Abstract
Gingipains are cysteine proteinases that are responsible for the virulence of Porphyromonas gingivalis. Recent studies have shown that P. gingivalis is trapped within autophagic compartments of infected cells, where it promotes survival. In this study we investigated the role of gingipains in the intracellular trafficking and survival of this bacterium in human aortic endothelial cells and any possible involvement of these enzymes in the autophagic pathway. Although autophagic events were enhanced by infection with either wild-type (WT) P. gingivalis strains (ATCC 33277, 381, and W83) or an ATCC 33277 mutant lacking gingipains (KDP136), we have found that more than 90% of intracellular WT and KDP136 colocalized with cathepsin B, a lysosome marker, and only a few of the internalized cells colocalized with LC3, an autophagosome marker, during the 0.5- to 4-h postinfection period. This was further substantiated by immunogold electron microscopic analyses, thus implying that P. gingivalis evades the autophagic pathway and instead directly traffics to the endocytic pathway to lysosomes. At the late stages after infection, WT strains in phagolysosomes retained their double-membrane structures. KDP136 in these compartments, however, lost its double-membrane structures, representing a characteristic feature of its vulnerability to rupture. Together with the ultrastructural observations, we found that the number of intracellular viable WT cells decreased more slowly than that of KDP136 cells, thus suggesting that gingipains contribute to bacterial survival, but not to trafficking, within the infected cells.
Porphyromonas gingivalis is a gram-negative anaerobic bacterium that is strongly implicated in the etiology of progressive periodontal disease. Many epidemiological studies have demonstrated a strong association of periodontal disease with atherosclerosis and coronary heart disease (5, 11), but the mechanism underlying this association remains poorly understood. Recent evidence shows that P. gingivalis can invade and infect epithelial (10, 22, 23, 41, 42) and endothelial (8, 9) cells, thus suggesting its possible involvement in the development of atherosclerosis. The connection between P. gingivalis infection and atherosclerosis is also supported by the finding that the bacterium is detectable in human carotid and coronary atheromas (7, 13, 21). Given the frequent access of periodontal pathogens to the systemic circulation of individuals with periodontal disease through events such as tooth brushing, scaling and root planing, tooth extraction, and periodontal surgery (24), the presence of P. gingivalis in both endothelial cells and atherosclerotic plaques strongly suggests its involvement in the development of atherosclerosis. Recently, we reported that P. gingivalis infection promotes atherosclerotic progression in apolipoprotein E-knockout mice through selective proteolysis of apo-B-100 protein, a major protein component of low-density lipoprotein particles (14).
P. gingivalis is well known to produce potent cysteine proteases, termed gingipains, in both secretory and cell-associated forms (6, 38, 46, 48). Gingipains consist of Arg- and Lys-specific cysteine proteinases, now termed Arg-gingipain (Rgp) and Lys-gingipain (Kgp), respectively (25, 30-32, 34, 37). In previous studies using various P. gingivalis mutants deficient in Rgp- and/or Kgp-encoding genes and proteinase inhibitors specific for each enzyme, we demonstrated that both enzymes are responsible for most of the virulence of the bacterium, as well as its survival (1, 3, 4, 17-19, 30, 35, 44). However, whether and how gingipains are involved in the bacterial invasion process and persistence of the bacterium in human aortic endothelial cells still remain speculative.
Recently, Dorn et al. (9) reported that P. gingivalis 381 was located within vacuoles morphologically resembling autophagosomes after infection into human coronary artery endothelial cells. Those authors also demonstrated that P. gingivalis-containing autophagosome-like vacuoles were positive for the endoplasmic reticulum marker Bip and the lysosomal membrane-associated protein LGP120. However, they were not positive for the lysosomal cysteine proteinase cathepsin L, thus suggesting that P. gingivalis evaded the endocytic pathway to lysosomes but instead trafficked to autophagosomes, thereby acquiring a beneficial environment for its survival and growth. Moreover, several pathogenic bacteria, including Brucella abortus (39), Listeria monocytogenes (20) and Streptococcus pyogenes (28), have also been shown to reside within autophagic compartments. Given the importance of autophagy in the degradation of undesirable cellular components and organelles, including invading microbes, autophagic events appear to be crucial for host defense against invading microbes. However, it still remains unclear how these bacteria within the infected host cells manage to resist the host defense mechanisms, including the autophagic mechanism.
In the present study, to define the trafficking pathways of P. gingivalis in human aortic endothelial cells (HAECs), we explored whether and how the autophagic pathway is actually involved in the intracellular fate of this bacterium. We also investigated whether gingipains play a role in the intracellular fate of P. gingivalis and in the establishment of a beneficial environment for bacterial survival and growth in infected HAECs. Here we found that the internalized wild-type (WT) strains, including strains ATCC 33277, W83, and 381, and the ATCC 33277 mutant lacking gingipains (KDP136) were confined mostly to cathepsin B-positive phagolysosomes and partly to LC3-positive autophagosomes during the period from 0.5 to 4 h postinfection. Our results thus indicate that the majority of internalized P. gingivalis organisms evade the autophagic pathway and instead directly traffic to the endocytic pathway to lysosomes. We also found that gingipains are important for P. gingivalis to acquire the resistance to lysosomal destruction within the infected cells.
MATERIALS AND METHODS
HAEC culture.
HAECs were obtained from Cell Applications Inc. (San Diego, CA). The cells were maintained through the use of an Endothelial Cell Growth Medium Kit (Cell Applications Inc.) or MCDB131 medium (Sigma, St. Louis, MO) supplemented with 0.1% NaHCO3, 10% fetal bovine serum (FBS) (ICN Biomedicals, Inc., Solon, OH), 10 ng/ml recombinant acid fibroblast growth factor (Sigma), 10 μg/ml heparin (Sigma), 60 μg/ml kanamycin sulfate (Gibco, Invitrogen Corp., Carlsbad, CA), 1 μg/ml hydrocortisone (Sigma), and 10 ng/ml epidermal growth factor (Sigma) in humidified 5% CO2 at 37°C. HAECs (1 × 105 cells per well in six-well tissue culture dishes) were infected with P. gingivalis WT strains or KDP136 for 20 min at a multiplicity of infection (MOI) of 104 (persistent assay) or 103 (confocal microscopy) in MCDB131 medium without kanamycin and then cultured in the same medium in the presence of kanamycin. In some experiments, HAECs were precultured for 30 min in the presence of either 100 nM wortmannin, a phosphatidylinositol 3-kinase inhibitor, or 100 nM bafilomycin A1, a vacuolar-type proton ATPase inhibitor, and then infected with P. gingivalis WT strains or KDP136 for 20 min in the absence of each agent. After washing, the cells were further incubated for the indicated times in the presence of each agent. Under these conditions, either wortmannin or bafilomycin A1 had little or no effect on internalization of bacterial cells into HAECs.
Bacterial strains and culture conditions.
P. gingivalis ATCC 33277, W83, and 381 were used as WT strains. The ATCC 33277 mutant lacking gingipains (rgpA-, rgpB-, and kgp-deficient triple mutant KDP136) was constructed as described previously (44). The bacterial cells were grown in enriched brain heart infusion broth (37 g/liter) (Difco, BD Biosciences, San Jose, CA) supplemented with yeast extract (5 g/liter), hemin (5 mg/liter), vitamin K1 (1 mg/liter), and cysteine (1 g/liter) under anaerobic conditions (10% CO2, 10% H2, 80% N2) at 37°C. Erythromycin (10 μg/ml) and tetracycline (1 μg/ml) were also included in the medium when necessary. Under these conditions, KDP136 as well as the WT strains did not exhibit a nonspecific growth defect in vitro, as described previously (44). Bacterial cells were cultured overnight, harvested by centrifugation, washed with phosphate-buffered saline (PBS), and resuspended in kanamycin- and FBS-free MCDB131 medium. The CFU were counted on CDC anaerobe blood agar plates (BD BBL, Franklin Lakes, NJ) after anaerobic incubation at 37°C for 2 weeks. The multiplicity of infection (MOI) was computed with reference to an optical density curve at 540 nm for the known CFU. The fit of the number of the infected bacterial cells to the MOI was further confirmed by determining CFU in the individual experiments. To determine the persistence of WT strains and KDP136 in HAECs, we used an MOI of 104 as a maximal effective dose because it is necessary to infect with as many bacteria as possible without damaging the cells. All the experiments were performed with saturating MOIs.
Antibodies.
Rabbit antibodies against microtubule-associated protein 1 light chain 3 (LC3), a specific marker of autophagosomes, were prepared as described previously (49). Rabbit antibodies against rat cathepsin B, a lysosomal marker, were also prepared as described previously (29). These antibodies were used as the primary antibodies. Tetramethylrhodamine isothiocyanate- or horseradish peroxidase-conjugated goat antibodies against rabbit immunoglobulin G (from Sigma and from Biosource International. Inc., Camarillo, CA, respectively) were used as the secondary antibodies. For immunoblot analysis of the cell lysate of HAECs, monoclonal antibodies to human cathepsin B (Oncogene Research Products, San Diego, CA) or monoclonal antibodies to human cathepsin D (Transduction Lab, Lexington, KY) were used. Polyclonal antibodies specific for phospho-Akt (Ser473) and Akt were purchased from Cell Signaling Technology (Beverly, MA).
Confocal laser scanning microscopy.
HAECs were grown on glass coverslips at 1 × 105 cells per well in six-well tissue culture dishes. They were grown in 2 ml of MCDB131 medium and incubated at 37°C. The HAECs were infected with WT P. gingivalis (strains ATCC 33277, 381 and W83) or KDP136 in kanamycin- and FBS-free MCDB131 medium at an MOI of 103 bacterial cells for 20 min. The HAECs were then washed with PBS and incubated with kanamycin-containing MCDB131 medium for 0.5 to 4 h. At each time point, the medium was removed and the infected HAECs were washed with PBS and then fixed in 4% paraformaldehyde-PBS for 30 min at room temperature, followed by washing and quenching in 50 mM NH4Cl-0.3% Tween 20-PBS for 10 min. After quenching, the HAECs were washed with PBS and incubated with diluted primary antibodies (against cathepsin B or LC3) in 1% bovine serum albumin-0.2% Tween 20-PBS overnight at 4°C. HAECs were then washed with PBS containing 0.2% Tween 20, incubated with the tetramethylrhodamine isothiocyanate-conjugated secondary antibodies against rabbit immunoglobulin G (1/100 dilution in wash buffer) for 3 h at room temperature, and then washed and mounted in Vectashield (Vector Laboratories, Inc., Burlingame, CA) onto glass microscope slides. Images were observed using confocal laser scanning microscopy (Leica TCS SP LCS, Germany). All immunofluorescence examinations were performed at least three times, and the most typical images were represented. The bacterial cells were fluorescently labeled with 5 μM 2′,7′-bis(2-carboxyethyl)-5-(ans-6)-carboxyfluorescein (BCECF) (Molecular Probes, Inc., Eugene, OR) in Hanks' balanced salt solution medium for 30 min prior to infection of HAECs. Immunofluorescence images were viewed using laser-scanning confocal microscopy (Leica TCS, Leica-Microsystems, Heidelberg, Germany). At each time point, 30 fields of view, each of which (62 μm2) contained on average 1.5 HAECs, were analyzed. To determine the quantitative colocalization of each bacterial cell (green) with cathepsin B (red) or LC3 (red), completely merged yellow spots, but not partially merged spots, were counted manually in entire fields of view and expressed as percentages of the total internalized bacteria.
Transmission electron microscopy.
HAECs were infected with P. gingivalis for 20 min, washed with PBS, and fixed in 4% paraformaldehyde-0.2% glutaraldehyde-0.01% CaCl2-0.2 M HEPES-KOH (pH 7.4) for 30 min at room temperature. The cells were then washed with PBS, collected in 10% ethanol with a cell scraper, and then sedimented by centrifugation at 1,000 × g for 3 min at 20°C. The cells were buried in low-melting-point agarose (Invitrogen Corp.) and embedded in Epon. Thin sections were cut with a diamond knife using an ultramicrotome (Reichert, Vienna, Austria). Sections were contrasted with 40 mM lead citrate for 5 min and then examined with a Hitachi H7500 electron microscope (Tokyo, Japan).
Immunoelectron microscopy.
Infected HAECs were fixed in 4% paraformaldehyde-0.2% glutaraldehyde-0.01% CaCl2-0.2 M HEPES-KOH (pH 7.4) for 30 min, washed with PBS, dehydrated at −20°C, and embedded in LR White resin. Thin sections were cut and mounted on nickel grids. Sections were incubated overnight with primary antibody at 4°C, followed by incubation with protein A-gold probe (15-nm gold). After contrasting with lead citrate and 2% uranyl acetate, sections were examined with an electron microscope.
SDS-PAGE and immunoblotting.
HAECs were washed with ice-cold PBS, incubated with PBS containing 2% Triton X-100 and an inhibitor cocktail (20 μg/ml antipain, 20 μg/ml chymostatin, 20 μg/ml leupeptin, 20 μg/ml pepstatin, and 20 μg/ml phenylmethylsulfonyl fluoride) for 30 min on ice, collected by scraping, and sonicated for 10 s. The soluble fraction was collected by centrifugation at 27,000 × g for 20 min at 4°C. The resulting cell extract (200 μg of protein) was denatured in sodium dodecyl sulfate (SDS) sample buffer and subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE) on 12.5% gels. The proteins in the gels were electrophoretically transferred onto nitrocellulose membranes. The membranes were incubated with anti-LC3 antibody (3 μg/ml) overnight at 4°C after using a blocking solution of 5% skim milk. The membrane was then incubated with the horseradish peroxidase-conjugated secondary antibody for 3 h at room temperature. Bands were visualized with an enhanced chemiluminescence detection system using ECL Western blotting detection reagents (Amersham Biosciences, Buckinghamshire, England). The data obtained were subjected to quantitative analysis with an LAS1000 luminescent image analyzer using Image Gauge software version 3.4 (Fuji Photo Film, Tokyo, Japan).
Persistence assay.
The numbers of intracellular bacterial cells at the various designated times were quantified as follows. HAECs (1 × 105 cells per well in six-well tissue culture dishes) were infected with P. gingivalis WT strains (ATCC 33277 and 381) or KDP136 for 20 min at an MOI of 104. This infection time was determined based on the observation that HAECs were largely damaged by infection for more than 60 min, while no detectable damage by infection was observed within 30 min. The cells were then washed and incubated in kanamycin-containing MCDB131 medium for 2-, 6-, and 15-h periods. Before harvesting the cells, gentamicin (300 μg/ml) and metronidazole (400 μg/ml) were added to the medium for 30 min in order to kill extracellular bacterial cells (50). The infected HAECs were washed four times with PBS and then lysed in sterilized water by aspiration with 10 strokes with a 29-gauge 1/2-in. (0.33- by 13-mm) syringe (Terumo Corp., Tokyo, Japan). The lysate was sequentially 10-fold diluted and plated on CDC anaerobe blood agar plates. The colonies on the plates were counted after anaerobic incubation at 37°C for 2 weeks. The values were obtained by duplicate assays in three independent experiments.
Statistical analysis.
Quantitative data are presented as means ± standard deviations (SD). The statistical significance of differences between mean values was assessed by Student's t test. P values of <0.05 were considered statistically significant.
RESULTS
Induction of autophagy by P. gingivalis infection.
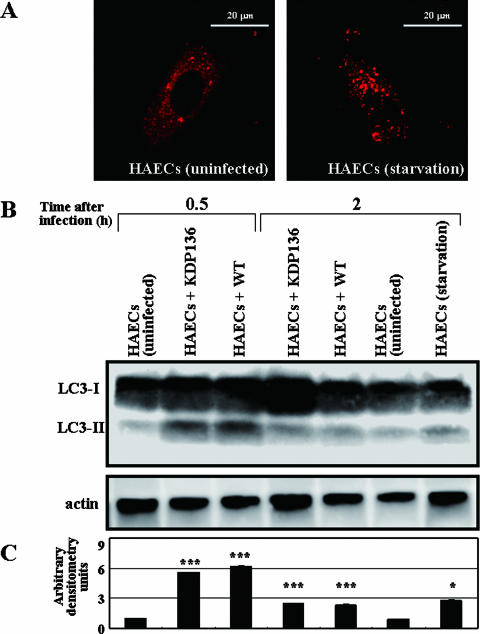
To directly examine the connection between P. gingivalis infection and autophagy stimulation, we first examined LC3-II formation in HAECs infected with P. gingivalis ATCC 33277 or its mutant lacking gingipains, KDP136. LC3 is known as a highly specific marker of autophagosomes (16, 54) and exists in two molecular forms, the 18-kDa cytosolic form (LC3-I) and the processed 16-kDa form (LC3-II) located on the autophagosomal membrane (16, 26). Thus, LC3-II formation can be used as a definitive marker of activation of the autophagic pathway. As shown in Fig. 1A, HAECs cultured under standard conditions showed mainly diffuse cytosolic staining for the anti-LC3 antibody and a partial granular staining, indicating the constitutive activation of autophagy at basal levels, even under normal culture conditions. Under conditions of starvation for 2 h, however, HAECs displayed an increased punctuate staining for the anti-LC3 antibody. This was substantiated by SDS-PAGE and immunoblot analysis (Fig. 1B). When infected with ATCC 33277 and KDP136, HAECs showed a marked increase in the LC3-II band, where amounts attained a peak value at 30 min after infection and then decreased within 2 h postinfection (Fig. 1B). Consistent with the previous finding of Dorn et al. (9), our results indicate that the autophagic machinery is obviously induced by the exposure of HAECs to WT P. gingivalis, as well as KDP136. However, the autophagic machinery induced by bacterial infection was down to basal levels at 4 h and 6 h postinfection, as evidenced by Western blotting (Fig. 1B). Our results thus suggest that the observed autophagy in HAECs at early stages of infection with either WT strains or KDP136 may simply function as a backup system for host defense mechanisms against invading pathogens.
FIG. 1.
Induction of autophagy in HAECs infected with P. gingivalis ATCC 33277 and KDP136. (A) Effect of starvation on LC3-staining patterns in HAECs. While the cells showed mainly diffuse staining for LC3, cells under starvation conditions for 2 h exhibited an increased punctuate staining for LC3, indicating activation of autophagy. (B) Immunoblot analysis of LC3-II in HAECs infected with ATCC 33277 or KDP136. Lysates of the respective infected cells were subjected to SDS-PAGE and immunoblot analysis with the anti-LC3 antibody. The LC3-II band significantly increased in HAECs infected with both ATCC 33277 and KDP136 at 0.5 h postinfection and then decreased at 2 h postinfection. The data are representative of four independent experiments. (C) A densitometric analysis for the quantification of LC3-II/LC3-I in the cell lysate of each cell type. The density was measured with an LAS1000 analyzer, and the arbitrary density unit was defined as the relative intensity of LC3-II/LC3-I obtained with uninfected HAECs. The data are the means ± SD from four independent experiments. ***, P < 0.001; *, P < 0.05 (compared with the value for the uninfected cells).
Intracellular localization of various P. gingivalis WT strains and KDP136 in infected HAECs.
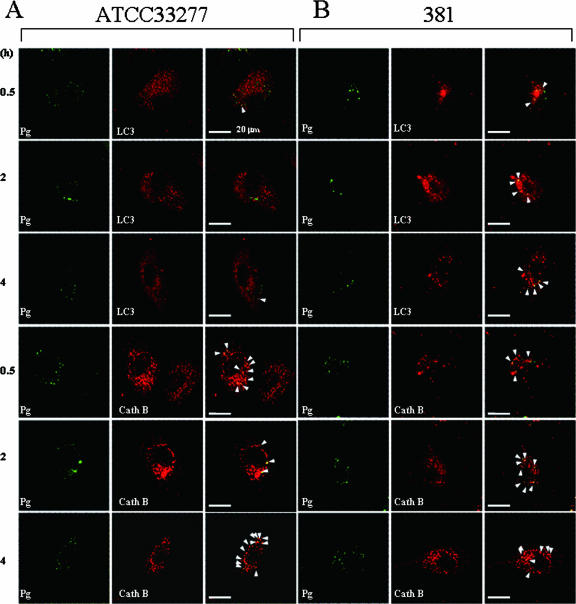
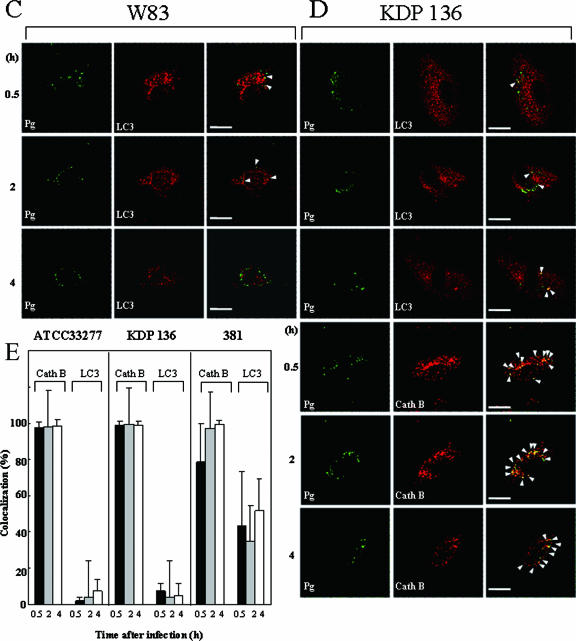
To fully understand the intracellular compartment occupied by internalized WT P. gingivalis or KDP136 in HAECs, we explored whether these bacteria directly entered the autophagic pathway or the endocytic pathway to lysosomes by using confocal microscopy. In addition to KDP136, three different WT P. gingivalis strains, ATCC 33277, 381, and W83, were used in order to test whether the different bacterial strains (possibly related to virulence) affect their trafficking and intracellular localization. We found that the internalized ATCC 33277 (Fig. 2A) was confined mainly (>90%) to cathepsin B-positive compartments and slightly (∼7%) to LC3-positive organelles in HAECs during the 0.5- to 4-h postinfection period. Similarly, the internalized 381 was confined mostly to the cathepsin B-positive compartments; however, in contrast to the ATCC 33277 and W83 strains, the intracellular 381 was to a considerable extent found in the LC3-positive compartments (Fig. 2B). On the other hand, the internalized W83, like ATCC 33277, was exclusively confined to the cathepsin B-positive organelles, and its colocalization with LC3 was very low during the 0.5- to 4-h postinfection period (Fig. 2C). Interestingly, most of the intracellular KDP136 was also found in cathepsin B-positive compartments (>90%), and only a few KDP136 organisms colocalized with LC3 in infected cells during the 0.5- to 4-h postinfection period (Fig. 2D). We further quantified the extent of colocalization between each bacterium and LC3 or cathepsin B (Fig. 2E). Within 30 min, more than 90% of the internalized ATCC 33277 and KDP136 colocalized with cathepsin B, while only a few of these bacteria colocalized with LC3. Even at 4 h postinfection, more than 90% of the intracellular ATCC 33277 strain and KDP136, as well as the internalized W83 strain (data not shown), was found in cathepsin B-positive compartments, and no significant increase in the colocalization with LC3 was observed. Although the internalized 381 strain, like ATCC 33277, W83, and KDP136, was exclusively confined to the cathepsin B-positive organelles, its colocalization with LC3 was more significant than those of the other bacterial strains during the 0.5- to 4-h postinfection period (Fig. 2E). Importantly, however, almost all the internalized 381 found in the LC3-positive compartments was also colocalized with cathepsin B, suggesting a rapid formation of autolysosomes in the infected cells. These results thus indicate that both WT strains (ATCC 33277 and W83) and KDP136, but not 381, do not directly enter the autophagic pathway in infected cells, although the autophagic machinery is effectively induced by infection with each bacterial type. However, instead of evading the autophagic pathway, they traffic exclusively through the endocytic pathway to the lysosomes directly. Our data also suggest that gingipains have little or no effect on the intracellular trafficking in HAECs infected with P. gingivalis.
FIG. 2.
Confocal microscopic images of intracellular P. gingivalis WT strains and KDP136 in infected cells. After infection with P. gingivalis (Pg) ATCC 33277 (A), 381 (B), W83 (C), and KDP136 (D) for 20 min, HAECs were washed and further cultured for the indicated times in the absence of extracellular bacteria and then stained with antibodies against LC3 or cathepsin B (CathB). Localization of each bacterial cell type (green) and LC3 or cathepsin B (red), was analyzed by confocal microscopy. Both the internalized ATCC 33277 and KDP136 were exclusively confined to cathepsin B-positive compartments, and only a few of them were colocalized with LC3 during the 0.5- to 4-h postinfection period. The WT 381 strain was also detectable mostly in cathepsin B-positive organelles and significantly in the LC3-positive compartments, most of which coincided with cathepsin B-positive organelles. The WT W83 strain was barely detectable in LC3-positive compartments during the 0.5- to 4-h postinfection period. Arrowheads indicate apparent colocalization. The data are representative of at least five independent experiments. (E) The percentages of colocalization of WT strains (ATCC 33277 and 381) and KDP136 with cathepsin B or LC3 were quantified from images obtained by confocal microscopy at each time point. Data are the means and SD from three independent experiments.
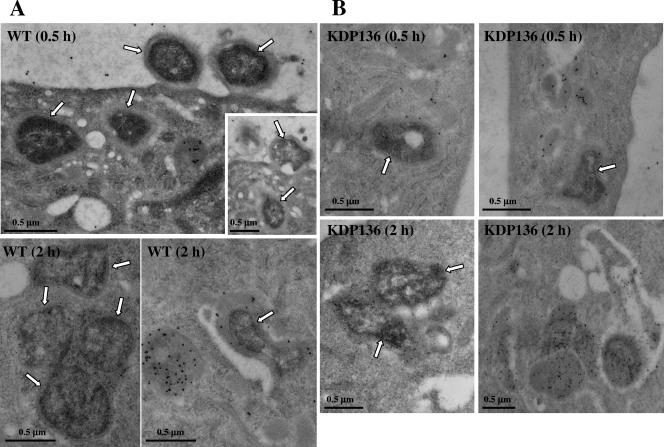
To fully understand that phagocytic membrane structures containing WT strains and KDP136 are phagolysosomes, HAECs infected with these bacteria were analyzed by immunogold electron microscopy with antibodies against cathepsin B. Within a 30-min infection, ATCC 33277 was entrapped by plasma membranes and internalized into the cells (Fig. 3A). The internalized bacteria were frequently enclosed by lamellar membrane structures. At 2 h postinfection, WT-containing compartments were mostly fused with or surrounded by cathepsin B-positive lysosomes (Fig. 3A). On the other hand, we observed KDP136-containing structures with features characteristic of phagolysosomes fused with cathepsin B-positive lysosomes within a 30-min postinfection period (Fig. 3B). At 2 h, KDP136 in cathepsin B-positive phagolysosomes exhibited loosely chaotic structures, suggesting their gradual degradation.
FIG. 3.
Immunoelectron microscopy of P. gingivalis-infected HAECs. After infection with ATCC 33277 and KDP136, HAECs were incubated in the absence of extracellular bacteria for the indicated times. The localization of cathepsin B, a lysosomal marker, was examined by immunogold electron microscopy with anti-cathepsin B antibody. (A) Within 30 min postinfection, ATCC 33277 was entrapped by plasma membranes, internalized, and frequently enclosed by lamellar membrane structures. At 2 h postinfection, WT-containing compartments mostly fused with or surrounded by cathepsin B-positive lysosomes. Arrows indicate ATCC 33277. (B) At 30 min postinfection, internalized KDP136 was found mostly in structures with features characteristic of phagolysosomes fused with cathepsin B-positive lysosomes. At 2 h, KDP136 in cathepsin B-positive phagolysosomes exhibited loosely chaotic structures, thus suggesting their gradual degradation. Arrows indicate KDP136.
Persistence of WT P. gingivalis and KDP136 in infected HAECs.
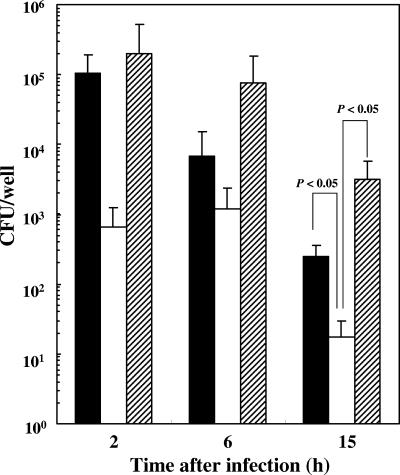
Given that the endolysosome system is responsible for the elimination of invading pathogens within cells, the efficiency of their sequestration and delivery to lysosomes is crucial for host defense. Therefore, we next examined how, and to what extent, the internalized bacteria were killed or survived after entering the phagolysosomal system. To address this question we directly scored bacterial viability by CFU. As demonstrated in Fig. 4, WT strains (ATCC 33277 and 381) appeared to be more resistant within phagolysosomes of HAECs than KDP136, although a time-dependent decrease in the extent of bacterial survival was observed with both WT strains and KDP136. We found a statistically significant difference between the numbers of intracellular ATCC 33277 or 381 and KDP136 at 15 h postinfection. The difference in internalization efficiency between WT strains and KDP136 followed by the exponential decrease appeared to make statistical analysis difficult. However, given the lower efficiency of KDP136 compared with WT strains in attachment to the cell surface of HAECs, probably due to the lack of adhesion molecules such as fimbriae on the cell surface (44), resulting in decreased internalization within the cells, the differences in the numbers of CFU between the two cell types observed at 2 and 6 h postinfection seemed to be statistically significant.
FIG. 4.
Viability of intracellular WT P. gingivalis (ATCC 33277 and 381) and KDP136 in HAECs. HAECs (1 × 105 cells per well) were infected with P. gingivalis ATCC 33277 (black bars), 381 (hatched bars), or KDP136 (white bars) for 20 min at an MOI of 104. After infection with each bacterium, HAECs were lysed at the indicated times, and the lysates were 10-fold serially diluted and plated onto CDC anaerobe blood agar plates. Data are the means ± SD of values from four independent experiments (P < 0.05 compared with the values for KDP136-infected cells).
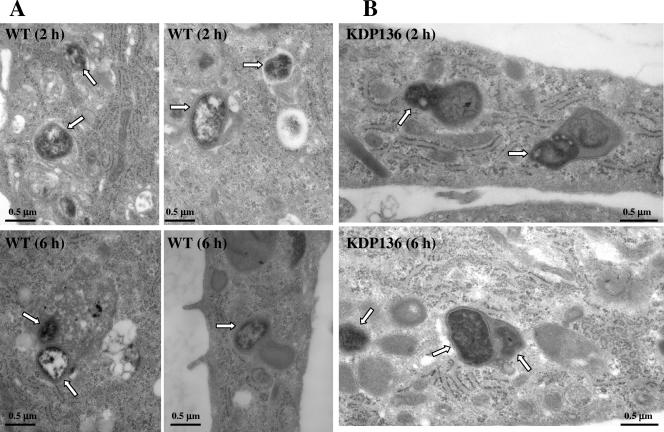
Since such differences in intracellular survival between WT strains and KDP136 were considered to be caused by differences in the ability to maintain their structural integrity within phagolysosomes, we further analyzed HAECs infected with each bacterial type by thin-section electron microscopy. At 2 h postinfection, most of the internalized WT organisms were enclosed by or fused with single or double membranous structures with features characteristic of phagolysosomes, and the bacterial double-membrane structures remained intact (Fig. 5A). Even at 6 h, the majority of WT organisms within these compartments retained intact membrane structures, although the bacterium-containing compartments became larger than those at 2 h postinfection. Some of the bacterium-containing compartments demonstrated morphology similar to that of lysosomal multivesicular bodies.
FIG. 5.
Ultrastructural observations of P. gingivalis ATCC 33277- and KDP136-infected HAECs by thin-section electron microscopy. (A) At 2 h postinfection, most of the internalized ATCC 33277 organisms were enclosed by or fused with single or double membranous structures with features characteristic of phagolysosomes, where the bacterial double-membrane structures retained intact. Even at 6 h postinfection, the majority of WT organisms within these compartments retained the intact membrane structures, some of which form morphology similar to that of lysosomal multivesicular bodies (bottom left). Arrows indicate the bacteria. (B) At 2 h postinfection, most of the internalized KDP136 organisms were entrapped by or fused with double or lamellar membranous structures with features characteristics of phagolysosomes containing some cellular components. At 6 h, KDP136-containing phagolysosomes fused with each other and became larger, where KDP136 did not show a clear bacterial structure. The arrows indicate the mutant.
In contrast, at 2 h postinfection, a larger amount of the internalized KDP136 was entrapped by or fused with double or lamellar membranous structures characteristic of phagolysosomes (secondary lysosomes) containing some cellular components, most of which lost the intact membrane structure (Fig. 5B). At 6 h postinfection, KDP136-containing phagolysosomes fused with each other and thereby became larger and chaotic structures. These observations were consistent with the results that WT strains showed longer persistence than KDP136 within the cells. Our data strongly suggest that gingipains are necessary for P. gingivalis to acquire resistance against destruction within the phagolysosomes.
Effects of wortmannin and bafilomycin A1 on the survival of WT P. gingivalis in infected HAECs.
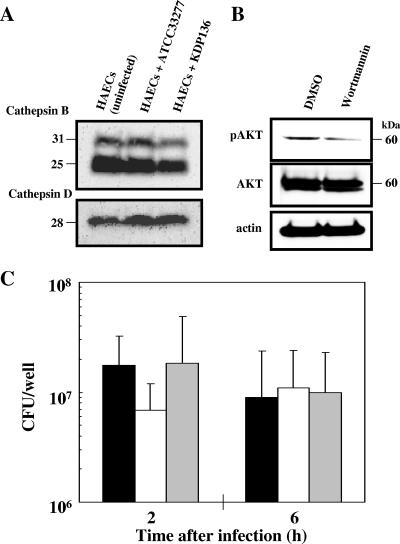
We initially observed that the protein levels of cathepsins B and D were not significantly different between control and infected HAECs with WT P. gingivalis and KDP136 (Fig. 6A). Similarly, the activity levels of V-ATPase and cathepsin L were not affected by either bacterial infection (data not shown), suggesting that the microenvironment of the phagolysosomal compartments is not affected by bacterial infection. Therefore, we next investigated how P. gingivalis managed to resist the lysosomal degradation within infected HAECs. To address this question, we examined the effects of wortmannin and bafilomycin A1 on the survival of WT P. gingivalis in infected HAECs. Since the progression of autophagy is sensitive to the phosphatidylinositol 3-kinase inhibitor wortmannin, we speculated that this agent had little effect on the survival of internalized WT bacteria, as well as the maturation of bacterium-engulfed phagosomes. Indeed, although this agent inhibited the phosphorylation of Akt in HAECs at a concentration of 100 nM (Fig. 6B), we found no significant difference in the survival of intracellular WT organisms between wortmannin-treated and nontreated cells throughout the 2- to 6-h postinfection period (Fig. 6C). This indicated that the suppression of autophagy had little effect on the intracellular fate and the viability of intracellular P. gingivalis. Meanwhile, the enhanced lysosomal pH upon treatment with bafilomycin A1 has been demonstrated to cause enhanced secretion of soluble lysosomal hydrolases (33), thereby resulting in a disruption of lysosomal functions. Therefore, we thought that the enhanced lysosomal pH caused by bafilomycin A1 might affect the trafficking and fate of the internalized bacterial cells in HAECs. As a control experiment, we confirmed that 100 nM bafilomycin A1 strongly inhibited intracellular vacuolar-type proton ATPase in HAECs. When HAECs were infected with WT strains in the presence and absence of 100 nM bafilomycin A1, it was also found that this agent had little or no effect on the viability of intracellular bacteria. These results suggest that P. gingivalis was killed by yet-unidentified mechanisms after entering the phagolysosome system of infected HAECs. These may include the reactive oxygen- and nitrogen-dependent pathways, as well as hydrolysis by neutral lysosomal hydrolases, rather than proteolysis by acidic cathepsins.
FIG. 6.
Effects of wortmannin and bafilomycin A1 on viability of intracellular P. gingivalis ATCC 33277 in HAECs. (A) SDS-PAGE and immunoblot analysis of the intracellular protein levels of cathepsins B and D in uninfected and infected HAECs with ATCC 33277 or KDP136. (B) Effect of wortmannin on phosphorylation of Akt in HAECs. After treatment with wortmannin (100 nM) for 2 h, the cell lysate was subjected to SDS-PAGE and immunoblot analysis. Phosphorylation of Akt was inhibited by this agent. (C) HAECs were cultured in the absence of agent (black bars) or in the presence of wortmannin (100 nM) (white bars) or bafilomycin A1 (100 nM) (gray bars) and then infected with ATCC 33277 for 20 min in the absence of each agent and further incubated in the presence of each agent for the indicated times. The colonies on CDC anaerobe blood agar plates after anaerobic incubation at 37°C for 2 weeks were counted. The data are the means ± SD of values from four independent experiments.
DISCUSSION
P. gingivalis has been reported to invade various human cell types, including gingival epithelial cells (10, 22, 41, 42) and endothelial cells (8, 9). Based on the previous findings that P. gingivalis is localized in various cellular compartments such as the cytoplasm (23, 36, 42, 43), endosomes (36, 42, 43), and autophagosomes (9), it has been suggested that the intracellular localization of this bacterium may be host cell specific. Dorn et al. (9) also reported that P. gingivalis 381 could evade the endocytic pathway to lysosomes and instead was targeted to the autophagosome shortly after invasion of human coronary artery endothelial cells, thereby suggesting the importance of this compartment as a site for the multiplication of the bacterium. In addition, several bacteria, including Brucella abortus (39) and Legionella pneumophila (47), were also shown to use the autophagic pathway for their replication. Therefore, the induction of autophagy in HAECs by the P. gingivalis infection may be crucial for bacterial survival in the infected cells. Given that the variable nature of the intracellular fate and survival of bacteria is compounded with virulence and structural/functional conditions, it is of particular importance for us to clarify the intracellular destination of various P. gingivalis strains, especially any possible involvement of autophagy, and the role of gingipains in this process. To clarify this process, we first investigated whether P. gingivalis WT strains and the mutant KDP136 lacking gingipains were targeted to the autophagosome in such a way as to evade the endocytic pathway to lysosomes. Our results demonstrate that not only WT P. gingivalis strains with various virulences (ATCC 33277, 381, and W83) but also KDP136 directly enter the endocytic pathway to lysosomes in the infected HAECs, but also that their trafficking to the autophagosome is minor. Therefore, the targeting of P. gingivalis to the phagolysosome is more likely to be independent of the bacterial strains and the presence of gingipains. Given the lack of the adhesion molecules of P. gingivalis, including fimbriae, in KDP136 (17, 31), the targeting to the phagolysosome is also considered to be independent of the bacterial cell structures, in spite of the fact that the efficiency of KDP136 in internalizing the host cells was markedly lower than that of WT strains. This is most probably due to P. gingivalis initiating its entry into host cells through an interaction between α5β1 integrin molecules of host cells and fimbriae (27, 53).
Nevertheless, the autophagic machinery was effectively induced in HAECs by infection with each type of P. gingivalis, as seen by the LC3-II formation that was visible during Western blotting and confocal microscopic analysis. To date, a number of studies have reported that several pathogenic bacteria, including Rickettsia conorii (51), Brucella abortus (39), Legionella pneumophila (47), and Listeria monocytogenes (40), can reside within autophagic compartments in infected host cells, although the significance of this localization remains unclear. It has also recently been suggested that the autophagic mechanism functions as an innate defense system against invading pathogens (28). Indeed, Listeria monocytogenes (12) and group A Streptococcus (28), which can escape from the phagosome into the cytoplasm with the aid of hemolysin and streptolysin, were shown to be captured and eliminated by the autophagosome system. Therefore, the observed activation of autophagy by P. gingivalis infection is likely to act as a backup system for host defense mechanisms against invading pathogens. Intriguingly, almost parallel to LC3-II formation, we also found that HAECs infected with either WT strains or KDP136 rapidly activated the Akt signaling pathway, which is thought to be triggered as a negative autophagy control system (2, 52). Considering that autophagy is crucial not only for cell survival but also for cell damage (45), including cancer, muscular disorders, neurodegeneration, and pathogen infection, activation of the Akt signaling pathway may serve to maintain cellular homeostasis by balancing between the beneficial and negative effects of autophagy. Although our results seem to be inconsistent with those found by Dorn et al. (9), this discrepancy may be due to the different experimental systems used, e.g., differences in host cell lines and culture and infection conditions.
To our knowledge, this is the first report demonstrating that the intracellular persistence of P. gingivalis WT is markedly greater than that of KDP136, indicating that gingipains appear to invest the bacteria with a strategy against killing by host cells. After entry into HAECs, either WT P. gingivalis or KDP136 resided in plasma membrane-derived vacuoles (immature phagosomes), which in turn are surrounded by and fused with lysosomes (mature phagolysosomes), thereby killing the internalized bacteria inside these compartments. By electron microscopy, however, we observed that the maturation of WT-containing phagosomes into phagolysosomes, thereby acquiring the cytolytic environment of lysosomes, appeared to be slower than that of KDP136-containing phagosomes. We also found that WT organisms inside phagolysosomes maintained the intact double-membrane structures much longer than KDP136. These observations were consistent with the finding that intracellular WT organisms persisted longer than KDP136. Cumulatively, our present data indicate that P. gingivalis gingipain deficiency enhances its susceptibility to destruction inside the phagolysosome compartments. This suggests that gingipains are critical for the bacterium to survive within the cells. To fully understand the identification and function of specific gingipain proteins (rgpA, rgpB, and kgp gene products), it is absolutely necessary to carry out experiments with genetically defined P. gingivalis mutants lacking rgpA and/or rgpB, as well as kgp, or with inhibitors specific for Rgp and Kgp.
The next logical step was to examine the mechanism by which P. gingivalis was killed after entering the phagolysosome system. To address this question, we examined the effects of wortmannin and bafilomycin A1 on viability of WT and KDP136 in these compartments. It is known that wortmannin blocks the P. gingivalis trafficking to autophagosomes and enhances phagolysosome biosynthesis (9), while bafilomycin A1 blocks the phagosome-lysosome fusion (15). Our results showed that the numbers of viable bacteria in cells treated with each agent did not differ from those in nontreated cells. Since there were no significant differences in the protein levels of cathepsins D and B and the activity levels of cathepsin L and V-ATPase between WT- and KDP136-infected HAECs, we speculate that gingipains have no effect on the microenvironment of the phagolysosome compartments. Accordingly, the exact mechanism by which gingipains render P. gingivalis resistant to phagolysosomal killing in infected cells still remains speculative. Once within the cell, P. gingivalis may use gingipains to reduce the cellular response to the pathogen. Considering the numerous observations of generation of reactive oxygen species linked with bacterial infection, gingipains may therefore play a role in the suppression of reactive oxygen species generation.
Editor: J. B. Bliska
Footnotes
Published ahead of print on 12 February 2007.
REFERENCES
- 1.Abe, N., A. Baba, R. Takii, K. Nakayama, A. Kamaguchi, Y. Shibata, Y. Abiko, K. Okamoto, T. Kadowaki, and K. Yamamoto. 2004. Roles of Arg- and Lys-gingipains in coaggregation of Porphyromonas gingivalis: identification of its responsible molecules in translation products of rgpA, kgp, and hagA genes. Biol. Chem. 385:1041-1047. [DOI] [PubMed] [Google Scholar]
- 2.Arico, S., A. Petiot, C. Bauvy, P. F. Dubbelhuis, A. J. Meijer, P. Codogno, and E. Ogier-Denis. 2001. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 276:35243-35246. [DOI] [PubMed] [Google Scholar]
- 3.Baba, A., N. Abe, T. Kadowaki, H. Nakanishi, M. Ohishi, T. Asao, and K. Yamamoto. 2001. Arg-gingipain is responsible for the degradation of cell adhesion molecules of human gingival fibroblasts and their death induced by Porphyromonas gingivalis. Biol. Chem. 382:817-824. [DOI] [PubMed] [Google Scholar]
- 4.Baba, A., T. Kadowaki, T. Asao, and K. Yamamoto. 2002. Roles of Arg- and Lys-gingipains in the disruption of cytokine responses and loss of viability of human endothelial cells by Porphyromonas gingivalis infection. Biol. Chem. 383:1223-1230. [DOI] [PubMed] [Google Scholar]
- 5.Beck, J., R. Garcia, G. Heiss, P. S. Vokonas, and S. Offenbacher. 1996. Periodontal disease and cardiovascular disease. J. Periodontol. 67:1123-1137. [DOI] [PubMed] [Google Scholar]
- 6.Bhogal, P. S., N. Slakeski, and E. C. Reynolds. 1997. A cell-associated protein complex of Porphyromonas gingivalis W50 composed of Arg- and Lys-specific cysteine proteinases and adhesins. Microbiology 143:2485-2495. [DOI] [PubMed] [Google Scholar]
- 7.Chiu, B. 1999. Multiple infections in carotid atherosclerotic plaques. Am. Heart J. 138:S534-S536. [DOI] [PubMed] [Google Scholar]
- 8.Deshpande, R. G., M. B. Khan, and C. A. Genco. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66:5337-5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 2001. Porphyromonas gingivalis traffics to autophagosomes in human coronary artery endothelial cells. Infect. Immun. 69:5698-5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duncan, M. J., S. Nakao, Z. Skobe, and H. Xie. 1993. Interactions of Porphyromonas gingivalis with epithelial cells. Infect. Immun. 61:2260-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Genco, R. J. 1998. Periodontal disease and risk for myocardial infection and cardiovascular disease. Cardiovasc. Rev. Rep. 19:34-40. [Google Scholar]
- 12.Gouin, E., H. Gantelet, C. Egile, I. Lasa, H. Ohayon, V. Villiers, P. Gounon, P. J. Sansonetti, and P. Cossart. 1999. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J. Cell Sci. 112:1697-1708. [DOI] [PubMed] [Google Scholar]
- 13.Haraszthy, V. I., J. J. Zambon, M. Trevisan, M. Zeid, and R. J. Genco. 2000. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 71:1554-1560. [DOI] [PubMed] [Google Scholar]
- 14.Hashimoto, M., T. Kadowaki, T. Tsukuba, and K. Yamamoto. 2006. Selective proteolysis of apolipoprotein B-100 by Arg-gingipain mediates atherosclerosis progression by bacterial exposure. J. Biochem. 140:713-723. [DOI] [PubMed] [Google Scholar]
- 15.Horwitz, M. A., and F. R. Maxfield. 1984. Legionella pneumophila inhibits acidification of its phagosome in human monocytes. J. Cell Biol. 99:1936-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kabeya, Y., N. Mizushima, T. Ueno, A. Yamamoto, T. Kirisako, T. Noda, E. Kominami, Y. Ohsumi, and T. Yoshimori. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadowaki, T., K. Nakayama, F. Yoshimura, K. Okamoto, N. Abe, and K. Yamamoto. 1998. Arg-gingipain acts as a major processing enzyme for various cell surface proteins in Porphyromonas gingivalis. J. Biol. Chem. 273:29072-29076. [DOI] [PubMed] [Google Scholar]
- 18.Kadowaki, T., and K. Yamamoto. 2003. Suppression of virulence of Porphyromonas gingivalis by potent inhibitors specific for gingipains. Curr. Prot. Pep. Sci. 4:451-458. [DOI] [PubMed] [Google Scholar]
- 19.Kadowaki, T., A. Baba, N. Abe, R. Takii, M. Hashimoto, T. Tsukuba, S. Okazaki, Y. Suda, T. Asao, and K. Yamamoto. 2004. Suppression of pathogenicity of Porphyromonas gingivalis by newly developed gingipain inhibitors. Mol. Pharmacol. 66:1599-1606. [DOI] [PubMed] [Google Scholar]
- 20.Kirkegaard, K., M. P. Taylor, and W. T. Jackson. 2004. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2:301-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozarov, E. V., B. R. Dorn, C. E. Shelburne, W. A. Dunn, Jr., and A. Progulske-Fox. 2005. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler. Thromb. Vasc. Biol. 25:e17-e18. [DOI] [PubMed] [Google Scholar]
- 22.Lamont, R. J., D. Oda, R. E. Persson, and G. R. Persson. 1992. Interaction of Porphyromonas gingivalis with gingival epithelial cells maintained in culture. Oral Microbiol. Immunol. 7:364-367. [DOI] [PubMed] [Google Scholar]
- 23.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 63:3878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lofthus, J. E., M. Y. Waki, D. L. Jolkovsky, J. Otomo-Corgel, M. G. Newman, T. Flemming, and S. Nachnani. 1991. Bacteremia following subgingival irrigation and scaling and root planing. J. Periodontol. 62:602-607. [DOI] [PubMed] [Google Scholar]
- 25.Mikolajczyk-Pawlinska, J., T. Kordula, N. Pavloff, P. A. Pemberton, W. C. Chen, J. Travis, and J. Potempa. 1998. Genetic variation of Porphyromonas gingivalis genes encoding gingipains, cysteine proteinases with arginine or lysine specificity. Biol. Chem. 379:205-211. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima, N., A. Yamamoto, M. Hatano, Y. Kobayashi, Y. Kabeya, K. Suzuki, T. Tokuhisa, Y. Ohsumi, and T. Yoshimori. 2001. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakagawa, I., A. Amano, H. Inaba, S. Kawai, and S. Hamada. 2005. Inhibitory effects of Porphyromonas gingivalis fimbriae on interactions between extracellular matrix proteins and cellular integrins. Microbes Infect. 7:157-163. [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa, I., A. Amano, N. Mizushima, A. Yamamoto, H. Yamaguchi, T. Kamimoto, A. Nara, J. Funao, M. Nakata, K. Tsuda, S. Hamada, and T. Yoshimori. 2004. Autophagy defends cells against invading group A Streptococcus. Science 306:1037-1040. [DOI] [PubMed] [Google Scholar]
- 29.Nakanishi, H., K. Tominaga, T. Amano, I. Hirotsu, T. Inoue, and K. Yamamoto. 1994. Age-related changes in activities and localizations of cathepsins D, E, B, and L in the rat brain tissues. Exp. Neurol. 126:119-128. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama, K., T. Kadowaki, K. Okamoto, and K. Yamamoto. 1995. Construction and characterization of arginine-specific cysteine proteinase (Arg-gingipain)-deficient mutants of Porphyromonas gingivalis. Evidence for significant contribution of Arg-gingipain to virulence. J. Biol. Chem. 270:23619-23626. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama, K., F. Yoshimura, T. Kadowaki, and K. Yamamoto. 1996. Involvement of arginine-specific cysteine proteinase (Arg-gingipain) in fimbriation of Porphyromonas gingivalis. J. Bacteriol. 178:2818-2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakayama, K. 1997. Domain-specific rearrangement between the two Arg-gingipain-encoding genes in Porphyromonas gingivalis: possible involvement of nonreciprocal recombination. Microbiol. Immunol. 41:185-196. [DOI] [PubMed] [Google Scholar]
- 33.Oda, K., Y. Nishimura, Y. Ikehara, and K. Kato. 1991. Bafilomycin A1 inhibits the targeting of lysosomal acid hydrolases in cultured hepatocytes. Biochem. Biophys. Res. Commun. 178:369-377. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto, K., Y. Misumi, T. Kadowaki, M. Yoneda, K. Yamamoto, and Y. Ikehara. 1995. Structural characterization of argingipain, a novel arginine-specific cysteine proteinase as a major periodontal pathogenic factor from Porphyromonas gingivalis. Arch. Biochem. Biophys. 316:917-925. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto, K., K. Nakayama, T. Kadowaki, N. Abe, D. B. Ratnayake, and K. Yamamoto. 1998. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis. J. Biol. Chem. 273:21225-21231. [DOI] [PubMed] [Google Scholar]
- 36.Papapanou, P. N., J. Sandros, K. Lindberg, M. J. Duncan, R. Niederman, and U. Nannmark. 1994. Porphyromonas gingivalis may multiply and advance within stratified human junctional epithelium in vitro. J. Periodontal Res. 29:374-375. [DOI] [PubMed] [Google Scholar]
- 37.Pavloff, N., J. Potempa, R. N. Pike, V. Prochazka, M. C. Kiefer, J. Travis, and P. J. Barr. 1995. Molecular cloning and structural characterization of the Arg-gingipain proteinase of Porphyromonas gingivalis. J. Biol. Chem. 270:1007-1010. [DOI] [PubMed] [Google Scholar]
- 38.Pike, R. N., J. Potempa, W. McGraw, T. H. Coetzer, and J. Travis. 1996. Characterization of the binding activities of proteinase-adhesin complexes from Porphyromonas gingivalis. J. Bacteriol. 178:2876-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pizarro-Cerda, J., S. Meresse, R. G. Parton, G. van der Goot, A. Sola-Landa, I. Lopez-Goni, E. Moreno, and J. P. Gorvel. 1998. Brucella abortus transits through the autophagic pathway and replicates in the endoplasmic reticulum of nonprofessional phagocytes. Infect. Immun. 66:5711-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rich, K. A., C. Burkett, and P. Webster. 2003. Cytoplasmic bacteria can be targets for autophagy. Cell. Microbiol. 5:455-468. [DOI] [PubMed] [Google Scholar]
- 41.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2001. Intracellular Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in buccal epithelial cells collected from human subjects. Infect. Immun. 69:2700-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandros, J., P. Papapanou, and G. Dahlen. 1993. Porphyromonas gingivalis invades oral epithelial cells in vitro. J. Periodontal Res. 28:219-226. [DOI] [PubMed] [Google Scholar]
- 43.Sandros, J., P. N. Papapanou, U. Nannmark, and G. Dahlen. 1994. Porphyromonas gingivalis invades human pocket epithelium in vitro. J. Periodontal Res. 29:62-69. [DOI] [PubMed] [Google Scholar]
- 44.Shi, Y., D. B. Ratnayake, K. Okamoto, N. Abe, K. Yamamoto, and K. Nakayama. 1999. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. Construction of mutants with a combination of rgpA, rgpB, kgp, and hagA. J. Biol. Chem. 274:17955-17960. [DOI] [PubMed] [Google Scholar]
- 45.Shintani, T., and D. J. Klionsky. 2004. Autophagy in health and disease: a double-edged sword. Science 306:990-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Slakeski, N., P. S. Bhogal, N. M. O'Brien-Simpson, and E. C. Reynolds. 1998. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology 144:1583-1592. [DOI] [PubMed] [Google Scholar]
- 47.Sturgill-Koszycki, S., and M. S. Swanson. 2000. Legionella pneumophila replication vacuoles mature into acidic, endocytic organelles. J. Exp. Med. 192:1261-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takii, R., T. Kadowaki, A. Baba, T. Tsukuba, and K. Yamamoto. 2005. A functional virulence complex composed of gingipains, adhesins, and lipopolysaccharide shows high affinity to host cells and matrix proteins and escapes recognition by host immune systems. Infect. Immun. 73:883-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanida, I., Y. S. Sou, J. Ezaki, N. Minematsu-Ikeguchi, T. Ueno, and E. Kominami. 2004. HsAtg4B/HsApg4B/Autophagin-1 cleaves the carboxyl termini of three human Atg8 homologues and delipidates microtubule-associated protein light chain 3- and GABAA receptor-associated protein-phsospholipid conjugates. J. Biol. Chem. 279:36268-36276. [DOI] [PubMed] [Google Scholar]
- 50.Ueshima, J., M. Shoji, D. B. Ratnayake, K. Abe, S. Yoshida, K. Yamamoto, and K. Nakayama. 2003. Purification, gene cloning, gene expression, and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 71:1170-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walker, D. H., V. L. Popov, P. A. Crocquet-Valdes, C. J. Welsh, and H. M. Feng. 1997. Cytokine-induced, nitric oxide-dependent, intracellular antirickettsial activity of mouse endothelial cells. Lab. Investig. 76:129-138. [PubMed] [Google Scholar]
- 52.Yilmaz, O., T. Jungas, P. Verbeke, and D. M. Ojcius. 2004. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect. Immun. 72:3743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yilmaz, O., K. Watanabe, and R. J. Lamont. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell. Microbiol. 4:305-314. [DOI] [PubMed] [Google Scholar]
- 54.Yoshimori, T. 2004. Autophagy: a regulated bulk degradation process inside cells. Biochem. Biophys. Res. Commun. 313:453-458. [DOI] [PubMed] [Google Scholar]