Abstract
Clostridium perfringens enterotoxin (CPE) is the etiological agent of the third most common food-borne illness in the United States. The enteropathogenic effects of CPE result from formation of large CPE-containing complexes in eukaryotic cell membranes. Formation of these ∼155- and ∼200-kDa complexes coincides with plasma membrane permeability changes in eukaryotic cells, causing a Ca2+ influx that drives cell death pathways. CPE contains a stretch of amino acids (residues 81 to 106) that alternates markedly in side chain polarity (a pattern shared by the transmembrane domains of the β-barrel pore-forming toxin family). The goal of this study, therefore, was to investigate whether this CPE region is involved in pore formation. Complete deletion of the CPE region from 81 to 106 produced a CPE variant that was noncytotoxic for Caco-2 cells and was unable to form CPE pores. However, this variant maintained the ability to form the ∼155-kDa large complex. This large complex appears to be a prepore present on the plasma membrane surface since it showed greater susceptibility to proteases, increased complex instability, and a higher degree of dissociation from membranes compared to the large complex formed by recombinant CPE. When a D48A mutation was engineered into this prepore-forming CPE variant, the resultant variant was unable to form any prepore ∼155-kDa large complex. Collectively these findings reveal a new step in CPE action, whereby receptor binding is followed by formation of a prepore large complex, which then inserts into membranes to form a pore.
Among the several potent toxins produced by the gram-positive, spore-forming anaerobe Clostridium perfringens is the C. perfringens enterotoxin (CPE). Despite being produced by less than 5% of global type A isolates, CPE remains biomedically relevant due to its role in enteric disease. CPE causes the symptoms of several human gastrointestinal diseases, one of which (C. perfringens type A food poisoning) is currently ranked as the third most prevalent food-borne illness in the United States (23).
CPE is a single polypeptide chain of 319 amino acids with a molecular weight of 35,346 (4). As a pore-forming toxin (12), CPE permeabilizes plasma membranes of sensitive cells to molecules up to 200 Da within a short period of time (18). CPE first binds via protein receptors which are thought to be certain members of the claudin family of proteins involved in tight junction structure and function (6, 15, 37). Under physiologic conditions, the enterotoxin rapidly forms a large CPE-containing complex that is resistant to sodium dodecyl sulfate (SDS) and has an approximate molecular mass of 155 kDa (40). Changes in cellular membrane permeability typically accompany formation of the ∼155-kDa large complex, suggesting that this complex functions as the transmembrane pore (or a subunit thereof). Upon longer CPE treatment, intoxicated cells can also form a second SDS-resistant, CPE-containing large complex with an estimated molecular mass of 200 kDa (31). This second large complex also includes another tight junction protein, named occludin (30). Calcium influx through CPE pores drives cell death via either the apoptotic or oncotic pathways, depending on the CPE dose (1, 2).
Previous studies using CPE variants blocked for one or more CPE functions have provided important insights into the action of this toxin (8, 10, 16, 17, 32). Binding activity has been mapped to the extreme C terminus, based upon the binding ability of recombinant CPE (rCPE) fragments containing C-terminal sequences (8-11) and the binding deficiencies of rCPE fragments lacking 5, 10, or 30 amino acids from the C terminus (17). In contrast, rCPE fragments lacking 36 or 44 amino acids at the N terminus were found to be approximately twofold more cytotoxic than full-length rCPE, indicating that sequences in the N terminus of CPE are mildly inhibitory for cytotoxicity (17). The same study also identified a region between amino acids 45 and 53 of rCPE that is essential for cytotoxicity, since rCPE fragments lacking these 9 amino acids were unable to form a large complex or induce pore formation. A subsequent site-directed mutagenesis study fine mapped this N-terminal CPE cytotoxicity region and demonstrated that the aspartic acid at position 48 is essential for large-complex formation and cytotoxicity (32).
While previous structure-function studies have mainly focused on regions of CPE important for binding and large-complex formation, the region (if any) involved in pore formation/membrane insertion has not yet been investigated. Despite lacking primary sequence homology with other bacterial pore-forming toxins, CPE amino acids 81 to 106 show a marked alternating side chain hydrophobicity pattern (Fig. 1) closely resembling transmembrane domains (TMDs) found in members of the β-barrel pore-forming toxin (β-PFT) family. In this family, one or two β-hairpins from each monomer in a toxin oligomer collectively insert into the membrane to form the phospholipid bilayer-penetrating channel (25, 35, 36). In efforts to begin ascertaining whether amino acids 81 to 106 in CPE might be important for membrane insertion/pore formation, the present study deleted this region in both the wild-type and D48A point mutant rCPE backgrounds. Careful characterization of these variants provided qualitative, as well as temporal, insights into the molecular mechanism of CPE action.
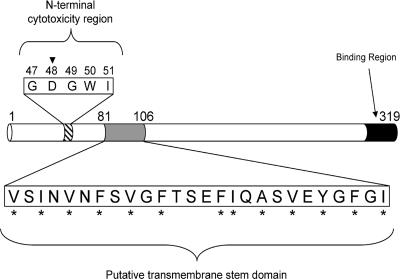
FIG. 1.
Functional map of CPE. Previous analysis of CPE identified two major functional regions (10, 17). Within the 5-amino-acid core cytotoxicity sequence at CPE's N terminus, the aspartic acid residue at position 48 (inverted triangle) is essential for cytotoxicity (32). At the C terminus, binding activity maps to the last 30 amino acids. The putative TMD examined in this study exists between residues 81 to 106 and contains alternating hydrophobic (asterisks) and hydrophilic amino acids.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
All mutagenesis and protein expression work was performed with the XL1-Blue strain of Escherichia coli. The plasmid pJKFLt-1 (17), which served as the template construct for all deletion and site-directed mutagenesis, contains the entire cpe open reading frame ligated into the multiple cloning site of the pTrcHis A vector (Invitrogen, Carlsbad, CA). Fused to the 5′ end of the cpe gene is a vector-encoded, 42-amino-acid hexahistidine tag, which enables the affinity enrichment of rCPE or its derivatives. All plasmids used or generated in this study can be found in Table 1. Bacterial cultures needed for DNA manipulation were grown with Luria-Bertani (solid or liquid) medium containing 100 μg/ml ampicillin. rCPE affinity enrichments were performed with cultures grown in liquid super optimal broth (SOB) media that contained 100 μg/ml ampicillin and 5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) when induction of rCPE expression was required. All cultures were grown at 37°C overnight unless otherwise indicated.
TABLE 1.
Plasmid constructs
| Plasmid | Description | Source or reference |
|---|---|---|
| pTrcHis A | His tag expression vector | Invitrogen |
| pJKFLt-1 | Complete cpe open reading frame inserted into pTrcHis A | 17 |
| pJSTM1 | pJKFLt-1 with codons for amino acids 81-106 of CPE deleted | This study |
| pJSD48A | pJKFLt-1 encoding a D48A point mutation in CPE | 32 |
| pJSTM1-D48A | pJSTM1 encoding a D48A point mutation in CPE | This study |
| pJKW226 | pJKFLt-1 with a W226Stop nonsense mutation resulting in a truncation of the C-terminal 93 amino acids | 16 |
Deletion mutagenesis of rCPE.
Splicing by overlap extension (14) was used to generate the internal deletion variants of CPE in this study. Regions upstream and downstream from each cpe sequence to be deleted were amplified by PCR, with the 3′ upstream reaction primer having complementarity with the 5′ downstream reaction primer. Products from these two PCRs were then used as the template in a third PCR, adding the 5′ upstream reaction primer and 3′ downstream reaction primer after three cycles to allow the amplification of the entire cpe gene (minus the sequence to be deleted). The product from the third PCR was excised from an agarose gel and purified with Freeze n' Squeeze spin columns (Bio-Rad, Hercules, CA). After simultaneous digestion with BamHI and EcoRI, the DNA was again gel purified, as described above, before it was precipitated using 0.1 volume of sodium acetate and 2.0 volumes of cold ethanol for 30 min at −20°C. The DNA was pelleted by centrifugation at 4°C for 30 min and then washed once with 70% ethanol, recentrifuged, and resuspended in the desired volume. This DNA was then ligated into a BamHI- and EcoRI-digested pTrcHis A vector, and this ligation mixture was used to transform XL1-Supercompetent Blue E. coli.
Site-directed mutagenesis.
Introduction of the D48A mutation into the TM1 construct, the rCPE internal deletion variant generated in this study, was performed using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers used in the reaction can be found in Table 2, and reaction parameters were in accordance with the manufacturer's instructions. Mutated plasmid DNA resulting from the site-directed mutagenesis reaction was used to transform XL1-Supercompetent Blue E. coli.
TABLE 2.
Primers used for deletion mutagenesis, site-directed mutagenesis, and sequencing
| Primer name | Sequence (5′-3′) | Purpose |
|---|---|---|
| TM1-A-F | CGCGGATCCGGCGATGTTAATTATAATATGCTTAGTAAC | TM1 deletion |
| TM1-D-R | CCGGAATTCTATATGGAAGGAGAAATTAATGC | TM1 deletion |
| TM1-B-R | TTGTTCTCCTATAGTTTCTTTAGATTTAGTTAATGATTGGCTAAA | TM1 deletion |
| TM1-C-F | ACTAAATCTAAAGAAACTATAGGAGAACAAAATACAATAGAAAG | TM1 deletion |
| D48A-F | GGATTATATGTAATAGATAAAGGAGCTGGTTGGATATTAGGGGAACCC | D48A point mutation |
| D48A-R | CCTAATATACATTATCTATTTCCTCGACCAACCTATAATCCCCTTGGG | D48A point mutation |
| TrcSeq-1F | GGACAGCAAATCGGTCGG | Sequencing |
| 2-F | GGTACCTTAAGCCAATCA | Sequencing |
| PH-310-F | GATTTAGCTGCTACAGAAAGA | Sequencing |
DNA sequencing.
The presence of all mutations in the cpe gene of pJKFLt-1 was confirmed by nucleotide sequencing. Plasmids containing the mutated cpe gene were isolated from overnight E. coli cultures with the QIAGEN Plasmid Mini Kit. Primers used for sequencing can be found in Table 2, and sequencing reactions were performed by the University of Pittsburgh Genomics and Proteomics Core Laboratories. BioEdit DNA analysis software (7) was used to confirm the presence of the desired mutation and the absence of unintended mutations.
Affinity enrichment of rCPE variants.
For use in this study, rCPE (or its derivatives) was affinity enriched (via its vector-encoded N-terminal hexahistidine tag) from lysates of E. coli transformed with the indicated construct (Table 1). One liter of SOB medium was inoculated with 20 ml of an overnight SOB culture of the desired rCPE-expressing E. coli strain. After vigorous shaking for 3 h at 37°C, rCPE expression was induced by adding IPTG to 5 mM. After an additional 4 h of incubation at 37°C, the bacteria were harvested by centrifugation. Final pellets were resuspended in HiTrap binding buffer (20 mM NaH2PO4, 500 mM NaCl, 10 mM imidazole, pH 7.4) and then stored at −80°C overnight. After the bacterial suspension was thawed, a Heat Systems ultrasonic sonicator (Misonix, Farmingdale, NJ) was used to lyse the bacteria. Cell debris was pelleted by centrifugation at 7,500 × g, and supernatant containing the His6-tagged rCPE was passed through a MILLEXHV 0.45-μm polyvinylidene difluoride filter (Millipore, Billerica, MA). This supernatant was then loaded on a cobalt-charged HiTrap chelating affinity column (Amersham Biosciences, Piscataway, NJ). The column was washed with HiTrap binding buffer, and rCPE was then eluted with a linear imidazole gradient from 0 to 100%. Protein-containing fractions from the elution were pooled and dialyzed overnight against sterile deionized H2O, pH 7.0. rCPE harvested from the each affinity enrichment was quantified with quantitative Western blotting, as described previously (32).
Competitive binding assay.
Binding activity of the rCPE variants was determined using a previously described (32) competition assay of 125I-labeled CPE binding to rabbit brush border membranes (BBMs). These membranes were isolated by a previously described protocol (29) from New Zealand White rabbits. This animal work was approved by Animal Care and Use Committee protocol 11594 from the University of California, Davis. To prepare 125I-labeled CPE, 0.5 mg of native CPE was labeled with 2.5 mCi of Na125I (specific activity, 17.0 mCi/mg; MP Biomedicals, Inc., Irvine, CA) using IODO-GEN precoated iodination tubes (Pierce, Rockford, IL). 125I-labeled CPE prepared by the Iodogen method retains significant specific binding and biologic activity (data not shown). As described previously (32), binding competition experiments were carried out by pretreating 100 μg of BBMs with the rCPE variant for 20 min at room temperature (RT) with gentle shaking. 125I-CPE was then added to a final concentration of 2.5 μg/ml, and this mixture was incubated again at RT for 20 min. BBMs were washed twice, and 125I-CPE bound to membranes was quantitated with a Cobra Quantum gamma counter (PerkinElmer). Data from these experiments are expressed as ng of competing rCPE required to inhibit 50% of specifically bound 125I-CPE. The binding-deficient point mutant W226Stop, which was generated in a previous study (16), was used as a negative binding control in these experiments.
Morphological damage assay.
Caco-2 cells were cultured in 100-mm dishes using Eagle minimal essential medium (Sigma, St. Louis, MO) with 10% fetal bovine serum (Mediatech, Herndon, VA), 1% minimal essential medium nonessential amino acids (Sigma), 100 units/ml ampicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine. Cells were kept in 5% atmospheric CO2 at 37°C. Once confluent, Caco-2 monolayers were washed once with prewarmed Hanks' balanced salt solution with Ca2+ and Mg2+ (HBSS2+; Mediatech) and then treated with 2.5 μg/ml of toxin in 2 ml of HBSS2+. Cellular morphological damage (nuclear condensation, cell rounding, detachment from the well bottom) was assayed after 1 h using a Zeiss Axiovert 25 inverted microscope at 10× magnification. A Powershot G5 digital camera (Canon, Inc., Lake Success, NY) and Canon Utilities RemoteCapture software were used to capture images of cells, and subsequent image processing was performed with Photoshop software (Adobe Systems, Inc., San Jose, CA).
86Rubidium release experiments.
Formation of a CPE membrane pore was determined by measuring 86Rb release from Caco-2 cells, as described previously (32). Briefly, confluent Caco-2 cell monolayers were preloaded with 4 μCi/well of 86RbCl (specific activity, 6.4 mCi/mg; PerkinElmer, Boston, MA) for 4 to 5 h at 37°C. After two washes with prewarmed HBSS2+, the radiolabeled cultures were treated with toxin for 15 min at 37°C. 86Rb released into the culture supernatant was collected and quantitated with a Cobra Quantum gamma counter (PerkinElmer). Spontaneous release of 86Rb from cells was measured by incubation of radiolabeled monolayers with HBSS2+ alone, and maximal release was determined by treating monolayers with 1 ml each of 0.5% saponin and 1.0 M citric acid. Data from 86Rb release experiments were converted and plotted as percent maximal release using the following formula: [(experimental release − spontaneous release)/(maximal release − spontaneous release)] × 100.
Large-complex formation.
The ability of each variant rCPE to form SDS-resistant large complexes was measured as described elsewhere (32). Briefly, isolated Caco-2 cells were treated with 2.5 μg/ml of toxin and incubated at 37°C for 20 min with gentle mixing. These cells were then pelleted by microcentrifugation, washed once with Hanks' balanced salt solution without Ca2+ or Mg2+ (Mediatech), and lysed with Laemmli buffer. All samples were treated for 10 min at RT with Benzonase nuclease (Novagen, Inc., Darmstadt, Germany) to degrade cellular DNA before loading on an SDS-containing, 4.25% acrylamide SDS-polyacrylamide gel (PAG). Electrophoresis of the samples was performed at 5 mA for ∼19 h, after which electrotransfer and CPE Western blotting (as described above) were used to detect CPE-containing complexes on the blot.
Pronase resistance of CPE large complex.
Experiments used to analyze the pronase susceptibility of large complexes formed by rCPE or its variants were adapted from a previously published procedure (39). BBMs (100 μg) were treated for 20 min at RT with 1.5 μg of rCPE or 4.5 μg of TM1, in 100 μl of phosphate-buffered saline containing Ca2+ and Mg2+ (PBS2+; Mediatech). The BBMs were then washed once with 100 μl of ice-cold PBS2+, before treatment for 5 or 60 min in 50 μl of PBS2+ containing pronase. BBMs were then washed three times with ice-cold PBS2+ containing the Complete protease inhibitor cocktail (Roche Applied Science, Penzberg, Germany). Final pellets were resuspended in Laemmli buffer without 2-mercaptoethanol, loaded on a 6% acrylamide SDS-PAG, and subsequently treated as a CPE Western blot as detailed above.
Dissociation of CPE large complex from membranes.
rCPE or its variants were added to 100 μg of BBMs at a final concentration of 2.5 μg/ml. Large-complex formation was allowed to occur at 37°C for 20 min, followed by a washing step with phosphate-buffered saline without Ca2+ or Mg2+ (Mediatech) containing the Complete protease inhibitor cocktail at 2× concentration (PBS-PI; Roche). After this wash, toxin-treated BBMs were resuspended in 200 μl of PBS-PI and inverted gently at 4°C for various time periods. At the end of this incubation, the toxin-treated BBMs were pelleted by centrifugation, and both the supernatants and pellets from this spin were separated on a 4% acrylamide SDS-PAG and Western immunoblotted for CPE as described above.
Heat denaturation of CPE large complex.
rCPE or TM1 was added to 100 μg of BBMs for 20 min at 37°C in PBS-PI. After centrifugation and one washing with 200 μl of PBS-PI, BBMs containing the large complex were extracted with Laemmli buffer containing 2-mercaptoethanol. Extraction samples were then heated at 90°C in a heating block for various time periods. After this incubation, samples were separated on a 4% acrylamide SDS-PAG and Western immunoblotted for CPE as described above.
RESULTS
Deletion mutagenesis of the CPE region from 81 to 106.
As mentioned in the introduction, CPE shares several structural similarities with toxins of the β-PFT family, including the presence of a 25-amino-acid region (residues 81 to 106) that strikingly alternates in side chain hydrophobicity (Fig. 1). Because similar regions serve as the TMDs for β-PFTs (25, 35, 36) and since no CPE region involved with membrane penetration has yet been identified, we targeted this putative TMD for deletion mutagenesis. A complete deletion in this CPE region was generated using “splicing by overlap extension” PCR (14), which amplifies sequences upstream and downstream from the intended deletion and splices these two products together in a third reaction (see Materials and Methods for a detailed description). This methodology has also been helpful with other β-PFTs to identify and characterize TMDs (3, 20, 22). TM1 had a 100% deletion of the CPE region from amino acids 81 to 106 (Fig. 2A).
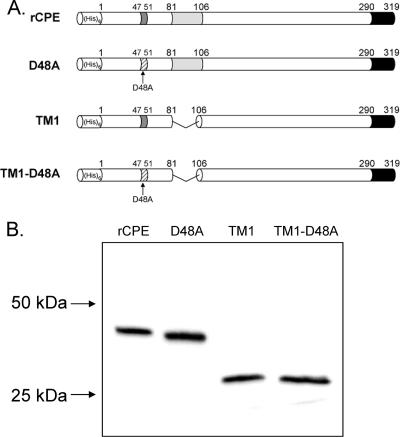
FIG. 2.
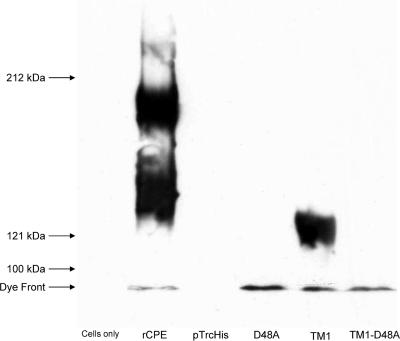
Constructs used in the present study. (A) Each of the four constructs in this study contains an N-terminal His6 for affinity enrichment from E. coli lysates. The D48A construct is a single point mutant generated previously (32). TM1 and TM1-D48A both have an internal deletion of 25 amino acids corresponding to the putative TMD; the TM1-D48A construct also contains the D48A point mutation described above. (B) Affinity-enriched samples (100 ng) of affinity enrichments of these constructs were electrophoresed on an SDS-containing 12% acrylamide gel and Western blotted with rabbit polyclonal anti-CPE antiserum to assess their stability. Arrows to the left of the blot represent the migration of molecular mass standards run with these samples.
To aid in identifying the temporal order of CPE functional regions during cytotoxicity, an additional variant was generated by introducing a point mutation into the TM1 deletion variant background. As mentioned above, D48 of CPE is a critical residue for forming the large complex and cytotoxicity. While the CPE point variant D48A is functional for receptor binding, it does not form the large complex and is unable to alter the plasma membrane permeability of Caco-2 cells (32). Therefore, to determine whether D48 contributes to CPE action before or after the involvement of the putative TMD in CPE, a D48A mutation was introduced into TM1, producing the combination variant TM1-D48A (Fig. 2A).
After DNA sequencing confirmed these mutations, affinity enrichments of rCPE, D48A, TM1, and TM1-D48A were performed, facilitated by the N-terminal His6 tag contributed by the pTrcHis A vector. While substantially enriched for rCPE or rCPE variants, as reported previously (17), these affinity-enriched preparations were not pure. Therefore, similar mock enrichments of pTrcHis A empty vector transformants were also performed and used throughout this study as a negative control. When the enriched samples containing rCPE or its variants were subjected to CPE Western blotting, each variant migrated to its expected molecular weight (Fig. 2B), confirming that these CPE species were stable and not proteolytically degraded.
Cytotoxicity of TM1 and TM1-D48A rCPE variants.
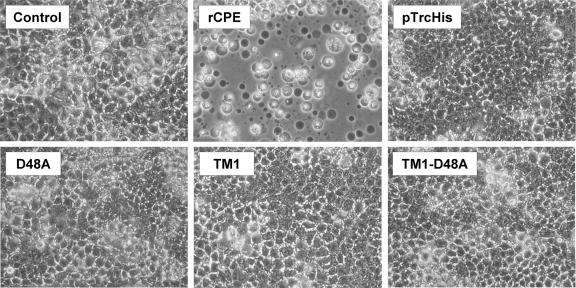
To initially assess whether deletion of the CPE region from 81 to 106 affects the ability of an rCPE variant to cause cytotoxicity, a qualitative Caco-2 morphological damage assay was used. Treatment of Caco-2 cells with 2.5 μg/ml of rCPE for 60 min caused the development of typical CPE cytopathology, including nuclear condensation, membrane blebbing, and cellular detachment from the plate (Fig. 3). No morphological damage was observed if Caco-2 cells were treated with HBSS2+ alone, an affinity-enriched empty vector negative control, or (consistent with previous studies [32]) the D48A point variant. Caco-2 monolayers also showed no observable morphological damage after 60 min (Fig. 3) or 24 h (data not shown) of treatment with 2.5 μg/ml of TM1 or TM1-D48A. These results provided an initial indication that the TM1 and TM1-D48A variants are deficient in some postbinding CPE activity.
FIG. 3.
Caco-2 morphological damage assay. Constructs were initially screened for cytotoxic activity by treating confluent Caco-2 cell monolayers with 2.5 μg/ml of each toxin for 60 min at 37°C. The control panel represents cells treated with HBSS2+ alone, while the pTrcHis panel depicts cells after treatment with a mock enrichment from E. coli transformed with an empty vector. After treatment, Caco-2 cells were photomicrographed at 200× total magnification.
Pore formation by the TM1 and TM1-D48A variants.
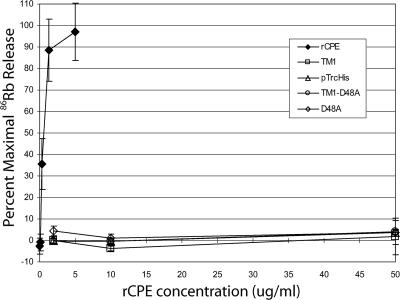
The variants were next assayed for their ability to elicit 86Rb release from Caco-2 cells via formation of a CPE pore. Using a standard quantitative assay (32), rCPE was measured to elicit 50% maximal 86Rb release at a concentration of ∼1.0 μg/ml, while the empty vector negative control and D48A point variant predictably did not elicit any 86Rb release (Fig. 4). When TM1 and TM1-D48A were similarly tested, neither was able to elicit any appreciable 86Rb release, even at artificially high toxin concentrations (up to 50 μg/ml). These results indicate that TM1 and TM1-D48A are both noncytotoxic to cells because they cannot alter the plasma membrane permeability of Caco-2 cells.
FIG. 4.
86Rubidium release from Caco-2 cells. Caco-2 cells grown to confluence in 24-well plates were radiolabeled with 4 μCi/well of 86Rb and, after a washing, treated with the indicated concentration of toxin for 15 min at 37°C. After treatment, 86Rb released into the culture supernatant was collected and gamma radiation was quantitated. Background release was corrected for by treating cells with HBSS2+ alone, and data were converted to percent maximal release (see Materials and Methods). All data points are the statistical means of three independent experiments, with error bars representing the standard deviations.
Assessment of TM1 and TM1-D48A competitive binding.
Since the TM1 and TM1-D48A variants appeared to be deficient at cytotoxicity, it was of interest to determine whether they were able to complete the first step in CPE action, receptor binding. To assess this, a competitive binding assay that measured the ability of the TM1 and TM1-D48A rCPE variants to inhibit binding of 125I-CPE was performed. Although Caco-2 cells are used for cytotoxic evaluations of rCPE or its variants, these cells demonstrate high levels of nonspecific binding of CPE, making them a nonoptimal model for binding analysis. Conversely, while BBMs isolated from rabbits obviously cannot be used to assess cytotoxicity, they are often used to examine CPE binding (16, 17, 19, 21, 31, 32, 40, 41) because they show much greater levels of specific binding versus nonspecific binding than Caco-2 cells.
A binding inhibition index, which represents the amount of competing rCPE species (in ng) required to cause a 50% inhibition of 125I-CPE specific binding to BBMs, was calculated for each construct. The binding-capable D48A variant (32) had a binding inhibition index similar to that of rCPE (240 ± 30 for D48A compared to 330 ± 80 for rCPE), while the C-terminally truncated variant W226Stop, as described previously (16), was incapable of competing with 125I-CPE for BBM binding (index of over 20,000). The affinity-enriched empty vector pTrcHis sample had a similar effect in these experiments, also having a competitive binding index of over 20,000. For the rCPE variants produced in this study, TM1 showed a binding inhibition index similar to that of rCPE (410 ± 120), while TM1-D48A showed only slightly less competitive binding activity, with an index of 650 ± 40. The similarity in competitive binding of TM1, TM1-D48A, and rCPE demonstrates that the introduced mutations do not exert a major effect on receptor binding. Thus, it appears that TM1 and TM1-D48A can complete the first step in CPE action.
Formation of SDS-resistant large complexes by the TM1 and TM1-D48A variants.
The data presented above indicated that, despite maintaining receptor binding activity, TM1 and TM1-D48A are both deficient at inducing cytotoxicity. Since CPE cytotoxicity requires the formation of an ∼155-kDa, SDS-resistant complex in the membranes of sensitive cells (19), we next determined whether TM1 or TM1-D48A can form this complex in mammalian cell membranes. Since Caco-2 cells have been shown to form two SDS-resistant large complexes of ∼155 and ∼200 kDa when treated with CPE (31), these cells were initially used to assess large-complex formation by the TM1 and TM1-D48A variants. As reported previously (32), treatment of isolated Caco-2 cells with 2.5 μg/ml of rCPE resulted in the formation of the ∼155-kDa large complex, as well as a second ∼200-kDa CPE complex (Fig. 5). As also reported previously (32), the D48A rCPE variant did not form either of the two large CPE complexes, although this rCPE variant can bind and is membrane associated (note the immunoreactivity at the dye front in Fig. 5).
FIG. 5.
Formation of SDS-resistant large complexes in Caco-2 cells. To assay for large-complex formation in Caco-2 cells, cultures were harvested from 100-mm plates by gentle scraping and treated with 2.5 μg/ml of toxin for 45 min at 37°C with inversion. After a washing, the treated cells were lysed and Western blotted with rabbit polyclonal anti-CPE antiserum. Lanes labeled “Cells only” and “pTrcHis” represent cells treated with Hanks' balanced salt solution without Ca2+ or Mg2+ alone or a mock enrichment from empty vector transformants, respectively. Arrows to the left of the blot represent the migration of molecular mass standards run with these samples.
Interestingly, the TM1 variant was able to form the ∼155-kDa large complex in Caco-2 cells, despite its complete loss of cytotoxicity depicted in earlier figures. However, the TM1 variant did not form any detectable ∼200-kDa complex in Caco-2 cells. This novel phenotype (formation of the ∼155-kDa large complex in the absence of cytotoxicity) did not extend to the TM1-D48A combination variant. Although this variant was membrane associated, no detectable large-complex formation was observed with this combination variant (Fig. 5).
Analysis of the TM1 prepore large complex.
The ability of the TM1 deletion variant to form the ∼155-kDa complex (Fig. 5) in the complete absence of cytotoxicity (Fig. 3) or pore formation (Fig. 4) led us to investigate whether the TM1 large complex could represent a previously unidentified prepore step in the mechanism of action of CPE. To this end, we examined the integrity of the TM1 large complex in membranes by assaying protease resistance, dissociation from the membrane, and disassembly of membrane-extracted complexes by heat denaturation. BBMs were used in these experiments since the long CPE treatment times and the large amount of proteases would lyse Caco-2 cells and thus cause sample loss. In addition, previous studies have demonstrated the stability of BBMs under similar conditions (39).
Pronase resistance of the TM1 large complex.
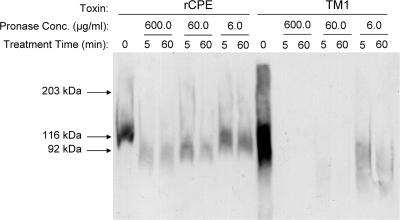
Previous work has shown that, when associated with BBMs, CPE in the ∼155-kDa large complex becomes resistant to pronase, a highly active protease cocktail from Streptomyces griseus (39). This same previous study showed that the ∼155-kDa large complex becomes susceptible to pronase when extracted from membranes. Those findings were interpreted as indicating that the membrane-associated CPE large complex gains pronase resistance because of its insertion into membranes, which is consistent with CPE being a membrane-penetrating, pore-forming toxin.
In order to test the hypothesis that the TM1 large complex might be trapped in a prepore stage unable to insert into BBMs, similar pronase resistance assays were performed with the TM1 large complex. While TM1 was shown in Fig. 5 to form only the ∼155-kDa large complex in Caco-2 cell membranes, it was first necessary to evaluate whether TM1 showed similar CPE complex formation in BBMs. When BBMs were treated with wild-type rCPE (Fig. 6, rCPE lane 0), the ∼155-kDa large complex, but no other complex, was observed. This was expected since the protein required for the formation of the ∼200-kDa large complex, occludin (31), is not present in BBM preparations (U. Singh and B. A. McClane, unpublished observations). As also seen in Caco-2 cells, the TM1 variant did form the ∼155-kDa large complex, but no ∼200-kDa large complex, in BBMs (Fig. 6, TM1 lane 0), thus demonstrating the consistency between the two model systems.
FIG. 6.
Pronase resistance of the CPE large complex. Isolated rabbit BBMs were treated with either rCPE or TM1 for 20 min and then washed to remove unbound toxin. Membranes were treated with the indicated concentrations of pronase and incubated at 4°C for the denoted amounts of time. After digestion, membranes were pelleted and resuspended in Laemmli buffer before separation and Western blotting with rabbit polyclonal anti-CPE antiserum. Arrows to the left of the blot represent the migration of molecular mass standards run with these samples.
In order to assay complex susceptibility to pronase, BBMs containing the ∼155-kDa large complex formed by rCPE were digested with 600, 60, or 6 μg/ml of pronase for 5 or 60 min. Significant amounts of rCPE large complex remained present in BBMs even after a 60-min treatment with 60 μg/ml of pronase (Fig. 6). In contrast, pronase treatment of membranes containing the TM1 large complex caused a nearly complete disappearance of this complex after only a 5-min treatment with 60 μg/ml of pronase. This effect is directly attributable to the addition of pronase since rCPE or TM1 complex levels did not similarly decrease when BBMs were incubated for 3 h in the absence of pronase (Fig. 7).
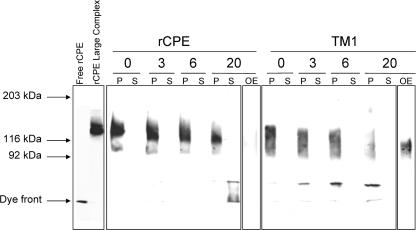
FIG. 7.
Dissociation of the large complex from membranes. BBMs were treated with rCPE or TM1 (2.5 μg/ml) for 20 min at 37°C to allow formation of the ∼155-kDa large complex. After a washing, the BBMs were incubated in PBS-PI at 4°C for the indicated periods of time (numbers at top of gel). After microcentrifugation, samples from both the supernatant and pellet were Western blotted with rabbit polyclonal anti-CPE antiserum. P, samples from the pellet; S, samples from the supernatants of the microcentrifugation; OE, overnight exposures of lanes containing the 20-h supernatant for either rCPE- or TM1-treated BBMs. Arrows to the left of the blot represent the migration of molecular mass standards run with these samples, along with the migration of free rCPE and rCPE-treated BBMs.
Enhancing these findings is the fact that the BBM samples in Fig. 6 were deliberately prepared to contain approximately threefold more TM1 large complex than rCPE large complex at the start of the experiment. Therefore, the ∼155-kDa large complex formed by TM1 is between 10- and 30-fold more sensitive to pronase digestion than is the ∼155-kDa large complex formed by wild-type rCPE. This finding is consistent with the TM1 variant forming the ∼155-kDa large complex but (due to the lack of the putative TMD) with the membrane insertion step required for pore formation and development of pronase resistance being blocked.
Membrane dissociation of the TM1 large complex.
Once sequestered in CPE large complexes, only limited dissociation of CPE from BBMs is observed (39). If the ∼155-kDa large complex made by TM1 remains on the membrane surface trapped in a prepore state, it might be anticipated that this TM1 large complex would exhibit dissociation from BBMs that was faster than dissociation of the large complex formed by rCPE. To test this hypothesis, isolated BBMs containing the ∼155-kDa large complex formed by either rCPE or TM1 were harvested by centrifugation at the indicated (Fig. 7) time intervals and samples from both the supernatants and pellets were Western blotted for CPE. These studies showed that the ∼155-kDa large complex formed by rCPE remained largely membrane associated, even after 20 h (Fig. 7). In contrast, little of the ∼155-kDa large complex formed by TM1 remained membrane associated after a similar 20-h incubation. In addition, a steady kinetic increase in lower-molecular-weight material accompanied the loss of large-complex immunoreactivity in the pellet lanes of the TM1-treated membranes (Fig. 7). This smaller complex may represent an intermediate species from the breakup of the TM1 large complex and is noticeably absent from the pellet lanes of similar experiments with rCPE-treated membranes.
Overnight film exposure of these blots also allowed the visualization of the intact TM1 large complex that had dissociated from the membrane (Fig. 7, TM1 lane OE). Very little (if any) large complex could be detected in similarly overexposed lanes containing rCPE-treated BBMs. Because these reactions were performed at 4°C and constantly in the presence of a strong protease inhibitor cocktail, the TM1 large-complex instability noted in Fig. 7 is unlikely to be the result of proteolytic activity. In support of this conclusion, no differences in total protein content were detected between rCPE-, TM1-, or control-treated BBMs after overnight dissociation experiments (data not shown).
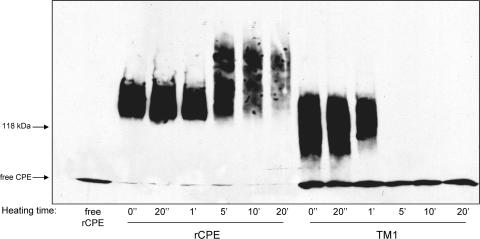
Heat denaturation of the TM1 large complex.
Since the ∼155-kDa large complex formed by TM1 appeared in Fig. 7 experiments to be less stable and more likely to disassemble than that formed by rCPE, a heat denaturation experiment was performed to test the stability of the TM1 large complex after its extraction from membranes. Heating of large complexes made by rCPE prior to SDS-PAG electrophoresis was previously shown to cause very limited complex disassembly, although it can promote complex aggregation (40). Therefore, it was of interest to assess the ability of the TM1 large complex to resist disassembly as a result of heating.
To perform these experiments, the large complex formed by rCPE or TM1 was extracted from BBMs and then heated for 0 s, 20 s, 1 min, 5 min, 10 min, or 20 min. While no decrease in the rCPE large complex could be seen at 1 min of heating, the TM1 large complex began to show obvious breakdown after only 1 min and was undetectable as a complex past this time point (Fig. 8). A qualitative change of the rCPE large complex, in the form of some kind of aggregation, started to appear at the 5-min time point; however, the immunoreactivity remaining as an intact complex was strikingly more heat resistant than that of the TM1 complex, as evident from TM1 lanes after 5 min of heating (Fig. 8). When urea was also included in similar heat denaturation experiments, it had no discernible effect on the stability of the large complex formed by either rCPE or TM1 (data not shown).
FIG. 8.
Heat denaturation of extracted large complexes. Heat sensitivity of the large complexes was assayed by treating BBMs with rCPE or TM1 (2.5 μg/ml) for 20 min at 37°C to allow formation of the ∼155-kDa large complex. After a washing, the BBMs were extracted with Laemmli buffer and heated at 90°C for the indicated time periods. After heat denaturation, samples were Western blotted with rabbit polyclonal anti-CPE antiserum. Arrows to the left of the blot represent the migration of molecular mass standards run with these samples, along with the migration of free rCPE.
DISCUSSION
While previous structure-function analyses of CPE have identified two major functional regions (Fig. 1), a stretch of amino acids between residues 81 and 106 of native CPE was investigated in this study for its potential role in membrane insertion. As mentioned in the introduction, the striking pattern of alternating side group polarity exhibited by these residues resembles that exhibited by the TMDs of the β-PFTs (25, 35, 36) and therefore makes amino acids 81 to 106 in CPE an ideal candidate for a membrane insertion domain. Complete deletion of this putative TMD led to a novel CPE variant phenotype: formation of the ∼155-kDa large complex in the absence of cytotoxicity (Fig. 3 and 5). In addition to not being toxic for Caco-2 cells, the TM1 deletion variant was defective for pore formation (Fig. 4) and appeared not to insert into membranes (Fig. 6). These observations collectively suggest that the TM1 large complex is trapped in a somewhat unstable (Fig. 7 and 8) prepore state on the surfaces of cells, unable to insert into the membrane to form a CPE pore.
Several possible explanations alone or in combination could explain the prepore phenotype of the TM1 deletion variant. First, amino acids 81 to 106 could form a beta hairpin secondary structure that CPE uses to penetrate the phospholipid bilayer, as is the case with members of the β-PFT family (25, 35, 36). Consistent with this explanation, secondary-structure analyses of CPE predict two beta strands between V81 and T92 and again between F95 and I106 (double-prediction method [5], Protein Sequence Analysis server [33, 34, 38], and PROF [27]). As mentioned above, the amino acid hydrophobicity pattern aligns very closely with those of many other β-PFTs, including several other toxins produced by C. perfringens. In addition, when the TMDs from the Staphylococcus aureus alpha hemolysin, Bacillus anthracis protective antigen, and Clostridium septicum alpha-toxin were deleted in previous studies, all resultant variant toxins were unaffected in binding and could form SDS-resistant oligomers but (like TM1) were impaired at membrane insertion/pore formation (3, 20, 22). These phenotypic similarities with the CPE TM1 variant, along with the shared distinctive amino acid pattern, are consistent with amino acids 81 to 106 in CPE acting as a TMD. Efforts are under way to probe the environments of these residues after pore formation using Cys scanning mutagenesis and labeling with environmentally sensitive fluorescent dyes, as performed with several other β-PFTs (20, 24, 28).
Alternatively, amino acids 81 to 106 of CPE could mediate some protein-membrane interactions other than direct membrane penetration. Perfringolysin O (PFO), a cholesterol-dependent cytolysin from C. perfringens in the β-PFT family, contains several small loops at the bottom of its membrane-interacting domain 4 (26). Although residues in these loops were implicated directly in pore formation in early models of PFO action (26), it was later shown that they do not insert deeply into the membrane but instead participate in membrane anchoring of PFO (13). Several hydrophobic residues are located in these small loops of PFO, similar to the many hydrophobic residues found between amino acids 81 and 106 of CPE. Therefore, the large complex formed by the TM1 variant could be deficient in the membrane anchoring necessary for subsequent insertion of CPE into the membrane. Also consistent with this possible explanation is the greater dissociation from BBMs of TM1 versus rCPE shown in Fig. 7.
A final possibility is that this CPE region mediates critical protein-protein interactions between CPE and other proteins, which then allow for membrane insertion/pore formation. Deletion of these residues may have removed crucial contact points between CPE and other proteins in the ∼155-kDa large complex, although it remains unclear whether other proteins are present in this CPE complex. The instability of the TM1 large complex (in addition to greater dissociation) seen in Fig. 7 and 8 could support this explanation, especially given the kinetic increase in a lower-molecular-weight complex on Fig. 7 blots immunoreactive for CPE. These TM1 species might represent pieces of the ∼155-kDa large complex that broke apart due to the loss of contacts normally provided by amino acids 81 to 106. It is also notable in Fig. 7 that the TM1 large complex disassembles while still membrane associated, while the rCPE complex breaks apart only after dissociating from BBMs. This effect is also consistent with the idea that TM1 forms a structurally compromised large complex on the membrane surface, whereas the rCPE complex is remarkably stable since it has undergone insertion into membranes.
Introduction of a D48A mutation into the TM1 background helped assign a temporal order to the cytotoxic contributions of known CPE functional regions. Interestingly, the TM1-D48A combination mutant paralleled the attenuated phenotype of the TM1 variant except that it could not form the ∼155-kDa large complex (Fig. 5). This result indicates that the D48 residue in CPE's N-terminal cytotoxicity domain acts prior to the involvement of amino acids 81 to 106 in CPE. This finding agrees with the idea that CPE first forms an SDS-resistant complex and then uses amino acids 81 to 106 as a TMD or membrane anchor or for some other function to insert and form a pore. It also demonstrates that the N-terminal cytotoxicity domain is not directly involved in membrane penetration or pore formation since the TM1 variant (containing native D48) formed the ∼155-kDa large complex but could not insert into the membrane or form a pore (Fig. 4, 5, and 6).
Another interesting finding from this study was that the TM1 variant formed the ∼155-kDa large complex but not the larger ∼200-kDa complex in Caco-2 cells (Fig. 5). This is particularly intriguing because all CPE variants generated in the past form both or neither of these two complexes in these cells. The ∼200-kDa large complex formed by Caco-2 cells, which has been shown to include the tight junction protein occludin (31), generally forms only upon longer CPE treatments or physical disruption of Caco-2 monolayers (30). However, the large-complex experiments in this study utilized isolated Caco-2 cells, a condition where occludin is exposed to CPE from the beginning of the experiment. In support of this claim, rCPE rapidly formed the ∼200-kDa large complex in the same experiments where TM1 was deficient for formation of this second large complex (Fig. 5). One interpretation from these results is that insertion of the ∼155-kDa large complex into membranes is a required precursor for ∼200-kDa large-complex formation. Another possible explanation for TM1's lack of ∼200-kDa large-complex formation is that amino acids 81 to 106 of CPE may directly or indirectly mediate CPE-occludin interactions. Therefore, deletion of this region could lead to a variant capable only of forming the ∼155-kDa large complex.
Taken together, results from this study have shed new light on both temporal and qualitative aspects of the molecular mechanism of action of CPE. A new model incorporating CPE's functional regions generated from these and previous data is presented in Fig. 9. CPE begins its mechanism of action by binding via its C-terminal residues to the second extracellular loops of certain claudins, which are transmembrane tight junction proteins (6). Next, residues (particularly D48) in the N-terminal cytotoxicity region of CPE directly mediate the formation of the SDS-resistant ∼155-kDa large complex, consisting of other CPE molecules and/or other cellular proteins. This large complex then undergoes a membrane insertion event (dependent on CPE's amino acids 81 to 106 as a TMD) that provides protection from proteases and forms a Ca2+-permeable pore. Additional biochemical analyses of CPE variants and large complexes are under way to test the validity of this model.
FIG. 9.
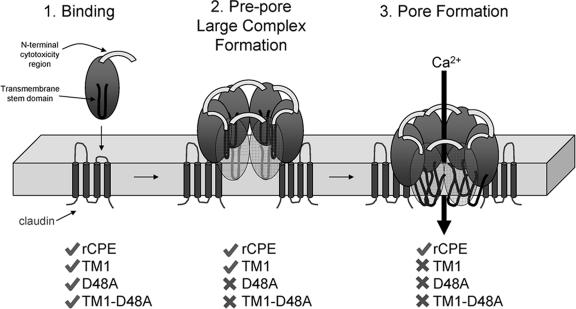
Updated model of CPE action. Stage 1 is binding. CPE action begins when CPE binds certain claudins, transmembrane tight junction proteins, using sequences at the CPE C terminus. Stage 2 is prepore large-complex formation. After membrane localization via receptor binding, CPE joins with other proteins (and/or additional molecules of CPE) to form an SDS-resistant complex. Formation of this complex is dependent upon the presence of an intact N terminus, particularly the aspartic acid residue at position 48. However, without the presence of CPE's putative TMD, this complex is trapped at the prepore stage and is unable to resist degradation by proteases. Stage 3 is membrane insertion/pore formation. With an intact N terminus and putative TMD, the CPE large complex can undergo the conformational changes necessary for insertion into the membrane. This stage not only protects the complex from proteases but also allows the penetration of the phospholipid bilayer of enterocytes, causing the influx of Ca2+ into the cell. Check marks and “x” indicate rCPE species which can or cannot, respectively, undergo this stage in CPE action.
Acknowledgments
This work was supported by Public Health Service grant R37-AI019844-24 from the National Institute of Allergy and Infectious Diseases.
We acknowledge helpful conversations with Rodney K. Tweten, Julian I. Rood, and Derek J. Fisher throughout this study.
Editor: D. L. Burns
Footnotes
Published ahead of print on 16 February 2007.
REFERENCES
- 1.Chakrabarti, G., and B. A. McClane. 2005. The importance of calcium influx, calpain and calmodulin for the activation of CaCo-2 cell death pathways by Clostridium perfringens enterotoxin. Cell. Microbiol. 7:129-146. [DOI] [PubMed] [Google Scholar]
- 2.Chakrabarti, G., X. Zhou, and B. A. McClane. 2003. Death pathways activated in CaCo-2 cells by Clostridium perfringens enterotoxin. Infect. Immun. 71:4260-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheley, S., M. S. Malghani, L. Song, M. Hobaugh, J. E. Gouaux, J. Yang, and H. Bayley. 1997. Spontaneous oligomerization of a staphylococcal alpha-hemolysin conformationally constrained by removal of residues that form the transmembrane beta-barrel. Protein Eng. 10:1433-1443. [DOI] [PubMed] [Google Scholar]
- 4.Czeczulin, J. R., P. C. Hanna, and B. A. McClane. 1993. Cloning, nucleotide sequencing, and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli. Infect. Immun. 61:3429-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deleage, G., and B. Roux. 1987. An algorithm for protein secondary structure prediction based on class prediction. Protein Eng. 1:289-294. [DOI] [PubMed] [Google Scholar]
- 6.Fujita, K., J. Katahira, Y. Horiguchi, N. Sonoda, M. Furuse, and S. Tsukita. 2000. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 476:258-261. [DOI] [PubMed] [Google Scholar]
- 7.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 8.Hanna, P. C., and B. A. McClane. 1991. A recombinant C-terminal toxin fragment provides evidence that membrane insertion is important for Clostridium perfringens enterotoxin cytotoxicity. Mol. Microbiol. 5:225-230. [DOI] [PubMed] [Google Scholar]
- 9.Hanna, P. C., T. A. Mietzner, G. K. Schoolnik, and B. A. McClane. 1991. Localization of the receptor-binding region of Clostridium perfringens enterotoxin utilizing cloned toxin fragments and synthetic peptides. The 30 C-terminal amino acids define a functional binding region. J. Biol. Chem. 266:11037-11043. [PubMed] [Google Scholar]
- 10.Hanna, P. C., E. U. Wieckowski, T. A. Mietzner, and B. A. McClane. 1992. Mapping of functional regions of Clostridium perfringens type A enterotoxin. Infect. Immun. 60:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanna, P. C., A. P. Wnek, and B. A. McClane. 1989. Molecular cloning of the 3′ half of the Clostridium perfringens enterotoxin gene and demonstration that this region encodes receptor-binding activity. J. Bacteriol. 171:6815-6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hardy, S. P., C. Ritchie, M. C. Allen, R. H. Ashley, and P. E. Granum. 2001. Clostridium perfringens type A enterotoxin forms mepacrine-sensitive pores in pure phospholipid bilayers in the absence of putative receptor proteins. Biochim. Biophys. Acta 1515:38-43. [DOI] [PubMed] [Google Scholar]
- 13.Heuck, A. P., E. M. Hotze, R. K. Tweten, and A. E. Johnson. 2000. Mechanism of membrane insertion of a multimeric beta-barrel protein: perfringolysin O creates a pore using ordered and coupled conformational changes. Mol. Cell 6:1233-1242. [DOI] [PubMed] [Google Scholar]
- 14.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 15.Katahira, J., N. Inoue, Y. Horiguchi, M. Matsuda, and N. Sugimoto. 1997. Molecular cloning and functional characterization of the receptor for Clostridium perfringens enterotoxin. J. Cell Biol. 136:1239-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokai-Kun, J. F., K. Benton, E. U. Wieckowski, and B. A. McClane. 1999. Identification of a Clostridium perfringens enterotoxin region required for large complex formation and cytotoxicity by random mutagenesis. Infect. Immun. 67:5634-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kokai-Kun, J. F., and B. A. McClane. 1997. Deletion analysis of the Clostridium perfringens enterotoxin. Infect. Immun. 65:1014-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClane, B. A. 2001. The complex interactions between Clostridium perfringens enterotoxin and epithelial tight junctions. Toxicon 39:1781-1791. [DOI] [PubMed] [Google Scholar]
- 19.McClane, B. A., and A. P. Wnek. 1990. Studies of Clostridium perfringens enterotoxin action at different temperatures demonstrate a correlation between complex formation and cytotoxicity. Infect. Immun. 58:3109-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melton, J. A., M. W. Parker, J. Rossjohn, J. T. Buckley, and R. K. Tweten. 2004. The identification and structure of the membrane-spanning domain of the Clostridium septicum alpha toxin. J. Biol. Chem. 279:14315-14322. [DOI] [PubMed] [Google Scholar]
- 21.Mietzner, T. A., J. F. Kokai-Kun, P. C. Hanna, and B. A. McClane. 1992. A conjugated synthetic peptide corresponding to the C-terminal region of Clostridium perfringens type A enterotoxin elicits an enterotoxin-neutralizing antibody response in mice. Infect. Immun. 60:3947-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller, C. J., J. L. Elliott, and R. J. Collier. 1999. Anthrax protective antigen: prepore-to-pore conversion. Biochemistry 38:10432-10441. [DOI] [PubMed] [Google Scholar]
- 23.Olsen, S. J., L. C. MacKinnon, J. S. Goulding, N. H. Bean, and L. Slutsker. 2000. Surveillance for foodborne-disease outbreaks—United States, 1993-1997. Morb. Mortal. Wkly. Rep. Surveill. Summ. 49:1-62. [PubMed] [Google Scholar]
- 24.Palmer, M., P. Saweljew, I. Vulicevic, A. Valeva, M. Kehoe, and S. Bhakdi. 1996. Membrane-penetrating domain of streptolysin O identified by cysteine scanning mutagenesis. J. Biol. Chem. 271:26664-26667. [DOI] [PubMed] [Google Scholar]
- 25.Parker, M. W., and S. C. Feil. 2005. Pore-forming protein toxins: from structure to function. Prog. Biophys. Mol. Biol. 88:91-142. [DOI] [PubMed] [Google Scholar]
- 26.Rossjohn, J., S. C. Feil, W. J. McKinstry, R. K. Tweten, and M. W. Parker. 1997. Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell 89:685-692. [DOI] [PubMed] [Google Scholar]
- 27.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 28.Shepard, L. A., A. P. Heuck, B. D. Hamman, J. Rossjohn, M. W. Parker, K. R. Ryan, A. E. Johnson, and R. K. Tweten. 1998. Identification of a membrane-spanning domain of the thiol-activated pore-forming toxin Clostridium perfringens perfringolysin O: an alpha-helical to beta-sheet transition identified by fluorescence spectroscopy. Biochemistry 37:14563-14574. [DOI] [PubMed] [Google Scholar]
- 29.Sigrist, H., P. Ronner, and G. Semenza. 1975. A hydrophobic form of the small-intestinal sucrase-isomaltase complex. Biochim. Biophys. Acta 406:433-446. [DOI] [PubMed] [Google Scholar]
- 30.Singh, U., L. L. Mitic, E. U. Wieckowski, J. M. Anderson, and B. A. McClane. 2001. Comparative biochemical and immunocytochemical studies reveal differences in the effects of Clostridium perfringens enterotoxin on polarized CaCo-2 cells versus Vero cells. J. Biol. Chem. 276:33402-33412. [DOI] [PubMed] [Google Scholar]
- 31.Singh, U., C. M. Van Itallie, L. L. Mitic, J. M. Anderson, and B. A. McClane. 2000. CaCo-2 cells treated with Clostridium perfringens enterotoxin form multiple large complex species, one of which contains the tight junction protein occludin. J. Biol. Chem. 275:18407-18417. [DOI] [PubMed] [Google Scholar]
- 32.Smedley, J. G., III, and B. A. McClane. 2004. Fine mapping of the N-terminal cytotoxicity region of Clostridium perfringens enterotoxin by site-directed mutagenesis. Infect. Immun. 72:6914-6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stultz, C. M., R. Nambudripad, R. H. Lathrop, and J. V. White. 1997. Predicting protein structure with probabilistic models, p. 447-506. In N. M. Allewell (ed.), Advances in molecular and cell biology, vol. 22B. Protein structural biology in biomedical research, part B. JAI Press, Greenwich, CT. [Google Scholar]
- 34.Stultz, C. M., J. V. White, and T. F. Smith. 1993. Structural analysis based on state-space modeling. Protein Sci. 2:305-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tilley, S. J., and H. R. Saibil. 2006. The mechanism of pore formation by bacterial toxins. Curr. Opin. Struct. Biol. 16:230-236. [DOI] [PubMed] [Google Scholar]
- 36.Tweten, R. K. 2005. Cholesterol-dependent cytolysins, a family of versatile pore-forming toxins. Infect. Immun. 73:6199-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Itallie, C. M., and J. M. Anderson. 2004. The molecular physiology of tight junction pores. Physiology (Bethesda). 19:331-338. [DOI] [PubMed] [Google Scholar]
- 38.White, J. V., C. M. Stultz, and T. F. Smith. 1994. Protein classification by stochastic modeling and optimal filtering of amino-acid sequences. Math. Biosci. 119:35-75. [DOI] [PubMed] [Google Scholar]
- 39.Wieckowski, E. U., J. F. Kokai-Kun, and B. A. McClane. 1998. Characterization of membrane-associated Clostridium perfringens enterotoxin following pronase treatment. Infect. Immun. 66:5897-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wnek, A. P., and B. A. McClane. 1989. Preliminary evidence that Clostridium perfringens type A enterotoxin is present in a 160,000-Mr complex in mammalian membranes. Infect. Immun. 57:574-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wnek, A. P., R. J. Strouse, and B. A. McClane. 1985. Production and characterization of monoclonal antibodies against Clostridium perfringens type A enterotoxin. Infect. Immun. 50:442-448. [DOI] [PMC free article] [PubMed] [Google Scholar]