Abstract
Five clinical isolates of Nocardia that showed ambiguous bases within the variable region of the 16S rRNA gene sequence were evaluated for the presence of multiple copies of this gene. The type strains of three Nocardia species, Nocardia concava, Nocardia ignorata, and Nocardia yamanashiensis, which also showed ambiguous bases in the variable region, were also examined. Cloning experiments using an amplified region of the 16S rRNA that contains the variable region showed that each isolate possessed 16S rRNA genes with at least two different sequences. In addition, hybridization studies using a 16S rRNA gene-specific probe and extracted genomic DNA of the patient isolates and of the type strain of N. ignorata showed that each isolate possessed at least three copies of the gene. These multiple differing copies of the 16S rRNA gene and the results of DNA-DNA hybridization studies indicate problems of species definition and identification for such isolates. A broader species concept than that currently in vogue may be required to accommodate such organisms.
The presence of multiple different copies of the 16S rRNA gene has been demonstrated for numerous genera and species of bacteria (3, 14, 16, 20); an analysis of the genomic sequence of a Nocardia farcinica clinical strain revealed the presence of three copies of the 16S rRNA operon (10). Recently, selected isolates of Nocardia nova have also been shown to contain multiple copies of this gene (8). The presence of multiple differing 16S rRNA gene copies is suggested by the observation of overlapping peaks at specific base loci on the sequence chromatogram, resulting in ambiguous base designations. In Nocardia spp., these multiple peaks most frequently occur within the variable region of the 16S rRNA gene (corresponding to bases 150 through 169 of the sequence of Nocardia asteroides ATCC 19247T; GenBank accession number X84850). These base ambiguities cannot be resolved on repeat sequence analysis of this region.
An analysis of five patient isolates which were determined to be nonidentifiable to the species level using multiple molecular approaches and three Nocardia type strains revealed base ambiguities within the variable regions of their 16S rRNA gene sequences; these patient isolates and type strains were evaluated for the presence of multiple differing 16S rRNA genes. Species level identification and definition problems for such isolates are discussed.
MATERIALS AND METHODS
Organisms.
The German Collection of Microorganisms and Cell Cultures type strains of Nocardia concava (DSM 44804), Nocardia ignorata (DSM 44496), and Nocardia yamanashiensis (DSM 44669) and five isolates from four patients being treated at the Clinical Center of the National Institutes of Health were examined. Single isolates from three patients, isolates 1, 4, and 5, were recovered from tissue biopsy, lung biopsy, and bronchoalveolar lavage samples, respectively; two isolates (isolates 2 and 3), one from sputum and one from lung biopsy, were recovered from a single patient on consecutive days. Each patient isolate was considered to be a significant pathogen for those patients. All organisms were grown on Sabouraud dextrose agar (Emmons modification; Hardy Diagnostics, Santa Monica, CA), were modified acid fast positive, and exhibited aerial hyphae. Molecular studies on all isolates were performed on subcultures derived from a single colony. DNA for sequencing studies was extracted from all isolates as previously described (7).
Direct 16S rRNA gene sequencing.
The 16S rRNA gene sequences (minimum of 1,386 bases) of each organism were determined as previously described (6, 7). The sequences of both the forward and the reverse strands of all isolates were determined.
HSP gene sequence.
A 441-bp region of the heat shock protein (HSP) gene of the five patient isolates was amplified using primers previously described by Telenti et al. (19) with tails containing M-13 binding sites attached. The sequences of the primers were as follows: 5′-GTA-AAA-CGA-CGG-CCA-GAC-CAA-CGA- TGG-TGT-GTC-CAT-3′ and 5′-CAG-GAA-ACA-GCT-ATG-ACC-TTG-TCG- AAC-CGC-ATA-CCC-T-3′ (sequences of the tail are indicated in bold type). Amplification was performed according to the method of Steingrube et al. (18). Amplification products were electrophoresed on 2% SeaKem agarose gels in Tris-borate-EDTA buffer, the resultant bands were excised, and the DNA was purified using the GFX PCR DNA and gel band purification kit (GE Healthcare, Fairfield, CT). Cycle sequencing was performed using M13 −20 forward (5′-GTA-AAA-CGA-CGG-CCA-G-3′) and M13 reverse (5′-CAG-GAA-ACA-GCT-ATG-AC-3′) primers and procedures as previously described (6).
secA1 gene sequence.
A 520-bp region of the five patient isolates was amplified and sequenced using primers and procedures as previously described (9).
Sequence analysis.
Sequences for all genes and clones were assembled using the Lasergene SeqMan II software (DNAStar, Inc., Madison, WI). Chromatograms were carefully analyzed to detect the presence of overlapping peaks within the variable region. Sequences were aligned using the Clustal W algorithm with the Lasergene MegAlign software (DNAStar, Inc.). Amino acid sequences were deduced from the HSP and secA1 gene sequences using the MegAlign software. Direct sequences and sequences of clones were subjected to BLAST analysis and were compared to the sequences of type strains in the NIH database. In all gene sequence comparisons, ambiguous bases were considered mismatches. Percent similarity was determined by counting the number of base differences and relating the number of these differences to sequence length.
Cloning.
A 531-bp region of the 16S rRNA gene closest to the 5′ terminus of the gene (bases 2 to 532 of N. asteroides X84850) was amplified for each patient isolate and type strain; the amplified product was cloned for sequence analysis as previously described (8). Briefly, amplified DNA was ligated into pCR2.1 (Invitrogen Corporation, Carlsbad, CA) using the TA cloning kit (Invitrogen Corporation) and plasmids were transformed into One Shot INVαF′ chemically competent Escherichia coli (Invitrogen Corporation). Transformants were plated on Luria-Bertani medium with ampicillin, X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and IPTG (isopropyl-β-d-thiogalactopyranoside) (K-D Medical, Columbia, MD), and colonies showing inserts were subcultured in Lennox L broth (Quality Biologicals, Gaithersburg, MD). Plasmids were recovered from between 17 and 22 colonies for each organism studied. Plasmids were checked for insertion of the appropriately sized insert using EcoRI digestion (New England Biolabs, Beverly, MA). The sequences of the clones were determined using the M13 −20 forward and M13 reverse primers as previously described (8); both the forward and the reverse strands were sequenced, and only clones with unambiguous sequences within the variable region were used in the analysis.
Preparation of genomic DNA.
Genomic DNA was extracted from the patient isolates and from the type strains of N. ignorata and N. concava using a procedure based on that of Loeffelholz and Scholl (13). Briefly, isolates were incubated at 28°C for 48 h in Middlebrook 7H9 broth (Remel, Lenexa, KS), transferred to Mueller-Hinton broth (BBL, Sparks, MD) with glycine (American Bioanalytical, Natick, MA) and glycerol (Sigma-Aldrich, Inc., St. Louis, MO), and incubated for another 48 h. Suspensions were concentrated, incubated overnight at 35°C in 150 mg lysozyme (Sigma-Aldrich) and 25% sucrose (ICN Biomedicals, Inc., Aurora, OH), and then incubated at 60°C in 25% sodium dodecyl sulfate (Phoenix Biotechnologies, Huntsville, AL), 50 μg/ml proteinase K (Sigma-Aldrich), and 1 M Tris (pH 9.5) (Molecular Biologicals, Columbia, MD) for 30 min. After centrifugation, the supernatant was washed once in phenol-chloroform-isoamyl alcohol (25:24:1) (Sigma-Aldrich), with two additional washes in chloroform-isoamyl alcohol (24:1) (Sigma-Aldrich). After concentration and drying, the pellet was resuspended in 0.5 M Tris-EDTA (pH 8.0) (Quality Biologicals) and incubated in 25 mg/ml RNase (Sigma-Aldrich), followed by incubation with 20 mg/ml proteinase K, both at 35°C for 30 min each. The DNA suspension was washed three times with chloroform-isoamyl alcohol and precipitated with 3 M sodium acetate (Quality Biologicals) and 95% ethanol (Warner-Graham Co., Cockeysville, MD). After concentration and drying, the pellet was washed with 70% ethanol, dried, and reconstituted with molecular-grade water. Extracted DNA was held at 4°C.
Southern blot analysis.
Genomic DNA from the patient isolates and from the type strain of N. ignorata was digested with SphI (isolates were determined to have no SphI recognition sites within their 16S rRNA sequence) and electrophoresed on an agarose gel. Resulting fragments were transferred to a nylon membrane, and hybridization was performed using an 89-bp probe (8) specific to a conserved region of the 16S rRNA gene. Chemiluminescence detection was performed (8).
DNA-DNA hybridization.
Genomic DNA of the type strain of N. concava was labeled with [32P]dCTP. The labeled N. concava was hybridized with the genomic DNA of patient isolate 2 as previously described (1, 2). Subsequently, the genomic DNA of patient isolate 2 was similarly labeled and hybridized with the unlabeled genomic DNA of the type strain of N. concava. Positive controls (labeled DNA and unlabeled DNA of the same species) and negative controls (labeled DNA only) were run with each analysis and gave acceptable results.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of the Nocardia type strains have been deposited in GenBank under accession numbers EF177464 (N. concava), DQ659907 (N. ignorata), and DQ659920 (N. yamanashiensis).
RESULTS
Analysis of the direct 16S rRNA gene sequences obtained for the five patient isolates and the three type strains showed between 6 and 13 ambiguous bases within the 20-base variable region (Table 1). BLAST analysis of the direct 16S rRNA gene sequences of the patient isolates resulted in no definitive identification for any of the isolates but confirmed their identification as Nocardia species, as only Nocardia species were listed as the most likely matches (Table 2). By the interpretive standards for identification of Nocardia isolates by 16S rRNA gene sequence currently in use in our institution, all patient isolates would be reported as Nocardia spp., as a comparison to related species showed <99.8% similarity to a type strain of any species (5).
TABLE 1.
Direct sequences of the variable region of the 16S rRNA genes, and sequences of the variable region of clones of patient isolates and the type strains of N. ignorata, N. concava, and N. yamanashiensis
| Isolate(s) or type strain | Source of sequence (no. of clones with sequence) | Base at positiona,b:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 150 | 151 | 152 | 153 | 154 | 155 | 156 | 162 | 163 | 164 | 165 | 166 | 167 | 168 | 169 | ||
| 1 | Direct sequence | A | R | G | G | R | Y | T | R | T | Y | T | Y | T | G | G |
| Clone A (10) | A | G | G | G | A | C | T | G | T | T | T | C | T | G | G | |
| Clone B (12) | A | A | G | G | G | T | T | A | T | C | T | T | T | G | G | |
| 2 and 3c | Direct sequence | A | S | S | V | D | W | Y | K | H | K | K | B | T | G | K |
| Clone A (3/3)d | A | G | G | G | G | A | T | T | C | T | T | C | T | G | G | |
| Clone B (6/8) | A | C | G | A | A | T | C | G | T | T | T | G | T | G | G | |
| Clone C (1/0)e | A | G | C | C | T | T | C | G | A | G | G | T | T | G | G | |
| Clone D (7/7) | A | G | C | C | T | T | C | G | A | G | G | T | T | G | T | |
| 4 | Direct sequence | M | K | Y | S | K | S | T | S | S | K | G | W | K | G | T |
| Clone A (6) | C | T | T | G | G | G | T | C | C | T | G | A | G | G | T | |
| Clone B (11) | A | G | C | C | T | C | T | G | G | G | G | T | T | G | T | |
| 5 | Direct sequence | M | K | C | M | K | T | C | G | M | K | G | K | K | G | T |
| Clone A (14) | C | T | C | A | G | T | C | G | C | T | G | G | G | G | T | |
| Clone B (4) | A | G | C | C | T | T | C | G | A | G | G | T | T | G | T | |
| N. ignorata | Direct sequence | T | Y | K | G | G | W | T | W | Y | Y | K | R | G | G | G |
| Clone A (10) | T | C | G | G | G | A | T | T | T | C | T | G | G | G | G | |
| Clone B (6) | T | T | T | G | G | T | T | A | C | T | G | A | G | G | G | |
| N. concava | Direct sequence | A | G | S | S | K | W | Y | K | M | K | K | Y | T | G | K |
| Clone A (16) | A | G | G | G | G | A | T | T | C | T | T | C | T | G | G | |
| Clone B (3) | A | G | C | C | T | T | C | G | A | G | G | T | T | G | T | |
| N. yamanashiensis | Direct sequence | W | B | Y | H | K | S | T | S | S | K | K | K | K | G | K |
| Clone A (8) | A | C | T | T | G | G | T | C | C | T | T | G | T | G | G | |
| Clone B (6) | T | T | C | A | G | G | T | C | C | T | G | G | G | G | T | |
| Clone C (5) | A | G | C | C | T | C | T | G | G | G | G | T | T | G | T | |
Compared to N. asteroides ATCC 19247T (GenBank accession no. X84850), bases at positions 157 through 161 are identical for all isolates.
Bold letters indicate ambiguous bases. Abbreviations: R represents A or G; Y represents C or T; S represents C or G; V represents A, C or G; D represents A, G, or T; W represents A or T; K represents G or T; H represents A, C, or T; B represents C, G, or T; M represents A or C.
Isolated from the same patient.
Indicates number of clones sequenced from isolates 2 and 3, respectively.
This clone not found in isolate 3.
TABLE 2.
Most closely related type strain to patient isolates by gene sequence of direct 16S rRNA, HSP, and secA1
| Patient isolate(s) | Most closely related type strain (% similarity) by sequence of:
|
||
|---|---|---|---|
| Direct 16S rRNAa | HSPb | secA1c | |
| 1 | Nocardia africana (97.8) | Nocardia niigatensis (96.6) | N. niigatensis (95.9) |
| 2 and 3 | N. concava (99.2) | N. concava (97.5) | N. concava (98.9) |
| 4 | N. yamanashiensis (97.8) | N. seriolae (99.4) | N. concava (98.2) |
| 5 | N. seriolae (97.8) | N. seriolae (97.2) | N. niigatensis (97.3) |
A total of 1,299 bases were analyzed.
A total of 356 bases were analyzed.
A total of 440 bases were analyzed.
Three of the five patient isolates (isolates 1, 4, and 5) showed various degrees of similarity to multiple different species when direct sequences of the three genes (16S rRNA, HSP, and secA1) were analyzed (Table 2), resulting in inconclusive identifications for these isolates. In no case were gene sequences most similar to a single species by all genes for these three isolates.
Isolates 2 and 3 (identical isolates recovered from the same patient on consecutive days) showed the most sequence similarity to N. concava by all three genes examined (Table 2), suggesting that these isolates may be clinical isolates of N. concava. In addition, the sequence of one of the 16S rRNA gene clones examined from patient isolates 2 and 3 (clone A) was identical to that of a clone derived from the type strain of N. concava (N. concava clone A) (Table 1). Repeated DNA-DNA hybridization experiments using isolate 2 and the N. concava type strain gave inconclusive results. Three experiments in which the patient isolate was labeled showed the isolate to be nonrelated to the N. concava type strain, with relative binding ratios of ≤58.6 and divergence values of ≤2.9. Three experiments in which the type strain of N. concava was labeled showed the isolates to be related to the N. concava type strain, with relative binding ratios of 89.6, 86.6, and 85.9 and divergence values of 4.7, 3.5, and 3.4, respectively.
Isolate 4 showed a high degree of HSP gene similarity to Nocardia seriolae (99.4%); however, sequence comparison of the 16S rRNA and secA1 genes to N. seriolae was inconclusive, showing 97.0% similarity to the N. seriolae type strain for both genes (data not shown).
Except for isolates 2 and 3, the deduced HSP amino acid sequence was not discriminatory enough to assign a species identification to the patient isolates on the basis of that sequence. The deduced HSP amino acid sequences for isolates 2 and 3 were identical to that of the N. concava type strain. For all patient isolates, the deduced SecA1 amino acid sequence was not discriminatory enough to assign a species identification (data not shown).
Sequence analysis of clones derived from an amplified region of the 16S rRNA gene showed that clones with between two and four different sequence patterns within the variable region were obtained for each of the five patient isolates and the three type strains (Table 1). These sequence patterns were determined for multiple clones from each isolate (with between 3 and 14 separate clones showing identical sequences), except for one sequence pattern found in a single clone derived from isolate 2. For each isolate, the sequences of all the clones corresponded to the ambiguous bases seen on analysis of the direct sequence of this region of the 16S rRNA gene (Table 1). Except for the one clone sequence (clone A from patient isolates 2 and 3) that showed significant similarity (99.6%) to the type strain of N. concava, BLAST analysis of all other cloned 16S rRNA genes showed ≤98.8% sequence similarity to the sequence of the closest type strain (data not shown).
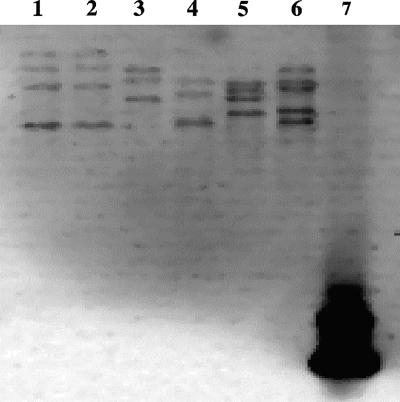
Hybridization of the digested genomic DNA with a 16S rRNA gene-specific probe verified the presence of multiple copies of the 16S rRNA gene in each of the patient isolates and in the type strain of N. ignorata. Patient isolates 1 and 5 showed at least three copies, isolates 2, 3, and 4 showed at least four copies, and the N. ignorata type strain showed at least five copies (Fig. 1).
FIG. 1.
DNA hybridization of SphI digests of genomic DNA with a probe complementary to an 89-bp region of the 16S rRNA gene. Lane 1, isolate 2; lane 2, isolate 3; lane 3, isolate 5; lane 4, isolate 1; lane 5, isolate 4; lane 6, N. ignorata DSM 44496T; lane 7, positive control.
DISCUSSION
The use of the 16S rRNA gene sequence has become the “gold standard” for the identification of Nocardia species due to the inadequacy of traditional biochemicals for the discrimination of an increasing number of clinically significant species in this genus. The presence of ambiguous bases in the direct 16S rRNA gene sequence of some Nocardia isolates has been noted (8, 15) and has been shown to be due to the presence of multiple copies of the 16S rRNA gene, at least for some isolates of N. nova (8). Data presented here show that some strains of Nocardia may possess up to five copies of the 16S rRNA gene, and cloning studies show that significant base pair substitutions exist within the variable regions of these copies. A determination of the transcriptional activity of these genes was not addressed in this study.
The variable region examined is near the 5′ terminus of the 16S rRNA gene (corresponding to bases 150 to 169 of the sequence of N. asteroides ATCC 19247T; GenBank accession number X84850) and has been shown to be specific for numerous species of Nocardia (7). Alignment of this region of the 16S rRNA gene sequence of the type or reference strains of 39 species or taxa shows unique sequences within this region for 24 species or taxa (data not shown). It is in part the variability of this region that allows identification of some species by partial sequencing of the 16S rRNA gene using the MicroSeq 500 system (4, 15).
Hybridization studies with a 16S rRNA gene-specific probe verified the presence of multiple 16S rRNA genes in all of the patient isolates and in the type strain of N. ignorata and, in most cases, demonstrated the presence of more 16S rRNA gene copies than the number of different sequences derived from clones suggested. It is possible that two or more gene copies share the same sequence or that an insufficient number of clones was evaluated to detect all sequence variations present.
We are confident that the data presented here represent multiple 16S rRNA gene copies, as all molecular work was performed using subcultures from a single colony. In addition, for seven of the eight isolates, multiple clones with identical sequences were obtained for all sequence patterns. Cloning experiments using isolates 2 and 3 showed multiple clones with three different patterns and a fourth pattern that was seen in only one clone from isolate 2. Except for the single clone found in isolate 2, the presence of multiple clones with the same sequence indicates that the sequence differences noted in the clones are not merely the result of transcription errors that occurred during PCR.
We do not think that any of the patient isolates examined in this study can be assigned to any currently recognized species because, in most cases, none of the multiple 16S rRNA gene sequences obtained from the clones is sufficiently similar to the sequences of any currently described species. In addition, for patient isolates 1, 4, and 5, an analysis of both the HSP gene and the secA1 gene sequences gave inconclusive identifications or gave identifications which were not in agreement with each other or with the identification obtained for the 16S rRNA gene sequence (Table 2). Although all genes examined for isolates 2 and 3 indicated a high degree of similarity to the type strain of N. concava and one 16S rRNA gene clone was identical to a clone from N. concava, DNA-DNA hybridization studies failed to show conclusive evidence of species identity. This may be due, in part, to the fact that the type strain of N. concava also possesses multiple different copies of the 16S rRNA gene.
In our hands, direct sequence of the type strain of N. ignorata showed numerous ambiguous bases within the variable region that are not present in the original sequence deposited in GenBank (21); some of these ambiguous bases correspond to deleted bases in that original sequence. Cloning experiments reported here show that these base ambiguities are the result of at least two different sequences among the multiple 16S rRNA genes present. Analysis of the entire sequences obtained from the clones of N. ignorata DNA revealed additional base ambiguities outside of the variable region (data not shown). These additional base ambiguities may indicate that more than two different 16S rRNA gene sequence patterns are present in the N. ignorata type strain. Hybridization using a 16S rRNA-specific probe with the genomic DNA of the N. ignorata type strain verified the presence of at least five copies of the 16S rRNA gene in this organism. Because only two different gene sequences were detected in the clone analysis, it is presumed that at least some gene copies possess the same sequence.
Recently, several clinical isolates of N. ignorata have been reported by Rodríguez-Nava et al. (17), some of which appear to be clinically significant. 16S rRNA gene sequences of these isolates deposited in GenBank have sequences identical to each other in their 16S rRNA gene variable regions; these variable region sequences differ by one base from the sequence of the type strain submitted by the same authors. All bases within the variable regions of the reported N. ignorata patient isolates correspond either to the unambiguous bases in the variable region of the direct sequence of the N. ignorata type strain sequenced in this study or to one of the two base possibilities for the ambiguous bases. While it is clear that the type strain of the species contains multiple copies of the 16S rRNA gene and that these copies have different sequences within the variable region, it is not known whether the patient isolates also possess multiple different copies.
Careful analysis of the 16S rRNA gene sequence of two additional Nocardia type strains, N. concava and N. yamanashiensis, revealed the presence of ambiguous bases within the variable regions of this gene which could not be resolved with repeat testing. Clones derived from the amplified 16S rRNA gene of the N. concava type strain showed two different gene sequences within this region. Two clinical isolates of N. concava have been reported; both were isolated from patients with cutaneous nocardiosis (12). One of these isolates is the type strain of the species. According to the sequences of these isolates submitted to GenBank, six base differences exist between the two isolates within the 19-base variable region. All of the bases within the variable regions of these reported N. concava clinical isolates that differ between the two strains occur at sites of ambiguous bases in our direct sequence of the N. concava type strain.
Clones derived from the amplified 16S rRNA gene of the N. yamanashiensis type strain showed three different gene sequences within the variable region. Only a single clinical isolate (from a skin abscess) of N. yamanashiensis has been reported (11); this isolate is the type strain of the species. In this study, direct sequencing of the 16S rRNA gene of the N. yamanashiensis type strain showed 12 ambiguous bases within the 19-base variable region; the bases in the variable region of the initially reported N. yamanashiensis type strain correspond to one of the two or three base possibilities of the ambiguous bases determined in this study.
The presence of multiple different 16S rRNA genes in at least some Nocardia type strains is problematic, as it is unclear whether the 16S rRNA gene(s) of any other isolate would be sufficiently similar to call the strains conspecific. Cloning studies might resolve this issue but are not practical in most circumstances. Results of the DNA-DNA hybridization performed in this study suggest that even isolates that have high degrees of gene similarity to such a type strain cannot be unambiguously identified as that species.
In our experience, the presence of multiple differing copies of the 16S rRNA gene sequence appears to be uncommon in patient isolates; careful analysis of the sequences of isolates that do not correspond to those of any described type strain may show the presence of numerous ambiguous bases and lead to the suspicion that multiple differing copies exist. Sequences that show single or very few ambiguous bases may be similar enough to a type strain to allow high levels of sequence similarity; only extensive sequencing and cloning could reveal whether these ambiguous bases are the result of multiple differing copies or of sequencing errors.
Detection of ambiguous bases (and, thus, isolates that may possess differing multiple gene copies) requires careful examination of the sequence chromatograms. Ambiguous bases appear as overlapping peaks that are not resolved with repeat testing. For Nocardia isolates, these base substitutions occur most frequently within the variable regions of the gene, so these regions should be routinely scrutinized by hand, as sequencing software may record only the largest peak at a specific locus on the chromatogram. In any case, bases flagged by sequence software as “ambiguous” should be carefully examined.
In our opinion, sequencing of the 16S rRNA gene still represents a reasonable method for the identification of many species of Nocardia as long as careful analysis of sequence data reveals no irresolvable ambiguous bases. The value of this gene results from the extensive amount of sequence information for all known Nocardia species in sequence databases. The usefulness of alternative targets would depend not only on the discriminative power of those targets but also on the ability to obtain and sequence the type strains of all newly described species and the deposition of the sequences of these alternative targets in the sequence databases. In our laboratory, we routinely sequence the secA1 gene if 16S gene sequence analysis of a clinical isolate shows less than 99.8% similarity to that of a type strain. Using these two genes, we have thus far been able to discriminate among all described species.
The Pandora's box of complexity and variation in organisms in the genus Nocardia that these isolates illustrate highlights several more general problems which need to be addressed. (i) The presence of ambiguous bases in 16S rRNA gene sequences which cannot be resolved by repeat testing should not be dismissed as the consequence of amplification or sequencing errors but should be taken as suggesting the presence of multiple differing copies of the 16S rRNA gene, the differing sequences of which require cloning studies for resolution. (ii) Authors describing new species should make every effort to resolve apparent sequence ambiguities in proposed type strains before publishing the sequences obtained or depositing them in gene databases. If such ambiguities are not resolved, the genomic complexity of such isolates is obscured. Furthermore, the likelihood of the misidentification of subsequent isolates as belonging to a species with unresolved sequence ambiguities is considerably increased, as ignoring the ambiguous bases may result in an erroneous overestimation of sequence similarity between such a type strain and subsequent isolates. (iii) Isolates of organisms with differing multiple gene copies of the 16S rRNA gene most probably are currently unidentifiable to the species level by studies sequencing only that gene, even if a full sequence is obtained for each differing gene. It also would hardly be practicable to assign each Nocardia isolate with multiple differing gene copies to its own species. (It is possible that future studies will show that many or all isolates with multiple differing gene copies have a definable range of variation of each copy within a given species, but this seems highly improbable at present.) (iv) Even when utilizing DNA-DNA hybridization (which is beyond the capabilities of nearly all diagnostic laboratories), it may be impossible for some Nocardia species isolates to be identified accurately to the species level. This problem is exemplified by the fact that we failed to obtain congruent degrees of DNA-DNA hybridization with some of our isolates and an apparently similar type strain, depending upon which organism DNA was labeled. (v) From our point of view, major problems lie with the current bacterial species concept and these problems are not restricted to the genus Nocardia. We argue that there currently exists no operationally useful such concept, and we think the concept needs careful reevaluation to stem the proliferation of newly described bacterial species, among which no significant biological differences and, particularly, no clinically significant differences may have been demonstrated. We think that a more useful concept needs to be developed by a consensus mechanism and also needed is some type of enforcement process so that the criteria determined will be adhered to rigidly. Perhaps the standards provided by the Clinical and Laboratory Standards Institute could constitute a useful model. The procedure to be employed needs to be carefully specified, with appropriate quality control measures identified and appropriate statistical measures selected (for example, acceptable ranges of variability in DNA-DNA hybridization results).
Without practicable criteria for the delineation of new species and the identification of clinical isolates, there will be continuing misidentification of patient isolates and continuing inability to associate clinically important data (such as antimicrobial susceptibility, geographic distribution, and relative pathogenicity) with particular species categories.
Acknowledgments
We thank Patrick R. Murray, Department of Laboratory Medicine, Warren G. Magnuson Clinical Center, NIH, for critically reviewing the manuscript.
The views expressed here are those of the authors and should not be construed as those of the U.S. Department of Health and Human Services.
This research was supported by the Intramural Research Program of the NIH, Warren G. Magnuson Clinical Center.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Brenner, D. J., F. W. Hickman-Brenner, J. V. Lee, A. G. Steigerwalt, G. R. Fanning, D. G. Hollis, J. J. Farmer III, R. E. Weaver, S. W. Joseph, and R. Seidler. 1983. Vibrio furnissii (formerly aerogenic biogroup of Vibrio fluvialis), a new species isolated from human feces and the environment. J. Clin. Microbiol. 18:816-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenner, D. J., A. C. McWhorter, J. K. Leete Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cilia, V., B. Lafay, and R. Christen. 1996. Sequence heterogeneities among 16S ribosomal RNA sequences, and their effect on phylogenetic analyses at the species level. Mol. Biol. Evol. 13:451-461. [DOI] [PubMed] [Google Scholar]
- 4.Cloud, J. L., P. S. Conville, A. Croft, D. Harmsen, F. G. Witebsky, and K. C. Carroll. 2004. Evaluation of partial 16S ribosomal DNA sequencing for identification of Nocardia species by using the MicroSeq 500 system with an expanded database. J. Clin. Microbiol. 42:578-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conville, P. S., J. M. Brown, A. G. Steigerwalt, J. W. Lee, V. L. Anderson, J. T. Fishbain, S. M. Holland, and F. G. Witebsky. 2004. Nocardia kruczakaie sp. nov., a pathogen in immunocompromised patients and a member of the “N. nova complex.” J. Clin. Microbiol. 42:5139-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conville, P. S., J. M. Brown, A. G. Steigerwalt, J. W. Lee, D. E. Byrer, V. L. Anderson, S. E. Dorman, S. M. Holland, B. Cahill, K. C. Carroll, and F. G. Witebsky. 2003. Nocardia veterana as a pathogen in North American patients. J. Clin. Microbiol. 41:2560-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conville, P. S., S. H. Fischer, C. P. Cartwright, and F. G. Witebsky. 2000. Identification of Nocardia species by restriction endonuclease analysis of an amplified portion of the 16S rRNA gene. J. Clin. Microbiol. 38:158-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conville, P. S., and F. G. Witebsky. 2005. Multiple copies of the 16S rRNA gene in Nocardia nova isolates and implications for sequence-based identification procedures. J. Clin. Microbiol. 43:2881-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conville, P. S., A. M. Zelazny, and F. G. Witebsky. 2006. Analysis of secA1 gene sequences for identification of Nocardia species. J. Clin. Microbiol. 44:2760-2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kageyama, A., K. Yazawa, K. Nishimura, and Y. Mikami. 2004. Nocardia inohanensis sp. nov., Nocardia yamanashiensis sp. nov. and Nocardia niigatensis sp. nov., isolated from clinical specimens. Int. J. Syst. Evol. Microbiol. 54:563-569. [DOI] [PubMed] [Google Scholar]
- 12.Kageyama, A., K. Yazawa, H. Taniguchi, H. Chibana, K. Nishimura, R. M. Kroppenstedt, and Y. Mikami. 2005. Nocardia concava sp. nov., isolated from Japanese patients. Int. J. Syst. Evol. Microbiol. 55:2081-2083. [DOI] [PubMed] [Google Scholar]
- 13.Loeffelholz, M. J., and D. R. Scholl. 1989. Method for improved extraction of DNA from Nocardia asteroides. J. Clin. Microbiol. 27:1880-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ninet, B., M. Monod, S. Emler, J. Pawlowski, C. Metral, P. Rohner, R. Auckenthaler, and B. Hirschel. 1996. Two different 16S rRNA genes in a mycobacterial strain. J. Clin. Microbiol. 34:2531-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel, J. B., R. J. Wallace, Jr., B. A. Brown-Elliott, T. Taylor, C. Imperatrice, D. G. B. Leonard, R. W. Wilson, L. Mann, K. C. Jost, and I. Nachamkin. 2004. Sequence-based identification of aerobic actinomycetes. J. Clin. Microbiol. 42:2530-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reischl, U., K. Feldmann, L. Naumann, B. J. M. Gaugler, B. Ninet, B. Hirschel, and S. Emler. 1998. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by presence of two different copies of 16S rRNA gene. J. Clin. Microbiol. 36:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodríguez-Nava, V., A. Couble, Z. U. Khan, M. Pérouse de Montclos, L. Brasme, C. Villuendas, C. Molinard, P. Boiron, and F. Laurent. 2005. Nocardia ignorata, a new agent of human nocardiosis isolated from respiratory specimens in Europe and soil samples from Kuwait. J. Clin. Microbiol. 43:6167-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steingrube, V. A., R. W. Wilson, B. A. Brown, K. C. Jost, Jr., Z. Blacklock, J. L. Gibson, and R. J. Wallace, Jr. 1997. Rapid identification of clinically significant species and taxa of aerobic actinomycetes, including Actinomadura, Gordona, Nocardia, Rhodococcus, Streptomyces, and Tsukamurella isolates, by DNA amplification and restriction endonuclease analysis. J. Clin. Microbiol. 35:817-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Telenti, A., F. Marchesi, M. Balz, F. Bally, E. C. Böttger, and T. Bodmer. 1993. Rapid identification of mycobacteria to the species level by polymerase chain reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang, Y., Z. Zhang, and N. Ramanan. 1997. The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J. Bacteriol. 179:3270-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yassin, A. F., F. A. Rainey, and U. Steiner. 2001. Nocardia ignorata sp. nov. Int. J. Syst. Evol. Microbiol. 51:2127-2131. [DOI] [PubMed] [Google Scholar]