Abstract
A need exists for the development of applicable surveillance tools to detect fluoroquinolone-resistant Neisseria gonorrhoeae (QRNG) in urine samples. We describe here a real-time PCR assay for detecting mutations in the Ser91 codon of the gyrA gene of N. gonorrhoeae in urine specimens. We tested 96 urine samples collected along with Gonorrhea Isolate Surveillance Project (GISP) urethral swab samples and compared the results with matched MICs of ciprofloxacin, as reported by the regional GISP laboratory. We then tested 100 urine specimens, known to be gonorrhea positive by nucleic acid amplification testing, provided by females to challenge the real-time PCR assay with urine specimens containing potentially less target DNA content than specimens from symptomatic males. With an MIC threshold of 0.125 μg of ciprofloxacin/ml, our assay correctly identified resistance in 41 of 44 (93.2%; 95% confidence interval [CI] = 81.3 to 98.6%) corresponding resistant culture specimens and correctly identified 51 of 51 (100%; 95% CI = 93.0 to 100%) susceptible specimens. One specimen did not amplify. The assay successfully amplified the gyrA amplicon and determined a susceptibility genotype in 72 of 100 (72%) urine specimens collected from female patients. We developed an assay for detecting QRNG in urine specimens that correlated well with MIC results of cultured specimens and had moderate sensitivity with urine specimens. This methodology might fulfill the need for a QRNG detection system for urine specimens, a useful characteristic in the age of nucleic acid amplification testing for gonococcal infection.
Neisseria gonorrhoeae urogenital infection is an important cause of morbidity, including pelvic inflammatory disease and infertility. Early detection and appropriate treatment of N. gonorrhoeae infection prevents a substantial proportion of this morbidity. Although fluoroquinolones have been used as an effective treatment of N. gonorrhoeae infection in the face of widespread penicillin resistance, a growing body of literature has documented the emergence and rapid increase in detecting fluoroquinolone-resistance in N. gonorrhoeae (QRNG) isolates in the United States, Asia, Western Europe, and Israel (1, 2, 8, 16, 17). In the United States, fewer than 1% of isolates were QRNG in 1996 compared to more than 7.5% in 2004. However, there are considerable geographic differences in the United States, with cities in the western regions reporting rates of resistance prevalence of more than 20% (6). In response to the worldwide dissemination of resistance, the Centers for Disease Control and Prevention has recently begun recommending that all local health clinics with the capacity to do so begin routine surveillance of fluoroquinolone resistance in gonorrhea isolates (15).
Although culture and susceptibility testing of N. gonorrhoeae isolates continues to be the gold standard method for the detection of antimicrobial resistance, decreases in the use of culture for diagnosis has precipitated the need for alternative methods to detect resistance. In response, methods for detecting QRNG have been developed and reported in the scientific literature that include sequencing (12), microarray biochip (5, 13, 19), and other molecular techniques (9, 11). However, each of these techniques has disadvantages with regard to detecting resistance in certain settings. The sequencing and biochip methods might be untenable in locales with limited resources. Real-time PCR techniques using TaqMan-style probes (Applied Biosystems, Foster City, CA) exclusively detect wild-type strains, with negative results interpreted as presumed resistant strains (9). The lack of internal control might therefore indicate resistance when, in actuality, laboratory error or other causes of failed wild-type amplification (e.g., low concentrations of DNA) has led to this result. Alternatively, another real-time PCR assay for QRNG detection using fluorescence resonance energy transfer (FRET) probes and melting-curve analysis addresses these problems (11). However, that technique was not demonstrably effective for use with urine specimens in our laboratory. Hence, such assays might be insensitive to the lower concentrations of DNA in urine specimens. We sought to develop a modified version of the melting-curve-based QRNG detection that might be applicable in settings with limited resources and with adequate sensitivity to survey for resistance in urine specimens.
We present here a real-time PCR assay for analysis of mutations in the Ser91 region of the gyrA gene by amplification and melting-curve analysis with gene-specific primers and probes. More than 99% of QRNG specimens have been shown to have mutations at this site of the gyrA gene (18, 19). This method allows for rapid detection of QRNG without complex laboratory techniques or extensive resources.
MATERIALS AND METHODS
Sample collection.
We first selected 96 urine samples collected from male patients of the San Francisco City Clinic who had also provided specimens that were analyzed as part of the Gonorrhea Isolate Surveillance Project (GISP) from 31 January to 30 November 2005. The goals and methodology of the GISP project have been described previously (6). Briefly, the protocol for the GISP study entails urethral swab specimen collection of the first 25 to 30 symptomatic patients with N. gonorrhoeae urethritis to arrive at the San Francisco City Clinic each calendar month. The swab specimens were isolated initially on Thayer-Martin agar plates and then were subcultured onto chocolate agar plates for 24 h. Colonies were collected from plates and placed into tryptic soy broth with 20% glycerol for freezer storage. Specimens were sent to the regional GISP laboratory in Seattle, WA, for culture and sensitivity testing.
We selected an additional 100 urine specimens from N. gonorrhoeae-infected female patients who were tested as part of routine clinical care. These samples were included in the study to assess the sensitivity of the assay to detect resistance in urine samples with lower levels of DNA content compared to urine samples from males with urethral discharge. All urine specimens tested positive for N. gonorrhoeae at the San Francisco Department of Public Health Laboratory by transcription-mediated amplification by using a TIGRIS system (GenProbe, San Diego, CA). Urine specimens were collected and stored in APTIMA Combo 2 (GenProbe) buffer at −35°C. Selected specimens were collected on the same day as the urethral swab collection for GISP analysis. All specimens in the study were collected and tested as part of routine public health surveillance in accordance with San Francisco Department of Public Health guidelines. Because we studied preexisting diagnostic specimens, the present study was not considered human subjects research in accordance with U.S. Department of Health and Human Services guidelines (14).
Real-time PCR analysis.
For real-time PCR analysis of the gyrA gene, urine specimens (200 μl) were extracted by using MagNAPure LC automated extraction techniques (Roche Diagnostics, Indianapolis, IN) with a MagNAPure LC total nucleic acid isolation kit. Extracted samples (5 μl) were used for real-time PCR analyses. Amplification and melting-curve analysis of specimens was performed by using a Roche LightCycler 2.0 (Roche Diagnostics). Amplified regions of the gyrA gene were probed for mutations in the Ser 91 codon through use of melting-curve analysis with probes specific to that region of the gyrA gene (Table 1) . We originally tested 10 urine specimens with the real-time PCR assay developed and published by Li et al. (11) but found crossing points to be suboptimal. The subsequent primers and probes (synthesized by SIGMA-Proligo, The Woodlands, TX) used in the present study were selected from prior published reports of similar assays and confirmed by using online BLAST analysis from the National Center for Biotechnology Information (National Institutes of Health, Bethesda, MD) (9, 11). The probes for the Ser91 amplicon (gyrA-ser-Flu and gyrA-ser-LC) are a paired set of FRET probes, including a LightCycler Red640 probe and a fluorescein-labeled probe, separated by 1 bp. The Red640 oligonucleotide has a maximal absorption and emissions of 622 and 638 nm, respectively.
TABLE 1.
Primers and real-time PCR FRET probe oligonucleotide sequences
| Primer or probea | Oligonucleotide sequence | Position |
|---|---|---|
| Primers | ||
| NG-GYRASER91-F* | 5′-CGC-GAT-GCA-CGA-GCT-GAA-AAA-3′ | 174-194 |
| NG-GYRASER91-R† | 5′-TTG-CGC-CAT-ACG-GAC-GAT-3′ | 289-306 |
| Probes | ||
| gyrA-ser-Flu* | 5′-GCA-TCG-TCG-GCG-ACG-TCA-TCG-GTA-AAT-ACC-ACC-C-3′-fluorescein | 227-261 |
| gyrA-ser-LC* | 5′-Red 640-ACG-GCG-ATT-CCG-CAG-TT-3′-phosphorylated | 263-279 |
Reactions were prepared (per reaction) by using 2 μl of LightCycler FastStart DNA Master HybProbe mix (Roche Diagnostics), 0.5 μmol (0.5 μl) each of forward and reverse primers, 0.2 μmol (0.2 μl) each of the fluorescein and Red 640 probes, 2.4 μl of 25 mmol MgCl2, 9.2 μl of H2O, and 5 μl of extracted, purified specimen for a final volume of 20 μl. The LightCycler amplification protocol included the following steps: denaturation at 95°C for 5 min, followed by 45 cycles of denaturation at 95°C for 5 s, annealing at 52°C for 5 s, and extension at 72°C for 10 s, with a ramp rate of 20°C/s. Next, we performed melting-curve analysis by denaturation at 95°C for 0 s, annealing probes at 40°C for 5 s, and heating the mixtures to 80°C with a ramp rate of 0.1°C/s.
Melting-curve analysis was performed by using LightCycler software (version 4.0; (Roche Diagnostics). We plotted the negative value of the first derivative of fluorescence per unit time to distinguish peak melting temperatures from melting-curve plots.
RESULTS
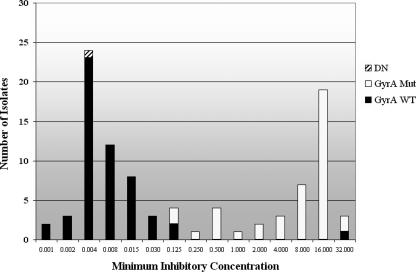
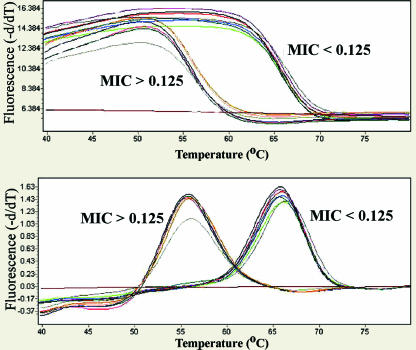
The 96 male urethral swab specimens tested had a range of MICs from 0.001 to 32.0 μg/ml (Fig. 1). The real-time PCR assay amplified product in 95 of 96 samples (99.0%). Of those that amplified, two distinct melting-curve peaks were identifiable at ca. 56 and 66°C corresponding to mutant and wild-type strains (Fig. 2), respectively. Using matched cultured urethral specimens with an MIC threshold of ≥0.125 μg of ciprofloxacin/ml to differentiate between susceptible and resistant specimens, the assay accurately detected resistance in 41 of 44 specimens for a sensitivity of 93.2% (95% confidence interval [CI] = 81.3 to 98.6%), and it identified 51 of 51 quinolone-susceptible samples for a specificity of 100% (95% CI = 93.0 to 100%). The single susceptible specimen that was not correctly analyzed demonstrated no amplification by PCR and therefore was not considered resistant or susceptible.
FIG. 1.
Ciprofloxacin MICs and melting-curve temperature for 96 N. gonorrhoeae-positive urine isolates. Of 96 specimens, 53 had a melting-curve temperature of 66°C, 42 had a melting-curve temperature of 56°C, and 1 did not amplify. DN, did not amplify; GyrA Mut, specimens with a melting-curve temperature of 56°C ± 1°C corresponding to a mutant genotype; GyrA WT, specimens with a melting-curve temperature of 66°C ± 1°C corresponding to a wild-type genotype. MICs are presented (in mg of ciprofloxacin/dl) as determined by the regional GISP laboratory in Seattle, WA.
FIG. 2.
Melting-curve analysis of 14 urine specimens with known MICs for ciprofloxacin, including six resistant isolates, seven susceptible isolates, and one negative control. Resistant isolates were defined as those with an MIC of ≥0.125 μg/ml by culture and susceptibility testing. Sensitive isolates were defined as those with an MIC of <0.125 μg/ml by culture and susceptibility testing.
The assay successfully amplified the selected portion of the gyrA gene and discerned a quinolone susceptibility genotype in 72 of 100 (72%) urine specimens from female patients. The remaining 28 specimens did not amplify, and therefore no genotype was determined.
We compared the real-time PCR assay described here to that published by Li et al. (11) with 10 urine samples. All 10 urine samples were amplified using our primers, and 9 of 10 amplified using those described by Li et al. Our assay had an average crossing point differential of 3.44 cycles less than the Li assay (Table 2).
TABLE 2.
Sensitivity analysis comparing crossing points for two real-time PCR assays for the detection of mutations in gyrA from urine specimens
| Specimen no. | % Sensitivitya according to:
|
Difference | |
|---|---|---|---|
| Li et al. (11) | This study | ||
| 1 | 29.62 | 27.09 | 2.53 |
| 2 | 32.17 | 28.01 | 4.16 |
| 3 | 30.86 | 27.85 | 3.01 |
| 4 | 30.69 | 27.70 | 2.99 |
| 5 | 30.30 | 27.79 | 2.51 |
| 6 | ND | >40.00* | NAb |
| 7 | >40.00* | 37.13 | NA |
| 8 | >40.00* | 34.65 | NA |
| 9 | 29.35 | 25.62 | 3.73 |
| 10 | 32.28 | 27.15 | 5.13 |
*, A result of >40.00 corresponds to an apparent log amplification that occurred at more than 40 cycles. ND, not detectable.
NA, not applicable.
DISCUSSION
This novel assay might prove valuable in settings in which rapid determination of quinolone resistance to N. gonorrhoeae specimens is desired. It is a faster method than that of culture and traditional agar dilution or disk susceptibility testing and does not require expensive sequencing equipment. Furthermore, in the age of nucleic acid amplification testing for gonococcal and chlamydial infection, using urine as a specimen for drug resistance genotyping is a valuable capability, precluding laborious culture and isolation methods and supplanting a requirement for urethral swab specimens. In summary, this novel assay provides a rapid and convenient method for detecting fluoroquinolone resistance in urine samples, enabling laboratories to respond to Centers for Disease Control and Prevention recommendations for increasing surveillance for QRNG (3).
The two primary regions in the N. gonorrhoeae genome associated with QRNG are the gyrA and parC genes (4). Other researchers have documented that parC mutations in conjunction with gyrA mutations are associated with higher levels of resistance (MIC of ≥1.0), whereas lower levels are almost exclusively associated with gyrA mutations (11, 12). Furthermore, parC mutations in isolation do not appear to be associated with antimicrobial resistance (19). On the basis of these data and reported estimates of clinically significant resistance for 500 mg of ciprofloxacin ranging between 0.125 and 1.0 mg (10), we restricted our assay to the detection of mutations in the Ser91 region of the gyrA gene. A recent study corroborating the necessary and sufficient role of Ser91 gyrA mutations in determining QRNG documented mutations in that region in 101 of 102 QRNG specimens (MIC of ≥0.125 mg of ciprofloxacin/ml) (18). Although our assay may not differentiate between intermediate and high level resistance, our results indicate that it detects clinically significant resistance.
A previously reported real-time PCR technique implementing melting-curve analysis for the discrimination of the quinolone resistance genotype also restricted detection to gyrA (11). However, when we attempted to mimic that assay by using DNA-extracted urine samples, we were unsatisfied with the sensitivity of the assay on urine specimens. We hypothesized that the limited sensitivity might have been the result of a large 225-bp amplicon size. In response, we restricted our assay to the detection of one codon (Ser91) and shortened our amplification length to 133 bp in an attempt to increase sensitivity in specimens with lower DNA concentrations. We accomplished this by using a primer already documented as effective in generating amplicons within gyrA (9). Pairwise comparison of the modified assay described here with that described by Li et al. revealed an average sensitivity increase of 3.44 crossing point values when generating the smaller amplicon (Table 2).
Overall, our assay detected a susceptibility genotype in 72% of female urine specimens and 99% of symptomatic male urine specimens. The most likely source of the decreased sensitivity of our real-time PCR assay in female urine samples was the use of the APTIMA-transcription-mediated amplification assay as a gold standard. Transcription-mediated amplification assays have been reported to be much more sensitive than standard PCR techniques for the detection of genetic content (7). Due to the single copy of gyrA in the N. gonorrhoeae genome, we are limited to the use of standard PCR for detection of the mutation. Low DNA concentration, DNA degradation, and PCR inhibition are other possible explanations for the lack of amplification in the remaining specimens. Overall, the assay exhibited 85.2% (167 of 196 samples) sensitivity to detect a susceptibility genotype in urine samples, a reasonable proportion for laboratory surveillance of QRNG. Although such a rate is unsuitable for clinical management, it might serve as an appropriate estimate for epidemiologic surveillance purposes when used on known positive specimens.
We documented a sensitivity and a specificity of 93.2% and 100.0%, respectively, for the detection of QRNG on the basis of correlated, cultured antimicrobial susceptibility testing. Considering the multiple mechanisms by which Neisseria attains resistance to quinolones, perfect correlation with culture and sensitivity techniques is not expected by using molecular methods. This assay is also limited by its inability to detect resistance to antibiotics other than fluoroquinolones. However, because of its applicability to urine specimens, this technique has advantages over the established culture- or sequencing-based methods of discrimination and may enable more widespread surveillance of QRNG.
Acknowledgments
We thank Tukisa Smith, the laboratory personnel at the Johns Hopkins Bayview Division of Allergy and Infectious Diseases, the staff of San Francisco City Clinic, and personnel at the San Francisco Department of Public Health Laboratory.
Financial support for this research was provided by San Francisco City and County and the California State Department of Public Health.
The findings and conclusions of this study are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Alcala, B., L. Arreaza, C. Salcedo, I. Antolin, N. Borrell, J. Cacho, C. De Las Cuevas, L. Otero, G. Sauca, F. Vazquez, H. Villar, and J. A. Vazquez. 2003. Molecular characterization of ciprofloxacin resistance of gonococcal strains in Spain. Sex. Transm. Dis. 30:395-398. [DOI] [PubMed] [Google Scholar]
- 2.Bala, M., K. Ray, and S. Kumari. 2003. Alarming increase in ciprofloxacin- and penicillin-resistant Neisseria gonorrhoeae isolates in New Delhi, India. Sex. Transm. Dis. 30:523-525. [DOI] [PubMed] [Google Scholar]
- 3.Bauer, H. M., K. E. Mark, M. Samuel, S. A. Wang, P. Weismuller, D. Moore, R. A. Gunn, C. Peter, A. Vannier, N. DeAugustine, J. D. Klausner, J. S. Knapp, and G. Bolan. 2005. Prevalence of and associated risk factors for fluoroquinolone-resistant Neisseria gonorrhoeae in California, 2000-2003. Clin. Infect. Dis. 41:795-803. [DOI] [PubMed] [Google Scholar]
- 4.Belland, R. J., S. G. Morrison, C. Ison, and W. M. Huang. 1994. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 14:371-380. [DOI] [PubMed] [Google Scholar]
- 5.Booth, S. A., M. A. Drebot, I. E. Martin, and L. K. Ng. 2003. Design of oligonucleotide arrays to detect point mutations: molecular typing of antibiotic resistant strains of Neisseria gonorrhoeae and hantavirus-infected deer mice. Mol. Cell Probes 17:77-84. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2005. Sexually transmitted disease surveillance 2004 supplement: gonococcal isolate surveillance (GISP) annual report-2004, Atlanta, Georgia. U.S. Department of Health and Human Services, Washington, DC.
- 7.Chernesky, M. A., and D. E. Jang. 2006. APTIMA transcription-mediated amplification assays for Chlamydia trachomatis and Neisseria gonorrhoeae. Expert Rev. Mol. Diagn. 6:519-525. [DOI] [PubMed] [Google Scholar]
- 8.Dan, M., F. Poch, and B. Sheinberg. 2002. High prevalence of high-level ciprofloxacin resistance in Neisseria gonorrhoeae in Tel Aviv, Israel: correlation with response to therapy. Antimicrob. Agents Chemother. 46:1671-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giles, J., J. Hardick, J. Yuenger, M. Dan, K. Reich, and J. Zenilman. 2004. Use of applied biosystems 7900HT sequence detection system and Taqman assay for detection of quinolone-resistant Neisseria gonorrhoeae. J. Clin. Microbiol. 42:3281-3283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knapp, J. S., J. A. Hale, S. W. Neal, K. Wintersheid, R. J. Rice, and W. L. Whittington. 1995. Proposed criteria for interpretation of susceptibilities of strains of Neisseria gonorrhoeae to ciprofloxacin, ofloxacin, enoxacin, lomefloxacin, and norfloxacin. Antimicrob. Agents Chemother. 39:2442-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Z., S. Yokoi, Y. Kawamura, S. Maeda, T. Ezaki, and T. Deguchi. 2002. Rapid detection of quinolone resistance-associated gyrA mutations in Neisseria gonorrhoeae with a LightCycler. J. Infect. Chemother. 8:145-150. [DOI] [PubMed] [Google Scholar]
- 12.Lindback, E., B. Gharizadeh, F. Ataker, A. Airell, S. Jalal, P. Nyren, and B. Wretlind. 2005. DNA gyrase gene in Neisseria gonorrhoeae as indicator for resistance to ciprofloxacin and species verification. Int. J. STD AIDS 16:142-147. [DOI] [PubMed] [Google Scholar]
- 13.Ng, L. K., P. Sawatzky, I. E. Martin, and S. Booth. 2002. Characterization of ciprofloxacin resistance in Neisseria gonorrhoeae isolates in Canada. Sex. Transm. Dis. 29:780-788. [DOI] [PubMed] [Google Scholar]
- 14.U.S. Department of Health and Human Services. 2005. Code of Federal Regulations: protection of human subjects, title 45, part 46. [Online.] http://www.hhs.gov/ohrp/humansubjects/guidance/45cfr46.htm.
- 15.World Health Organization. 2000. Fluoroquinolone-resistance in Neisseria gonorrhoeae, Hawaii, 1999, and decreased susceptibility to azithromycin in N. gonorrhoeae, Missouri, 1999. Morb. Mortal. Wkly. Rep. 49:833-837. [PubMed] [Google Scholar]
- 16.World Health Organization. 2004. Increases in fluoroquinolone-resistant Neisseria gonorrhoeae among men who have sex with men-United States, 2003, and revised recommendations for gonorrhea treatment. Morb. Mortal. Wkly. Rep. 53:335-338. [PubMed] [Google Scholar]
- 17.World Health Organization. 2003. Surveillance of antibiotic resistance in Neisseria gonorrhoeae in the WHO Western Pacific Region, 2002. Commun. Dis. Intell. 27:488-491. [DOI] [PubMed] [Google Scholar]
- 18.Yang, Y., M. Liao, W. M. Gu, K. Bell, L. Wu, N. F. Eng, C. G. Zhang, Y. Chen, A. M. Jolly, and J. A. Dillon. 2006. Antimicrobial susceptibility and molecular determinants of quinolone resistance in Neisseria gonorrhoeae isolates from Shanghai. J. Antimicrob. Chemother. 58:868-872. [DOI] [PubMed] [Google Scholar]
- 19.Zhou, W., W. Du, H. Cao, J. Zhao, S. Yang, W. Li, Y. Shen, S. Zhang, W. Du, and X. Zhang. 2004. Detection of gyrA and parC mutations associated with ciprofloxacin resistance in Neisseria gonorrhoeae by use of oligonucleotide biochip technology. J. Clin. Microbiol. 42:5819-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]