Abstract
Human β-defensin 2 (hBD-2) is an inducible antimicrobial peptide synthesized by the epithelium to counteract bacterial adherence and invasion. Proinflammatory cytokines, as well as certain bacterial strains, have been identified as potent endogenous inducers. Recently, we have found that hBD-2 induction by probiotic Escherichia coli Nissle 1917 was mediated through NF-κB- and AP-1-dependent pathways. The aim of the present study was to identify the responsible bacterial factor. E. coli Nissle 1917 culture supernatant was found to be more potent than the pellet, indicating a soluble or shed factor. Chemical analysis demonstrated the factor to be heat resistant and proteinase digestible. Several E. coli Nissle 1917 deletion mutants were constructed and tested for their ability to induce hBD-2 expression in Caco-2 cells. Deletion mutants for flagellin specifically exhibited an impaired immunostimulatory capacity. Reinsertion of the flagellin gene restored the induction capacity to normal levels. Isolated flagellin from E. coli Nissle 1917 and from Salmonella enterica serovar Enteritidis induced hBD-2 mRNA significantly in contrast to the flagellin of the apathogenic E. coli strain ATCC 25922. H1 flagellin antiserum abrogated hBD-2 expression induced by flagellin as well as E. coli Nissle 1917 supernatant, confirming that flagellin is the major stimulatory factor of E. coli Nissle 1917.
Probiotics are microbial organisms beneficial to health (3, 15). The spectrum of probiotics, including Escherichia coli, lactobacilli, bifidobacteria, and streptococci, as well as yeasts, is as complex as their clinical applications. For example, the duration of traveler's diarrhea and other self-limited gastrointestinal infections is shortened by probiotics (24, 34), especially in infants (9, 33). Allergies seem to be reduced, and the incidence of infections is decreased after administration of the probiotic E. coli strain PZ 720 to newborns (18). However, the best evidence-based indication is the remission maintenance of ulcerative colitis by E. coli Nissle 1917 (16, 17, 31). Using this strain experimental murine colitis has been ameliorated (36). Also, a probiotic mixture composed of eight different strains, named VSL#3, is effective in primary and secondary prevention of pouchitis after proctocolectomy in ulcerative colitis (11, 12). The numerous effects of probiotics are difficult to explain by a unifying hypothesis that is based on a single quality or mechanism. The mechanisms of action proposed thus far include an alteration of the hosts’ cytokine repertoire (21), increased immunoglobulin A secretion (22, 26), and mucus formation (20), lymphocyte or macrophage activation (13, 26, 35), or an inhibition of epithelial adhesion and invasion (4, 21).
The genome of E. coli Nissle 1917 (serotype O6:K5:H1) has recently been sequenced (14, 39) and contains many characteristic fitness factors that promote its competitiveness (e.g., six iron uptake systems) and highly effective colonization (e.g., several types of adhesins) of the host (19). Recent in vitro data have demonstrated that E. coli Nissle 1917 induces intestinal epithelial cells to block adherence and inhibit invasion of various pathogenic strains (1, 5). These observations proposed a soluble factor responsible for the inhibition of the pathogen itself or engagement of an epithelial defense mechanism (1) apart from anti-inflammatory influences on T-cell proliferation and function (38). We have recently provided a possible explanation for the probiotic effect of E. coli Nissle 1917 as it induces human β-defensin 2 (hBD-2) expression in cell culture in a time- and dose-dependent manner (41). Since this antimicrobial peptide, produced by the mucosal epithelium, has a broad antibiotic spectrum against gram-negative and -positive bacteria, as well as against fungi and viruses, it may reinforce the mucosal barrier, thereby limiting bacterial adherence and invasion. E. coli Nissle 1917, in contrast to most other E. coli strains, induces hBD-2 expression via nuclear factor-κB (NF-κB)- and, to a smaller extent. activator protein-1 (AP-1)-dependent pathways. The aim of the present study was to identify the factor(s) of E. coli Nissle 1917 responsible for hBD-2 induction.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in the present study are shown in Table 1. Bacteria were grown overnight at 37°C under gentle agitation at 200 rpm in Trypticase soy broth (TSB). To obtain bacteria in a linear growth phase, 100 μl of the bacterial suspension was added to 10 ml of fresh TSB medium and grown under permanent shaking for 5 h. Heat inactivation was carried out in a water bath at 65°C for 1 h. Bacterial culture supernatants were collected by centrifugation at 4,000 × g for 10 min. Bacteria were adjusted to a density of 3 × 108 cells/ml, and the supernatant was diluted equally in fetal calf serum (FCS)- and antibiotic-free culture medium.
TABLE 1.
Bacterial strains used in this study
| Strain | Serotype | Relevant characteristic | Sourceb |
|---|---|---|---|
| E. coli Nissle 1917 wild type | O6:K5:H1 | Probiotic strain | ACS |
| E. coli Nissle mutant strain | IMBI | ||
| E. coli CFT073Δhlya | O6:K2:H1 | Intestinal isolate | IMBI |
| E. coli JM109 ATCC 53323 | Orough:H48 | Reference strain | IMBI |
| E. coli PZ 720 | O83:K24:H31 | Probiotic strain | ACS |
| E. coli W536 | O6:K15:H31 | Uropathogenic strain | IMBI |
| E. coli DH5α | Reference strain | IMBI | |
| E. coli PZ 830 | O4:H− | Clinical isolate | ACS |
| E. coli ATCC 25922 | Reference strain | DSMZ | |
| S. enterica serovar Enteritidis | Intestinal isolate | RBS |
Deletion of toxin alpha-hemolysin.
ACS, Ardeypharm Collection of Strains (fecal isolates); DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; IMBI, strain collection of the Institute of Molecular Biology of Infection, University of Würzburg, Würzburg, Germany; RBS, clinical isolate from the Robert Bosch Hospital, Stuttgart, Germany.
Cell culture.
Caco-2 cells (German Collection of Microorganisms and Cell cultures [DSMZ] ACC 169) and SW-620 (American Type Culture Collection [ATCC] CCL-227) were grown in Dulbecco modified Eagle medium (DMEM) containing 25 mM HEPES and 2 mM glutamine supplemented with 10% FCS, 50 μg of gentamicin/ml, and 5% nonessential amino acids. T84 cells (ATCC CCL-248) were maintained in a 1:1 mixture of DMEM and Ham F-12 medium supplemented with 15 mM HEPES, 14 mM NaHCO3, 5% FCS, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. For stimulation experiments, cells were seeded into 12-well culture plates (Becton Dickinson GmbH, Heidelberg, Germany). After an overnight culture in FCS- and antibiotic-free DMEM, ∼70% confluent cells were incubated with bacterial culture supernatant, pellet, or flagellin for 6 h at 37°C. For signaling pathway studies, specific mitogen-activated protein (MAP) kinase inhibitors AG126 (Calbiochem, Darmstadt, Germany) SB203580 and SP600125 (Tocris, Ellisville, MO) were resuspended in dimethyl sulfoxide (DMSO). Cells were pretreated with the specific ERK1/2 (for extracellular signal-regulated kinase 1/2), p38, and JNK (for c-Jun N-terminal kinase) inhibitors 1 h prior to stimulation experiments. Cell viability was assessed by the MTT cell proliferation assay (ATCC, Manassas, VA) according to the manufacturer's protocol.
Construction of bacterial mutants.
The E. coli Nissle 1917 (EcN) mutant strains, i.e., the ΔHPI (yersiniabactin-gene cluster), Δfim (type 1 pili), Δfoc (F1C pili), ΔfimΔfoc (type 1 pili plus F1C pili), ΔcsgBA (curli-negative), Δbcs (cellulose-synthesis-gene cluster), ΔmcmDAB (microcin-negative), ΔK5 (capsule 5-gene cluster), ΔfliA (sigma factor of flagellum genes), ΔfliC (flagellin filament protein), and ΔflgE (hook) mutant strains, were constructed according to a previously described method (6). EcNΔc (Nissle 1917 cured of both plasmids) was generated according to the German patent 103 28 669 and lacks its strain-specific plasmids. The single gene clusters were replaced by a cat antibiotic resistance cassette, which was part of a PCR product generated with plasmid pKD3 as the template and the primer pair: p1 (5′-AAGATAGCGGCTTAATGGCCGTGTGTAGGCTGGAGCTGCTT-3′) and p2 (5′-CGTTCATCCCTGGGGGCTATCCCATATGAATATCCTCCTTAGTTCCTA-3′) for fliA, p3 (5′-GGCAATTTGGCGTTGCCGTCAGGTGTAGGCTGGAGCTGCTT-3′) and p4 (5′-ACGGCGATTGAGCCGACGGGTGCATATGAATATCCTCCTTAGTTCCTA-3′) for fliC, or p5 (5′-ACGCTGGATCTCGGCACTTACGGTGTAGGCTGGAGCTGCTT-3′) and p6 (5′-TGTTGATTCAGCGTCTGGCTGGCATATGAATATCCTCCTTAGTTCCTA-3′) for flgE. From the resulting chloramphenicol-resistant E. coli Nissle 1917 derivate the cat cassette was not deleted after transformation for better selection of the derivates. The E. coli Nissle 1917 ΔfliC and ΔflgE flagellin mutant strains were complemented with a PCR product of the lacking gene cloned into the pGEM-T Easy vector (Promega, Madison, WI), and the corresponding complementants were labeled ΔfliCpDB2 and ΔflgEpDB3, respectively. The primers used for PCR corresponded to those for the deletion method in the underlined sequences.
Western blotting of whole bacteria.
Heat-killed bacterial samples from overnight cultures were used to analyze flagellin expression by Western blotting. Briefly, the resultant sample was separated in a modular mini-electrophoresis system (Bio-Rad, Hercules, CA) on a 12.5% polyacrylamide gel and then transferred to a 0.2-μm-pore-size nitrocellulose membrane (Bio-Rad). Membranes were washed thrice with Tris-buffered salt solution (TBS) and subsequently incubated with anti-H7 E. coli rabbit polyclonal antibody (gift from T. K. Korhonen, General Microbiology, Faculty of Biosciences, University of Helsinki, Helsinki, Finland) diluted 1:5,000 in blocking buffer overnight at 4°C under gentle agitation. After removal of the primary antibody and extensive washing with TBS, the blot was incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-rabbit immunoglobulin (Calbiochem, San Diego, CA) diluted 1:5,000 in blocking buffer. The blot was washed afterward, covered with ECL light detection reagent (Amersham Biosciences, Buckinghamshire, United Kingdom), and wrapped to cover the blot before exposing it to film for 60 s (Kodak, Rochester, NY).
Purification of flagellin.
Flagellin of diverse bacterial strains was isolated as previously described (25). Briefly, 1 liter of TSB was inoculated with 10 ml of the bacteria and cultured for 16 h at 37°C, pelleted by centrifugation (4,400 × g, 30 min, 4°C), and resuspended in 20 ml of phosphate-buffered saline (PBS). For the flagellin isolation from bacterial supernatant, bacteria were heat-killed, and the supernatant was separated by centrifugation. The pelleted bacteria suspension or the supernatant was adjusted to pH 2 with 1 M HCl and maintained at that pH under constant stirring at room temperature for 30 min. After centrifugation (100,000 × g, 4°C, 1 h), the pH of the supernatant containing soluble monomeric flagellin was adjusted to 7.2 with 1 M NaOH. Solid (NH4)2SO4 was added slowly with constant stirring to obtain 65% saturation. After an overnight incubation at 4°C, the precipitate was pelleted by centrifugation (15,000 × g, 4°C, 15 min). The precipitate was dissolved in distilled water and dialyzed under constant stirring against distilled water for 16 h with three changes of water. To remove heat-labile proteins, the dialysate was heated at 65°C for 15 min, placed on ice, and centrifuged (100,000 × g, 4°C, 1 h). Then, 0.7 M solid (NH4)2SO4 was added to the supernatant, which contained depolymerized flagellin. After incubation overnight at 4°C, polymerized flagellin was collected by centrifugation (100,000 × g, 4°C, 1 h) and dissolved in PBS. The protein concentration of isolated flagellin was determined by using the Bradford assay (Bio-Rad) and bovine serum albumin as a standard. The molecular size of the flagellin was identified by mass spectrometry. The samples were diluted 1:10 in 50% acetonitrile containing 0.02% formic acid and analyzed by electrospray mass spectrometry in the positive ionization mode with a quadrupole-orthogonal accelerating-time-of-flight mass spectrometer (Micromass, Manchester, United Kingdom). The spectra were evaluated by using the program MassLynx 3.5. (Micromass).
Coomassie staining and Western blotting of flagellin.
To verify the purity of the flagellin, the isolated protein was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12% gel (Invitrogen, Carlsbad, CA). The gels were stained for protein with Coomassie brilliant blue. For Western blotting, isolated protein was transferred to 0.45-μm-pore-size nitrocellulose membranes (Schleicher & Schuell, Keene, NH). Membranes were incubated with 5% skimmed milk powder in TBST (20 mM Tris-Base [pH 7.4], 0.14 M NaCl, 0.1% Tween 20) for 1 h. E. coli H1-antiserum (Statens Serum Institute, Copenhagen, Denmark) served as primary antibody and was diluted 1:500 in TBST. After overnight incubation, strips were washed and incubated with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (Dianova, Hamburg, Germany) diluted 1:5,000 in TBST for 1 h. The membranes were washed in TBST, followed by incubation for 5 min in SuperSignal West Dura extended duration substrate (Pierce, Rockford, IL). Developing bands were detected with a chemiluminescence camera charge-coupled device LAS-1000 (Fuji), and analysis was performed with the software AIDA 2.1 (Raytest, Straubenhardt, Germany).
Luciferase reporter gene assay.
To assess hBD-2 promoter activity, Caco-2 cells were seeded into 12-well culture plates (2.8 × 105 cells/ml) and transfected upon 70% confluence. The luciferase reporter constructs for hBD-2-2338-luc, NF-κB-mut1+2-luc, AP-1-mut-luc, and AP-1+NF-κB-mut-luc have been described recently (48). Cells were transfected with 0.5 μg of hBD-2 reporter plasmid and 0.05 μg of an internal control Renilla luciferase expression plasmid (phRG-TK; Promega) by using 1 μl of transfection reagent FuGENE 6 (Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's protocol. At 24 h after transfection, cells were stimulated for 4.5 h with either flagellin, the bacterial pellet, or the supernatant, respectively. Cells were washed, lysed, and harvested by using 250 μl of passive lysis buffer (Promega) per well. Firefly luciferase activity from the hBD-2-pGL3 reporter vector and Renilla luciferase activity were analyzed with the Dual-Luciferase reporter assay system (Promega) using a luminometer (Berthold). Promoter activity was normalized to the activity of the internal Renilla luciferase control.
RNA isolation and cDNA synthesis.
At the end of the stimulation experiment, cells were washed twice with PBS and harvested with TRIzol reagent (Invitrogen), and RNA was isolated according to the supplier's protocol. RNA quality and quantity were determined by gel electrophoresis and photometry. Subsequently, 1 μg of total RNA was reverse transcribed into cDNA with oligo(dT) primers and 15 U of avian myeloblastosis virus reverse transcriptase (Promega)/μg according to standard procedures.
Real-time RT-PCR.
Real-time reverse transcription PCR (RT-PCR) analyses were performed in a fluorescence temperature cycler (LightCycler; Roche Diagnostics GmbH, Mannheim, Germany) according to the manufacturer's instructions. This technique continuously monitors the cycle-by-cycle accumulation of fluorescently labeled PCR product. cDNA corresponding to 10 ng of RNA served as a template in a 10-μl reaction mixture containing 3 mM MgCl2, 0.5 μM concentrations of each primer, and 1× LightCycler-FastStart DNA Master SYBR green I mix (Roche Diagnostics GmbH). Samples were loaded into capillary tubes and placed in the fluorescence thermocycler (LightCycler). Initial denaturation at 95°C for 10 min was followed by 45 cycles of 95°C for 15 s, the primer-specific annealing temperature for 5 s, and elongation at 72°C for 15 s. For hBD-2 (sense, 5′-ATCAGCCATGAGGGTCTTGT-3′; antisense, 5′-GAGACCACAGGTGCCAATTT-3′) the annealing temperature was set at 62°C. Amplification using these primers resulted in a 172-bp fragment. For the housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase; sense, 5′-CCAGCCGAGCCACATCGCTC-3′; antisense, 5′-ATGAGCCCCAGCCTTCTCCAT-3′), we used a touchdown protocol with a primary temperature of 66°C and a target temperature of 60°C. At the end of each run melting-curve profiles were achieved by cooling the sample to 65°C for 15 s and then heating the sample slowly at 0.20°C/s up to 95°C with continuous measurement of the fluorescence to confirm the amplification of specific transcripts. Cycle-to-cycle fluorescence emission readings were monitored and analyzed by using LightCycler software (Roche Diagnostics GmbH). Melting curves were generated after each run to confirm amplification of specific transcripts. The specificity of the amplification products was verified by subjecting the amplification products to electrophoresis on a 2% agarose gel. Obtained fragments were visualized by ethidium bromide staining. All quantifications were normalized to the housekeeping GAPDH gene. Relative expression is given as a ratio between target gene and GAPDH gene expression.
Swarm plate assay.
Swarm agar (0.3%; containing 10 g of NaCl, 5 g of yeast extract, 10 g of peptone, and 3 g of agar per liter) was used to evaluate the motility of E. coli Nissle 1917 wild type and the ΔfliC, ΔflgE, ΔfliA, and complemented ΔfliCpDB2 and ΔflgEpDB3 mutant strains. Small wells were punched out in semisolid LB agar plates, loaded with 5 μl of each strain, and incubated for 10 h at 37°C.
IL-8 ELISA.
Interleukin-8 (IL-8) secretion in response to IL-1β and E. coli Nissle 1917 supernatant was measured in cell culture supernatants from Caco-2, T84, and SW620 cells by enzyme-linked immunosorbent assay (ELISA; OptEIA human IL-8 ELISA kit II; BD Biosciences, San Diego, CA) according to the supplier's protocol.
Statistics.
Statistical evaluation was performed with the software package GraphPad Instat (version 3.1 for Windows; GraphPad Software, San Diego, CA). For the description of random samples, the arithmetic mean ± the standard deviation (SD) or the median with minimum and maximum values was used. To test for normality, we used the Kolmogorov-Smirnov test. When data failed to follow a Gaussian distribution, nonparametric analysis of variance was performed by using Kruskal-Wallis or Mann-Whitney test.
RESULTS
Caco-2 cells have the highest potential to express hBD-2.
In order to identify the most suitable model for studying the hBD-2-inducing effect of E. coli Nissle 1917, various intestinal epithelial cell lines (Caco-2, T84, and SW620 cells) were stimulated with IL-1β and E. coli Nissle 1917 supernatant. SW620 cells did not express hBD-2 mRNA after stimulation with IL-1β and expressed only a small amount of hBD-2 after stimulation with the bacterial supernatant (Table 2) . In contrast, IL-1β as well as E. coli Nissle 1917 supernatant strongly induced hBD-2 in Caco-2 cells. This effect was less pronounced in T84 cells especially regarding IL-1β as stimulant. The spread of GAPDH mRNA expression levels of the different cell types and between the experiments was narrow, indicating a low variability. IL-8 served as a control response gene measured in culture supernatants by ELISA after 6 h of stimulation. As shown in Table 2, IL-1β as well as E. coli Nissle 1917 stimulation induced the secretion of IL-8 in Caco-2 cells, whereas SW620 cells were expressing IL-8 only after incubation with the bacterial supernatant. The degree of differentiation might also influence hBD-2 expression; hence, we tested hBD-2 transcription levels of cells grown for different time intervals before stimulation. The relative expression of hBD-2 mRNA upon treatment with IL-1β was inferior in confluent versus subconfluent Caco-2 cells (data not shown). Thus, we preferred subconfluent cell layers for our experiments.
TABLE 2.
Response of intestinal cell lines to IL-1β (5 ng/ml) and E. coli Nissle supernatant exposure
| Cell line | Treatment | Mean (ng/ml) ± SD
|
|
|---|---|---|---|
| hBD-2 mRNAb | IL-8 protein | ||
| Caco-2 | Unstimulated | 0 ± 0 | 39.5 ± 3.2 |
| IL-1β | 584.5 ± 82.0 | 5,510.2 ± 1,865.1 | |
| EcNsna | 52.1 ± 6.2 | 2,852.4 ± 572.7 | |
| T84 | Unstimulated | 0 ± 0 | 295.1 ± 24.,1 |
| IL-1β | 48.4 ± 18.7 | 336.0 ± 167.5 | |
| EcNsn | 28.6 ± 17.2 | 981.7 ± 426.3 | |
| SW620 | Unstimulated | 0 ± 0 | 122.5 ± 10.5 |
| IL-1β | 0 ± 0 | 141.0 ± 13.0 | |
| EcNsn | 13.8 ± 12.0 | 4,441.9 ± 617.5 | |
EcNsn, E. coli Nissle supernatant.
Relative expression.
Nonpathogenic E. coli Nissle 1917 induces hBD-2 by a supernatant factor.
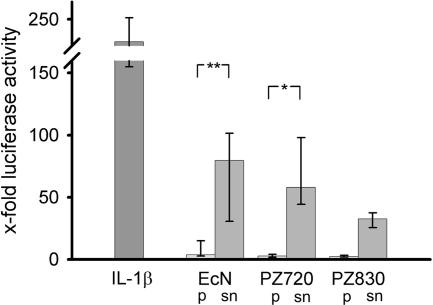
To analyze the origin of the unknown hBD-2-inducing factor of E. coli Nissle 1917, Caco-2 cells were incubated with either resuspended bacterial pellet or supernatant. hBD-2 promoter enhancing activity was predominant in the supernatant (92.7%) compared to the bacterial pellet (7.3%). The other strains tested also displayed most of the activity in the supernatant (Fig. 1). Induction of hBD-2 mRNA by E. coli Nissle 1917 was distributed similarly (84.1% activity in the supernatant). In addition, we tested the stability of the hBD-2-inducing factor in the supernatant of E. coli Nissle 1917. Exposure to 100°C for 20 min was unable to abrogate hBD-2 induction compared to E. coli Nissle 1917 heated to 65°C (the hBD-2 promoter activity of E. coli Nissle supernatant at 100°C was 91.1% ± 18.2% of that of the E. coli Nissle supernatant at 65°C). However, the supernatant from unheated bacteria exhibited a diminished promoter inducing capacity (38.1% ± 8.3%). Treatment of the supernatant for 1 h with proteinase K (QIAGEN, Hilden, Germany) reduced its activation of the hBD-2 promoter to 28.4% ± 3.4% (n = 3) of that of untreated controls. These results indicate that the bacterial factor in the supernatant is heat resistant and proteinase sensitive. The incubation of bacterial supernatant with polymyxin B (Detoxi-Gel endotoxin removing gel [Pierce]) to remove lipopolysaccharide (LPS) did not abrogate the supernatant-mediated hBD-2 promoter activation (115.5% ± 28.2% activity compared to the untreated supernatant set at 100%), indicating that LPS is not a stimulatory factor for hBD-2 transcription.
FIG. 1.
Effect of crude bacterial pellet versus supernatant on hBD-2 reporter gene activity. Transfection of Caco-2 cells with an hBD-2 promoter construct (2,338 bp) for 24 h was followed by incubation for 4.5 h with 5 ng of IL-1β or bacterial preparations/ml. The probiotic fecal E. coli isolates Nissle 1917 (EcN), PZ720, and E. coli PZ830 were adjusted to a density of 3 × 108 bacteria/ml and tested either as crude bacterial pellets (p) or corresponding supernatants (sn) in the same amount of volume. hBD-2 promoter activation was determined as a ratio between firefly and Renilla luciferase activities. The data represent the median with minimum and maximum values of three to five experiments performed in triplicate. The median was normalized to the basal luminescence of unstimulated controls, set at 1. IL-1β served as a positive stimulatory control (left column). A statistical comparison between the crude pellet and the supernatant was performed by using the Mann-Whitney test. *, P < 0.05; **, P < 0.01.
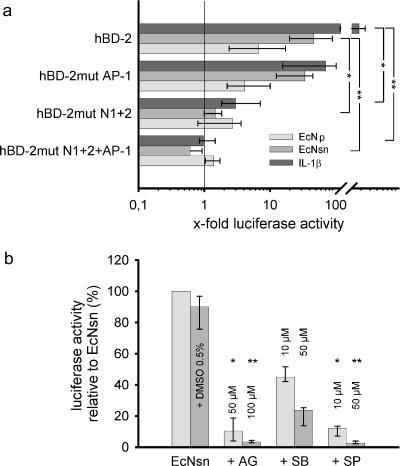
E. coli Nissle 1917 supernatant-mediated hBD-2 induction depends on NF-κB and AP-1.
The importance of NF-κB and AP-1 binding sites in the promoter region of hBD-2 was analyzed by transfecting Caco-2 cells with hBD-2 promoter luciferase expression constructs encompassing the 5′-untranslated region upstream of the start of gene transcription (2,338 bp). Binding sites for the two 5′ upstream NF-κB sites (positions −205 to −186 and −596 to −572) and AP-1 (positions −127 to −121) were mutated separately or in combination. Consistent with previous findings regarding the bacterial pellet (41), promoter activation by the hBD-2-inducing factor in the supernatant was both NF-κB and AP-1 dependent (Fig. 2). This was confirmed by the complete abrogation of hBD-2 promoter activity after the mutation of both NF-κB and the AP-1 binding sites. Mutation of the AP-1 site alone decreased supernatant-mediated hBD-2 promoter activation by ca. 30%. Promoter activity was almost completely lost after the deletion of both NF-κB sites.
FIG. 2.
Contribution of NF-κB and AP-1 binding sites and MAP kinases for E. coli Nissle 1917 mediated hBD-2 promoter activation. (a) Various mutant promoter constructs were tested for their ability to respond to IL-1β, as well as E. coli Nissle 1917 culture supernatant (EcNsn) or crude bacterial pellet (EcN), in luciferase reporter gene assays as outlined in Fig. 1. NF-κB (positions −205 to −186 and −596 to −572) and AP-1 (positions −127 to −121) were mutated separately (hBD-2mut N1+2 or hBD-2mut AP-1) or in combination (hBD-2mut N1+2+AP-1). Maximal activity was observed with the complete promoter encompassing all known NF-κB and AP-1 sites. Sequential loss of transcription factor binding sites within the hBD-2 promoter resulted in a stepwise decrease in activation upon IL-1β, as well as pellet (EcN) and supernatant (EcNsn) treatment. The data represent the median with minimum and maximum values of duplicate samples of three to four experiments. Statistical evaluation was performed by using Kruskal-Wallis analysis with post hoc Dunn's test. *, P < 0.05; **, P < 0.01. (b) Specific MAP kinase inhibitors (diluted in DMSO) at the indicated concentrations were added 1 h prior to 4.5 h stimulation with bacterial supernatant or the corresponding supernatant, including 0.5% DMSO. Blocking of ERK1/2 by AG126 (AG) and JNK by SP600125 (SP) resulted in almost complete abrogation of E. coli Nissle 1917 supernatant-mediated hBD-2 promoter activation. Inhibition of p38 MAP kinase by SB203580 (SB) had the slightest effect. The data represent the median with minimum and maximum values of duplicate determinations of three experiments. Each inhibitor was compared to the DMSO control by Kruskal-Wallis analysis and post hoc testing. *, P < 0.05; **, P < 0.01.
ERK1/2, JNK, and p38 are essential for the E. coli Nissle 1917 supernatant-mediated induction of hBD-2.
To gain more insight into the signaling events of the hBD-2-inducing supernatant factor of E. coli Nissle 1917 via the three main MAP kinase pathways, we pretreated Caco-2 cells with specific inhibitors: AG126 for ERK1/2, SB203580 for p38, and SP600125 for JNK. The inhibition of p38 by SB203580 reduced hBD-2 promoter activation by at least 50%; higher doses of the inhibitor were more effective (Fig. 2). Inhibition of ERK1/2 and JNK by AG126 or SP600125, respectively, blocked hBD-2 induction almost completely. An MTT cytotoxicity assay revealed that the decrease in hBD-2 induction was not related to cytotoxic side effects of the inhibitors (data not shown).
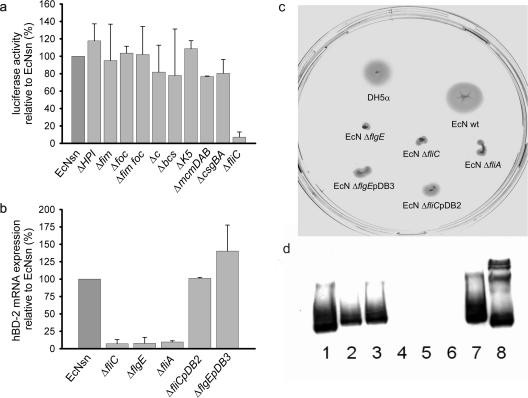
Flagellin mutants lose their capacity to induce hBD-2.
To clarify which factor in the bacterial culture supernatant is responsible for hBD-2 induction, several E. coli Nissle 1917 strains with mutations of known fitness factors (e.g., adhesins, microcins, siderophores, and flagellin) were constructed and added to Caco-2 cells. The three different mutants of the flagellin gene (Fig. 3a and b [the ΔfliC, ΔfliE, and ΔfliA mutants]) lost their inducing capacity completely, while the other mutants displayed no change compared to the wild-type E. coli Nissle 1917. In order to verify that this loss of hBD-2 induction by the flagellin mutants was specifically attributable to the lack of flagellin, deleted genes were complemented in trans by transforming the respective mutant with a recombinant plasmid harboring the corresponding gene. After complementation, the ΔfliC and ΔflgE mutants regained their capacity to induce hBD-2 mRNA (Fig. 3b). As further proof for the loss of flagellin synthesis, LB swarm agar plates were inoculated with the different strains. The nonmotile colonies of the E. coli Nissle 1917 ΔfliA, ΔfliC, and ΔflgE mutant strains appeared as dots, whereas the wild-type colonies spread out in larger spots. Complementation of the ΔfliC and ΔflgE mutants only partially restored their ability to swarm (Fig. 3c). This shows that the complementation with the flagellin or the hook gene, respectively, was successful regarding the regain of hBD-2 inducing capacity but failed to accomplish the full motility function of the flagella. The lack of flagellin synthesis after the mutation was also confirmed by Western blotting. Anti-H7 flagellin antiserum showed only a positive band of the proteins of wild-type or complemented E. coli Nissle 1917 strains (Fig. 3d).
FIG. 3.
Effect of E. coli Nissle 1917 fitness factor mutants on hBD-2 expression and analysis of flagellin-mutant and complemented E. coli Nissle strains. (a) Transfected Caco-2 cells were incubated for 4.5 h with supernatants from various E. coli Nissle 1917 mutant strains (i.e., the iron siderophore yersiniabactin-negative ΔHPI mutant strain; the fimbria-negative Δfim, Δfoc, ΔfimΔfoc, and ΔcsgBA mutant strains; the cellulose-synthesis-negative Δbcs mutant strain; the microcin-negative ΔmcmDAB mutant strain; the capsule-negative ΔK5 mutant strain; the plasmid-negative Δc mutant strain; and the flagellin filament protein-negative ΔfliC mutant strain). hBD-2-inducing activity was determined by the luciferase reporter gene assay. Only the supernatant of the flagellin mutant (EcNsn ΔfliC) showed a decrease in activation ability. The data are means ± the SD of three independent experiments assayed in duplicate. (b) Induction of hBD-2 mRNA expression by flagellin-deficient E. coli Nissle 1917. Stimulation of Caco-2 cells with supernatants of different flagellin mutants (i.e., the ΔfliC, ΔflgE, and ΔfliA mutant strains) for 6 h resulted in a similar decrease in hBD-2 mRNA expression. Reinsertion of the gene into the corresponding E. coli Nissle 1917 mutant [ΔfliCpDB2 and ΔflgEpDB3] completely restored mRNA induction by these strains compared to that induced by the supernatant from wild-type E. coli Nissle 1917 (EcNsn). Gene expression was analyzed by real-time PCR. The data represent means ± the SD of four experiments in duplicate. (c) Characterization of motility by swarming in semisolid agar. Cultures of the wild-type E. coli Nissle 1917 and the flagellar mutants and their complements were plated into wells punched into 0.3% LB agar plates and incubated. Visually determined swarming was normal in the case of the DH5α and wild-type E. coli Nissle 1917 (EcN) strains. All flagellin mutants (i.e., the ΔflgE, ΔfliC, and ΔfliA mutant strains) showed an abolished swarming. The complemented mutants displayed an intermediate swarming [ΔfliCpDB2 and ΔflgEpDB3]. (d) Flagellin expression assessed by Western blot analysis. Heat-killed overnight bacterial cultures were subjected to SDS-PAGE electrophoresis, and flagellin expression was analyzed with H7 flagellin antiserum. Lanes: 1 and 7, EcN (wild type); 2, EcNΔflgE(pDB3) (complemented ΔflgE mutant); 3, EcNΔfliC(pDB2) (complemented ΔfliC mutant); 4, EcNΔflgE; 5, EcNΔfliC; 6, EcNΔfliA; 8, E. coli W536. Flagellin mutants (i.e., the ΔflgE, ΔfliC, and ΔfliA mutant strains) lacked the specific bands of flagellin, whereas complemented mutants (ΔfliCpDB2 and ΔflgEpDB3 mutants) displayed immunoreactivity.
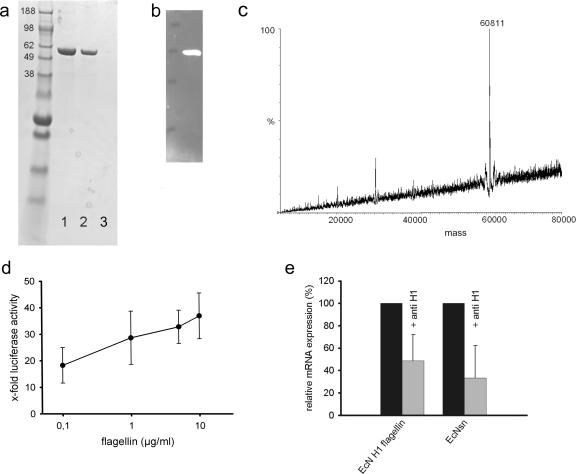
Isolated E. coli Nissle 1917 flagellin induces hBD-2 mRNA expression in Caco-2 cells.
To determine whether E. coli Nissle 1917 H1 flagellin induces hBD-2 expression in intestinal epithelial cells, Caco-2 cells were stimulated with flagellin isolated from wild-type E. coli Nissle 1917 (Fig. 4a and b). The molecular mass of the flagellin, determined by mass spectrometry, was 60.81 kDa (Fig. 4c). As shown in Fig. 4d, flagellin induced a dose-dependent increase of hBD-2 promoter activation. Moreover, the preincubation of E. coli Nissle 1917 culture supernatants with H1 flagellin antiserum inhibited hBD-2 mRNA expression by almost 70%. Consistent with this observation, the hBD-2 induction by isolated H1 flagellin was inhibited by 50% in the presence of the corresponding antiserum (Fig. 4e).
FIG. 4.
Analysis of isolated E. coli Nissle flagellin. (a) Coomassie blue staining of flagellin isolated from E. coli Nissle 1917 pellets. SDS-PAGE analysis of the purified flagellin preparation by Coomassie brilliant blue. The relevant positions of the molecular mass standard (SeeBlue Plus 2; Invitrogen) are marked on the left. Lanes 1 and 2, flagellin isolate (1 and 0.5 μg/μl); lane 3, blank. (b) Western blot of flagellin isolated from the E. coli Nissle 1917 culture supernatant. (c) Mass spectrometric analysis of isolated E. coli Nissle 1917 flagellin. The sample was diluted 1:10 in 50% acetonitrile containing 0.02% formic acid and analyzed by electrospray mass spectrometry. The determined size was 60.81 kDa. (d) Dose response of hBD-2 promoter activity by E. coli Nissle 1917 flagellin stimulation. Caco-2 cells were stimulated for 4.5 h with different concentrations of isolated flagellin. The hBD-2-inducing activity was determined by using a luciferase reporter gene assay. The relative luciferase activity is shown with the unstimulated activity of the promoter set at 1. The data are means ± the SD of three independent experiments in duplicate. (e) Effect of anti-H1 flagellin antiserum on activity of flagellin and E. coli Nissle 1917 supernatant. Isolated flagellin (0.1 μg) or E. coli Nissle 1917 supernatant (□) were mixed with H1-flagellin antiserum (1:100) in FCS-antibiotic-free DMEM and added to Caco-2 cells. The untreated flagellin and supernatant were used as a reference (▪). After 6 h of exposure, hBD-2 mRNA expression was determined by real-time PCR. Treatment of isolated flagellin or E. coli Nissle 1917 supernatant with anti-H1 flagellin antiserum reduced hBD-2 mRNA expression substantially. The data are means ± the SD of three experiments in duplicate.
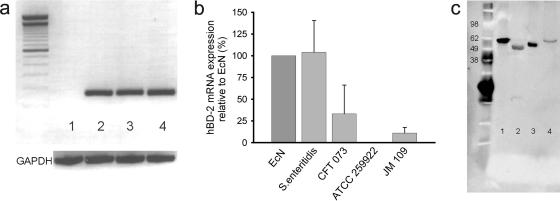
Since flagellins from different Salmonella species have been shown to induce hBD-2 expression in epithelial cells (25, 27), we isolated flagellin from Salmonella enterica serovar Enteritidis, as well as from the apathogenic E. coli strain ATCC 25922 and the E. coli strain JM109 as controls. The genome structure of E. coli Nissle 1917 was shown to resemble closely that of the uropathogenic E. coli CFT073Δhly strain (14). Hence, we compared the hBD-2-inducing capacity of the serotype-identical CFT073Δhly with that of the E. coli Nissle 1917 flagellin. Only flagellin from serovar Enteritidis induced hBD-2 mRNA significantly (Fig. 5a and b) in contrast to the flagellin of the apathogenic E. coli strain ATCC 25922 that lacked an induction. As expected, the E. coli JM109 isolate hardly induced hBD-2, whereas CFT073Δhly was able to induce hBD-2 but not as strongly as the flagellin of E. coli Nissle 1917 (Fig. 5b). The isolated flagellins were also subjected to Western blot analysis with an anti-H1 antiserum. Since serologically distinct flagella possess a highly conserved primary sequence of flagellin monomers within the exposed central region, all flagellins could be detected by Western blotting despite their different flagellin serotypes (Fig. 5c).
FIG. 5.
Flagellin isolates induce hBD-2 mRNA expression. (a) Qualitative hBD-2 mRNA expression induced by E. coli Nissle 1917 flagellin. The results of agarose gel electrophoresis (2%) of RT-PCR products from Caco-2 cells incubated for 6 h with medium (lane 1), E. coli Nissle 1917 supernatant (lane 2), E. coli Nissle 1917 flagellin (lane 3), or serovar Enteritidis flagellin (lane 4) are shown. hBD-2 fragments are given in the upper panel, GAPDH controls of each sample are shown in the bottom panel (molecular mass marker, 100 bp). The product sizes were as follows: hBD-2, 172 bp; and GAPDH, 360 bp. (b) Quantitative hBD-2 mRNA expression induced by flagellins from different bacterial strains. Caco-2 cells were stimulated for 6 h with flagellin isolates from E. coli Nissle 1917, S. enterica serovar Enteritidis, CFT073Δhly, E. coli ATCC 25922, and E. coli JM109. hBD-2 mRNA expression was assayed by RT-PCR. The data are means ± the SD of three experiments in duplicate. (c) Western blot of flagellins from different bacterial strains. Flagellins from E. coli Nissle 1917 (lane 1), serovar Enteritidis (lane 2), CFT073Δhly (lane 3), and E. coli ATCC 25922 (lane 4) were separated by electrophoresis on SDS-PAGE gels and incubated with H1 flagellin antiserum overnight.
DISCUSSION
The present study suggests that flagellin is the major factor expressed by the probiotic bacterium E. coli Nissle 1917 responsible for the induction of hBD-2 based on the following findings: (i) the factor is released into bacterial supernatant and is digestible by proteinase, (ii) genetically manipulated E. coli deficient in flagellin or the hook protein fails to induce the defensin expression, (iii) isolated flagellin from the E. coli Nissle 1917 strain is effective through the same signaling pathway as intact E. coli Nissle 1917, and (iv) the effect of isolated flagellin and of E. coli Nissle 1917 supernatant on hBD-2 induction is abrogated by H1 flagellin antiserum. Thus, the Nissle 1917 flagellin appears to be unique among most E. coli strains and mimics the similar induction capacity of flagellin from pathogenic strains, including Salmonella sp. (25).
In order to characterize the responsible factor for hBD-2 induction, we tested known genes, which code for surface expressed or secreted proteins. The genes of known fitness factors, differing from other strains, were therefore deleted to evaluate their relevance for hBD-2 expression. Among the constructed deletion mutants were flagellin-negative mutants, which turned out to be the only strains with a diminished capacity for hBD-2 promoter activation. Reinsertion of the flagellin gene reconstituted normal hBD-2 induction, strengthening the evidence that this bacterial component alone exhibits already a strong defensin-stimulating trait.
Isolation of the E. coli Nissle 1917 flagellin by previously reported methods revealed a unique protein of 60.81 kDa as determined by mass spectrometry which was detectable also with anti-H1 flagellin antiserum.
Expanding our findings regarding hBD-2 induction in Caco-2 cells by E. coli Nissle 1917 (41), we determined a higher induction potential of bacterial suspension supernatant compared to the pellet. The flagellin might be shed in the supernatant; this was probably triggered by heat treatment.
The next step was the investigation of the signaling pathway used by the stimulatory factor in the E. coli Nissle 1917 supernatant to induce hBD-2 expression. The transcription factors NF-κB and AP-1 appeared to both be necessary for full hBD-2 promoter activation by the bacterial supernatant, similar to our previous findings with the bacterial pellet (41). AP-1 might serve thereby in principal as a synergistic factor, not being able to elicit hBD-2 gene activation without the contribution of NF-κB. An NF-κB-dependent hBD-2 induction through flagellin as the assumed main stimulatory factor in the supernatant seems to be most probable since Salmonella flagellin was shown to induce excessive NF-κB activation in Caco-2BBe cells (8).
Previous experiments regarding signaling events upstream of NF-κB mediated by whole bacteria (41) revealed the explicit involvement of the JNK, whereas the supernatant factor transmitted its signal also via ERK 1/2 and to a smaller extent via p38. Our results indicate that E. coli Nissle 1917 shares with several bacterial pathogens, including Helicobacter pylori and enteropathogenic and enterohemorrhagic E. coli, the ability to activate MAP kinase signaling pathways and epithelial proinflammatory responses such as IL-8 but in a noninvasive manner. In contrast to pathogens, E. coli Nissle 1917 might induce a protective response, including hBD-2 as well as IL-8 but just below the threshold level of an active inflammation.
The isolated flagellin protein was able to induce hBD-2 in a dose-dependent fashion, further substantiating that flagellin alone is a sufficient stimulant. This effect is also not a secondary artifact due to LPS contamination, since this endotoxin lacks any hBD-2-inducing effect in Caco-2 cells (41). Moreover, different flagellin mutant strains failed to show hBD-2 gene induction. The anti-H1 flagellin antiserum displayed also immunoreactivity against the flagellin isolates of the reference strains. This serological cross-reaction might be explained by the remarkable conservation of the sequences that mediate filament assembly, which indicates that all bacterial flagellins are likely to be packed into filaments in a comparable way (2). The reduced activity of CFT073Δhly H1 flagellin to induce hBD-2 expression compared to that of E. coli Nissle 1917 may be based on posttranslational modifications, i.e., glycosylation resulting in a different electrophoretic mobility of the CFT073Δhly flagellin in the Western blot.
Similar to our observations, the filament protein flagellin (fliC gene), isolated from Salmonella, has been demonstrated to induce hBD-2 (25). However, the missing pathogenicity of E. coli Nissle 1917 favors positive effects by this strain rather than by Salmonella, despite the defensin induction characteristic of the latter. Finally, the addition of anti-H1 to Caco-2 cells suppressed flagellin-mediated hBD-2 induction by ca. 50%. This abrogation was probably incomplete because the polyclonal H1 flagellin antibody likely had a relatively low binding affinity. We conclude that flagellin is the responsible factor of E. coli Nissle 1917-mediated hBD-2 induction in Caco-2 cells. This finding is remarkable since in another probiotic bacterial mix (VSL#3) the active factor was bacterial CpG-DNA (deoxycytidylate-phosphate-deoxyguanylate), which has been demonstrated elegantly in various experimental animal models (29). Interestingly, the oral route of irradiated bacteria, as well as isolated CpG-DNA from VSL#3, was equally effective (30). Thus, the relevant factors in probiotics appear to differ from strain to strain.
Next to its main function in providing bacterial motility, the flagellum also plays a role in adhesion and biofilm formation on the mucosa (28). Flagellin, the flagellum filament structural protein, triggers further native immune responses since flagellins from Pseudomonas aeruginosa (7), S. enterica serovar Typhimurium (10), and enteroaggregative E. coli (37) have been shown to stimulate epithelial cells to express IL-8 or nitric oxide (8). Further, flagellin-deficient enteropathogenic E. coli failed to induce IL-8 secretion (43), and this capacity was restored after complementation of the fliC gene.
The signal might be transmitted by TLR5 (Toll-like receptor), which is expressed in Caco-2 cells, but other pathways may be relevant, too. In a recent investigation, we determined that NOD2, the intracellular receptor for muramyldipeptide, a fragment of bacterial peptidoglycan, is involved in the induction of hBD-2 (40). Ren et al. (32) suggests further that in macrophages which did not express TLR5 the murine Naip5 (for neuronal apoptosis inhibitory protein 5) (42) might be responsible for flagellin recognition. In addition, Ipaf, the closest homologue of Naip, has been proposed as an intracellular microbial sensor involved in Salmonella-initiated caspase-1 pathway activation (23). Whether Ipaf might also be involved in flagellin signal transduction in intestinal epithelial cells is questionable. However, the scope of the present study was the identification of the bacterial factor rather than the elucidation of the recognition mechanism(s) by intestinal epithelial cells.
In the present study, we provide the first evidence that flagellin from an apathogenic, probiotic bacterial strain specifically stimulates the expression of an antimicrobial peptide by epithelial intestinal cells. Although IL-8 is also induced by this strain in vitro, it fails to cause any inflammation in the patient but exerts clinical benefit instead (16). However, the precise mechanisms involved in preferentially triggering the protective (defensin) rather than the inflammatory (cytokine) pathways are still unclear.
Editor: A. D. O'Brien
Footnotes
Published ahead of print on 5 February 2007.
REFERENCES
- 1.Altenhoefer, A., S. Oswald, U. Sonnenborn, C. Enders, J. Schulze, J. Hacker, and T. A. Oelschlaeger. 2004. The probiotic Escherichia coli strain Nissle 1917 interferes with invasion of human intestinal epithelial cells by different enteroinvasive bacterial pathogens. FEMS Immunol. Med. Microbiol. 40:223-229. [DOI] [PubMed] [Google Scholar]
- 2.Beatson, S. A., T. Minamino, and M. J. Pallen. 2006. Variation in bacterial flagellins: from sequence to structure. Trends Microbiol. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 3.Bengmark, S. 2001. Pre-, pro-, and synbiotics. Curr. Opin. Clin. Nutr. Metab. Care 4:571-579. [DOI] [PubMed] [Google Scholar]
- 4.Bernet-Camard, M. F., V. Lievin, D. Brassart, J. R. Neeser, A. L. Servin, and S. Hudault. 1997. The human Lactobacillus acidophilus strain LA1 secretes a nonbacteriocin antibacterial substance(s) active in vitro and in vivo. Appl. Environ. Microbiol. 63:2747-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boudeau, J., A. L. Glasser, S. Julien, J. F. Colombel, and A. Darfeuille-Michaud. 2003. Inhibitory effect of probiotic Escherichia coli strain Nissle 1917 on adhesion to and invasion of intestinal epithelial cells by adherent-invasive E. coli strains isolated from patients with Crohn's disease. Aliment. Pharmacol. Ther. 18:45-56. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DiMango, E., H. J. Zar, R. Bryan, and A. Prince. 1995. Diverse Pseudomonas aeruginosa gene products stimulate respiratory epithelial cells to produce interleukin-8. J. Clin. Investig. 96:2204-2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eaves-Pyles, T., K. Murthy, L. Liaudet, L. Virag, G. Ross, F. G. Soriano, C. Szabo, and A. L. Salzman. 2001. Flagellin, a novel mediator of Salmonella-induced epithelial activation and systemic inflammation: IκBα degradation, induction of nitric oxide synthase, induction of proinflammatory mediators, and cardiovascular dysfunction. J. Immunol. 166:1248-1260. [DOI] [PubMed] [Google Scholar]
- 9.Gaon, D., H. Garcia, L. Winter, N. Rodriguez, R. Quintas, S. N. Gonzalez, and G. Oliver. 2003. Effect of Lactobacillus strains and Saccharomyces boulardii on persistent diarrhea in children. Medicina 63:293-298. [PubMed] [Google Scholar]
- 10.Gewirtz, A. T., T. A. Navas, S. Lyons, P. J. Godowski, and J. L. Madara. 2001. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167:1882-1885. [DOI] [PubMed] [Google Scholar]
- 11.Gionchetti, P., F. Rizzello, U. Helwig, A. Venturi, K. M. Lammers, P. Brigidi, B. Vitali, G. Poggioli, M. Miglioli, and M. Campieri. 2003. Prophylaxis of pouchitis onset with probiotic therapy: a double-blind, placebo-controlled trial. Gastroenterology 124:1202-1209. [DOI] [PubMed] [Google Scholar]
- 12.Gionchetti, P., F. Rizzello, A. Venturi, P. Brigidi, D. Matteuzzi, G. Bazzocchi, G. Poggioli, M. Miglioli, and M. Campieri. 2000. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology 119:305-309. [DOI] [PubMed] [Google Scholar]
- 13.Goldin, B. R. 1998. Health benefits of probiotics. Br. J. Nutr. 80:S203-S207. [PubMed] [Google Scholar]
- 14.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isolauri, E. 2001. Probiotics in human disease. Am. J. Clin. Nutr. 73:1142S-1146S. [DOI] [PubMed] [Google Scholar]
- 16.Kruis, W., P. Fric, J. Pokrotnieks, M. Lukas, B. Fixa, M. Kascak, M. A. Kamm, J. Weismueller, C. Beglinger, M. Stolte, C. Wolff, and J. Schulze. 2004. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 53:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kruis, W., E. Schutz, P. Fric, B. Fixa, G. Judmaier, and M. Stolte. 1997. Double-blind comparison of an oral Escherichia coli preparation and mesalazine in maintaining remission of ulcerative colitis. Aliment. Pharmacol. Ther. 11:853-858. [DOI] [PubMed] [Google Scholar]
- 18.Lodinova-Zadnikova, R., B. Cukrowska, and H. Tlaskalova-Hogenova. 2003. Oral administration of probiotic Escherichia coli after birth reduces frequency of allergies and repeated infections later in life (after 10 and 20 years). Int. Arch. Allergy Immunol. 131:209-211. [DOI] [PubMed] [Google Scholar]
- 19.Lodinova-Zadnikova, R., and U. Sonnenborn. 1997. Effect of preventive administration of a nonpathogenic Escherichia coli strain on the colonization of the intestine with microbial pathogens in newborn infants. Biol. Neonate 71:224-232. [DOI] [PubMed] [Google Scholar]
- 20.Mack, D. R., S. Michail, S. Wei, L. McDougall, and M. A. Hollingsworth. 1999. Probiotics inhibit enteropathogenic Escherichia coli adherence in vitro by inducing intestinal mucin gene expression. Am. J. Physiol. 276:G941-G950. [DOI] [PubMed] [Google Scholar]
- 21.Madsen, K., A. Cornish, P. Soper, C. McKaigney, H. Jijon, C. Yachimec, J. Doyle, L. Jewell, and C. De Simone. 2001. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology 121:580-591. [DOI] [PubMed] [Google Scholar]
- 22.Majamaa, H., E. Isolauri, M. Saxelin, and T. Vesikari. 1995. Lactic acid bacteria in the treatment of acute rotavirus gastroenteritis. J. Pediatr. Gastroenterol. Nutr. 20:333-338. [DOI] [PubMed] [Google Scholar]
- 23.Mariathasan, S., K. Newton, D. M. Monack, D. Vucic, D. M. French, W. P. Lee, M. Roose-Girma, S. Erickson, and V. M. Dixit. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430:213-218. [DOI] [PubMed] [Google Scholar]
- 24.McFarland, L. V., C. M. Surawicz, R. N. Greenberg, R. Fekety, G. W. Elmer, K. A. Moyer, S. A. Melcher, K. E. Bowen, J. L. Cox, and Z. Noorani. 1994. A randomized placebo-controlled trial of Saccharomyces boulardii in combination with standard antibiotics for Clostridium difficile disease. JAMA 271:1913-1918. [PubMed] [Google Scholar]
- 25.Ogushi, K., A. Wada, T. Niidome, N. Mori, K. Oishi, T. Nagatake, A. Takahashi, H. Asakura, S. Makino, H. Hojo, Y. Nakahara, M. Ohsaki, T. Hatakeyama, H. Aoyagi, H. Kurazono, J. Moss, and T. Hirayama. 2001. Salmonella enteritidis FliC (flagella filament protein) induces human beta-defensin-2 mRNA production by Caco-2 cells. J. Biol. Chem. 276:30521-30526. [DOI] [PubMed] [Google Scholar]
- 26.Olivares, M., M. P. Diaz-Ropero, N. Gomez, F. Lara-Villoslada, S. Sierra, J. A. Maldonado, R. Martin, J. M. Rodriguez, and J. Xaus. 2006. The consumption of two new probiotic strains, Lactobacillus gasseri CECT 5714 and Lactobacillus coryniformis CECT 5711, boosts the immune system of healthy humans. Int. Microbiol. 9:47-52. [PubMed] [Google Scholar]
- 27.O'Neil, D. A., E. M. Porter, D. Elewaut, G. M. Anderson, L. Eckmann, T. Ganz, and M. F. Kagnoff. 1999. Expression and regulation of the human β-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718-6724. [PubMed] [Google Scholar]
- 28.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 29.Rachmilewitz, D., F. Karmeli, K. Takabayashi, T. Hayashi, L. Leider-Trejo, J. Lee, L. M. Leoni, and E. Raz. 2002. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology 122:1428-1441. [DOI] [PubMed] [Google Scholar]
- 30.Rachmilewitz, D., K. Katakura, F. Karmeli, T. Hayashi, C. Reinus, B. Rudensky, S. Akira, K. Takeda, J. Lee, K. Takabayashi, and E. Raz. 2004. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 126:520-528. [DOI] [PubMed] [Google Scholar]
- 31.Rembacken, B. J., A. M. Snelling, P. M. Hawkey, D. M. Chalmers, and A. T. Axon. 1999. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: a randomised trial. Lancet 354:635-639. [DOI] [PubMed] [Google Scholar]
- 32.Ren, T., D. S. Zamboni, C. R. Roy, W. F. Dietrich, and R. E. Vance. 2006. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenfeldt, V., K. F. Michaelsen, M. Jakobsen, C. N. Larsen, P. L. Moller, P. Pedersen, M. Tvede, H. Weyrehter, N. H. Valerius, and A. Paerregaard. 2002. Effect of probiotic Lactobacillus strains in young children hospitalized with acute diarrhea. Pediatr. Infect. Dis. J. 21:411-416. [DOI] [PubMed] [Google Scholar]
- 34.Salminen, S., C. Bouley, M. C. Boutron-Ruault, J. H. Cummings, A. Franck, G. R. Gibson, E. Isolauri, M. C. Moreau, M. Roberfroid, and I. Rowland. 1998. Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80(Suppl. 1):S147-S171. [DOI] [PubMed] [Google Scholar]
- 35.Schiffrin, E. J., F. Rochat, H. Link-Amster, J. M. Aeschlimann, and A. Donnet-Hughes. 1995. Immunomodulation of human blood cells following the ingestion of lactic acid bacteria. J. Dairy Sci. 78:491-497. [DOI] [PubMed] [Google Scholar]
- 36.Schultz, M., U. G. Strauch, H. J. Linde, S. Watzl, F. Obermeier, C. Gottl, N. Dunger, N. Grunwald, J. Scholmerich, and H. C. Rath. 2004. Preventive effects of Escherichia coli strain Nissle 1917 on acute and chronic intestinal inflammation in two different murine models of colitis. Clin. Diagn. Lab. Immunol. 11:372-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steiner, T. S., J. P. Nataro, C. E. Poteet-Smith, J. A. Smith, and R. L. Guerrant. 2000. Enteroaggregative Escherichia coli expresses a novel flagellin that causes IL-8 release from intestinal epithelial cells. J. Clin. Investig. 105:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sturm, A., K. Rilling, D. C. Baumgart, K. Gargas, T. Abou-Ghazale, B. Raupach, J. Eckert, R. R. Schumann, C. Enders, U. Sonnenborn, B. Wiedenmann, and A. U. Dignass. 2005. Escherichia coli Nissle 1917 distinctively modulates T-cell cycling and expansion via Toll-like receptor 2 signaling. Infect. Immun. 73:1452-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun, J., F. Gunzer, A. M. Westendorf, J. Buer, M. Scharfe, M. Jarek, F. Gossling, H. Blocker, and A. P. Zeng. 2005. Genomic peculiarity of coding sequences and metabolic potential of probiotic Escherichia coli strain Nissle 1917 inferred from raw genome data. J. Biotechnol. 117:147-161. [DOI] [PubMed] [Google Scholar]
- 40.Voss, E., J. Wehkamp, K. Wehkamp, E. F. Stange, J. M. Schroder, and J. Harder. 2006. NOD2/CARD15 mediates induction of the antimicrobial peptide human beta-defensin-2. J. Biol. Chem. 281:2005-2011. [DOI] [PubMed] [Google Scholar]
- 41.Wehkamp, J., J. Harder, K. Wehkamp, B. Wehkamp-Von Meissner, M. Schlee, C. Enders, U. Sonnenborn, S. Nuding, S. Bengmark, K. Fellermann, J. M. Schroder, and E. F. Stange. 2004. NF-κB- and AP-1-mediated induction of human beta defensin-2 in intestinal epithelial cells by Escherichia coli Nissle 1917: a novel effect of a probiotic bacterium. Infect. Immun. 72:5750-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wright, E. K., S. A. Goodart, J. D. Growney, V. Hadinoto, M. G. Endrizzi, E. M. Long, K. Sadigh, A. L. Abney, I. Bernstein-Hanley, and W. F. Dietrich. 2003. Naip5 affects host susceptibility to the intracellular pathogen Legionella pneumophila. Curr. Biol. 13:27-36. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, X., J. A. Giron, A. G. Torres, J. A. Crawford, E. Negrete, S. N. Vogel, and J. B. Kaper. 2003. Flagellin of enteropathogenic Escherichia coli stimulates interleukin-8 production in T84 cells. Infect. Immun. 71:2120-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]