Abstract
The immunogenic iron-regulated FetA outer membrane protein of Neisseria meningitidis is one of various outer membrane proteins that have been considered potential meningococcal vaccine candidates. In this report, we describe the characterization of three meningococcal isolates that have deleted fetA sequences through genetic recombination at repetitive elements.
Neisseria meningitidis is a leading cause of bacterial meningitis and other serious infections worldwide. Most current meningococcal vaccines rely on capsular polysaccharide-based approaches. Serogroup B meningococcal disease is common in many parts of the world, yet the development of an effective vaccine for sporadic strains of this serogroup is hampered by immunologic tolerance to serogroup B capsular sialic acid residues (3). Thus, serogroup B vaccine strategies have focused on immunogenic noncapsular outer membrane proteins, including FetA, an iron-regulated enterobactin receptor (1, 2, 7). Substantial antigenic variation of a surface-exposed loop of the mature FetA protein is well documented and may therefore limit the utility of this protein as the sole component of a serogroup B vaccine (5, 6, 9, 10). Outer membrane protein (OMP) sequence profiling can provide antigenic information that is useful in characterizing emergent clones (4, 10). We have performed multilocus sequence typing (MLST) and OMP profiling with ∼1,000 invasive isolates of N. meningitidis collected from the United States from 1990 to 2006 (4; unpublished data). This analysis identified 2/439 (0.46%) serogroup B isolates and 1/329 (0.30%) serogroup C isolates that have deleted fetA gene sequences (Table 1). The purpose of this study was to further characterize the fetA deletions in these isolates.
TABLE 1.
Description of invasive N. meningitidis isolates
| Isolate | Serogroup | Sequence type | Clonal complexa | OMP typing resultb | Location | Date of isolation | Clinical syndrome |
|---|---|---|---|---|---|---|---|
| NM0005 | C | 11 | ST-11 | 2-59:P1.5,2:F.1-30 | Maryland | Jan 1992 | Meningitis |
| NM0160 | B | 2971 | 3-84:P1.17,16-3:F.del | Maryland | Apr 1997 | Meningitis | |
| NM1042 | C | 5762 | ST-35 | 3-39:P1.22-1,14:F.del | Maryland | Mar 2005 | Cellulitis/tenosynovitis |
| NM1630 | B | 2588 | ST-35 | 3-39:P1.22-1,14:F.del | Ohio | Aug 2004 | Meningitis |
ST, sequence type.
OMP typing results are expressed as porB allele:P1.porA VR1 allele,porA VR2 allele:F.fetA allele; del, deleted.
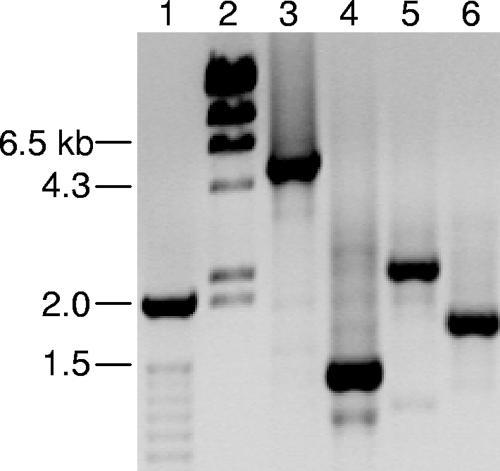
DNA templates prepared by boiling were amplified by PCR with fetA primers S1 and S8 (9). PCR amplification of fetA from strains NM0160, NM1042, and NM1630 consistently failed, while amplification of MLST and other OMP loci was successful (data not shown). Consensus primers (primer FetAoutF, 5′-GAAAGCATCGTGGYGGTGTCC-3′; primer FetAoutR, 5′-CGTTCGGGGTTTTGCGGTATTT-3′) that reside outside the FetA-coding region were designed on the basis of the fetA sequences from reference strains of serogroup B (strain MC58) and serogroup C (strain FAM18) (www.sanger.ac.uk) (8). These primers are predicted to amplify ∼4.5-kb products from serogroup B and C meningococcal strains bearing full-length fetA. Genomic DNA from strains NM0160, NM1042, and NM1630 and a serogroup C control strain (strain NM0005) containing fetA was prepared by DNeasy column purification (QIAGEN, Valencia, CA). Long-range PCR amplification was performed by using a GeneAmp high-fidelity PCR system (Applied Biosystems, Foster City, CA). The reaction conditions were 94°C for 2 min, followed by 10 cycles of 94°C for 15 s, 62°C for 30 s, and 68°C for 4 min, followed by 20 cycles of 94°C for 15 s, 62°C for 30 s, and 68°C for 4 min plus an additional 5 s per cycle, with a final incubation at 72°C for 7 min. The resulting PCR products were resolved on a 0.8% agarose gel containing ethidium bromide. The PCR products for strains NM0160, NM1042, and NM1630 were significantly smaller and of various sizes compared to the PCR product generated with the control strain (Fig. 1). These results suggest that the fetA gene is deleted in strains NM0160, NM1042, and NM1630.
FIG. 1.
Long-range PCR amplification of fetA region from N. meningitidis genomic DNA. Lane 1, 100-bp ladder; lane 2, bacteriophage λ HindIII ladder; lane 3, strain NM0005 (serogroup C); lane 4, strain NM0160 (serogroup B); lane 5, strain NM1042 (serogroup C); lane 6, strain NM1630 (serogroup B).
In order to explore potential mechanisms of fetA deletion in these strains, the PCR products were sequenced by using the FetAout primers. The PCR products were treated with ExoSap-It (USB, Cleveland, OH) and were sequenced with a BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems). The resulting electropherograms were analyzed by using LaserGene (version 7.0) SeqMan software (DNAstar, Madison, WI). Sequence analysis did not identify fetA gene sequences in the PCR products generated from NM0160, NM1042, or NM1630. The fetA nucleotide sequence identified from control isolate NM0005 corresponded to the full-length gene. These results demonstrate that fetA gene sequences have been deleted from these three clinical N. meningitidis isolates.
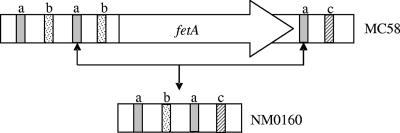
Analysis of the DNA sequence from the isolates bearing the fetA deletion identified multiple repetitive palindromic RS3 core sequences (ATTCCC-8 nucleotides-GGGAAT) similar to those previously described for porA deletions (11). NM0160 contains three RS3 core elements that are homologous to the RS3 elements located upstream of fetA in the MC58 reference strain. One RS3 element in this strain is homologous to an RS3 core sequence located downstream of fetA in the MC58 genome sequence. These data and the additional nucleotide identities upstream and downstream of the MC58 fetA gene indicate that the fetA gene in NM0160 was deleted through homologous recombination at RS3 elements (Fig. 2). Deletion of fetA and its flanking regions is reflected by the size of the NM0160 PCR product (Fig. 1, lane 4).
FIG. 2.
Depiction of the recombination event between core RS3 palindromic repetitive elements in serogroup B N. meningitidis isolates which results in the deletion of fetA from the genome of isolate NM0160. Letters illustrate multiple RS3 core elements. The connected arrows indicate the region of homologous recombination.
Sequence analysis of isolate NM1630 identified nine RS3 core elements. Three elements are homologous to the RS3 core elements located upstream of fetA in the isolate MC58 genome and to the three RS3 core elements identified in isolate NM0160. One of the RS3 core elements in isolate NM1630 shows homology to an RS3 element located downstream of fetA in the MC58 genome. Further analysis of the NM1630 sequence revealed 96% identity to a 367-bp region downstream of fetA in MC58. These homologies and the size of the NM1630 PCR product (Fig. 1, lane 6) suggest that deletion of fetA occurred through recombination at RS3 core elements in isolate NM1630.
Sequence analysis of isolate NM1042 revealed 10 RS3 core elements. Four of these elements are identical to the RS3 core elements located upstream of fetA in the serogroup C isolate FAM18 reference sequence. In addition, the isolate NM1042 sequence shows 96% identity to a 367-bp region downstream of fetA in FAM18. Interestingly, no RS3 core sequences are located downstream of fetA in FAM18. However, analysis of downstream fetA sequences from serogroup C isolate NM0005 identified several RS3 core elements flanked by a region of duplication. Homologous recombination between these duplicated regions would result in the deletion of RS3 elements and would explain the absence of these elements in fetA downstream sequences of FAM18. The identification of RS3 repetitive elements in serogroup C control strain NM0005 suggests that homologous recombination through these elements resulted in the deletion of fetA in NM1042. None of these deletions affected coding regions upstream or downstream of fetA (i.e., thdF and NMB1989 or fetB).
Taken together, these data indicate that fetA can be deleted in clinical meningococcal isolates by genetic recombination through RS3 repetitive elements, albeit rarely. This finding and the observation that other vaccine candidates such as PorA can be deleted in invasive meningococcal strains have major implications for serogroup B vaccine design (4). If natural immunity promotes the emergence of meningococcal clones with OMP deletions, vaccine-induced immunologic pressure could lead to the emergence and clonal expansion of strains from which the OMP is deleted. Thus, accumulating evidence suggests that a multiprotein approach will be required in the development of meningococcal vaccines that rely on OMPs as primary antigens.
Nucleotide sequence accession numbers.
The nucleotide sequences of the fetA region from the four isolates have been deposited in the GenBank database under accession numbers EF153762 (strain NM0160), EF153764 (strain NM1042), EF153764 (strain NM1630), and EF157665 (strain NM0005).
Acknowledgments
We thank Dan Granoff and Sheldon Kaplan for sharing the isolate from Ohio.
This work was supported in part by a career development award to L. H. Harrison (National Institute of Allergy and Infectious Diseases grant K24 AI52788).
L. H. Harrison receives consulting fees and speaking honoraria from Sanofi Pasteur, Chiron Vaccines, and GlaxoSmithKline and receives research funding from Sanofi Pasteur.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Ala'Aldeen, D. A., H. A. Davies, and S. P. Borriello. 1994. Vaccine potential of meningococcal FrpB: studies on surface exposure and functional attributes of common epitopes. Vaccine 12:535-541. [DOI] [PubMed] [Google Scholar]
- 2.Beucher, M., and P. F. Sparling. 1995. Cloning, sequencing, and characterization of the gene encoding FrpB, a major iron-regulated, outer membrane protein of Neisseria gonorrhoeae. J. Bacteriol. 177:2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finne, J., M. Leinonen, and P. H. Makela. 1983. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet ii:355-357. [DOI] [PubMed] [Google Scholar]
- 4.Harrison, L. H., K. A. Jolley, K. A. Shutt, J. W. Marsh, M. O'Leary, L. T. Sanza, and M. C. Maiden. 2006. Antigenic shift and increased incidence of meningococcal disease. J. Infect. Dis. 193:1266-1274. [DOI] [PubMed] [Google Scholar]
- 5.Kortekaas, J., S. A. Muller, P. Ringler, M. Gregorini, V. E. Weynants, L. Rutten, M. P. Bos, and J. Tommassen. 2006. Immunogenicity and structural characterisation of an in vitro folded meningococcal siderophore receptor (FrpB, FetA). Microbes Infect. 8:2145-2153. [DOI] [PubMed] [Google Scholar]
- 6.Kortekaas, J., A. Pettersson, J. van der Biezen, V. E. Weynants, P. van der Ley, J. Poolman, M. P. Bos, and J. Tommassen. 2007. Shielding of immunogenic domains in Neisseria meningitidis FrpB (FetA) by the major variable region. Vaccine 25:72-84. [DOI] [PubMed] [Google Scholar]
- 7.Pettersson, A., B. Kuipers, M. Pelzer, E. Verhagen, R. H. Tiesjema, J. Tommassen, and J. T. Poolman. 1990. Monoclonal antibodies against the 70-kilodalton iron-regulated protein of Neisseria meningitidis are bactericidal and strain specific. Infect. Immun. 58:3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 9.Thompson, E. A., I. M. Feavers, and M. C. Maiden. 2003. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology 149:1849-1858. [DOI] [PubMed] [Google Scholar]
- 10.Urwin, R., J. E. Russell, E. A. Thompson, E. C. Holmes, I. M. Feavers, and M. C. Maiden. 2004. Distribution of surface protein variants among hyperinvasive meningococci: implications for vaccine design. Infect. Immun. 72:5955-5962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Ende, A., C. T. Hopman, and J. Dankert. 1999. Deletion of porA by recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect. Immun. 67:2928-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]