic DNA and protein sequence analysis. Gene. 1996;167:GC1–GC10. doi: 10.1016/0378-1119(95)00714-8. [DOI] [PubMed] [Google Scholar]
- Young JM, Hope IA. Molecular markers of differentiation in Caenorhabditis elegans obtained by promoter trapping. Dev Dyn. 1993;196:124–132. doi: 10.1002/aja.1001960206. [DOI] [PubMed] [Google Scholar]
- Zorio DAR, Cheng NN, Blumenthal T, Spieth J. Operons as a common form of chromosomal organization in C. elegans. Nature. 1994;372:270–272. doi: 10.1038/372270a0. [DOI] [PubMed] [Google Scholar]