Abstract
A Caulobacter sp. isolate was recovered from the dialysis fluid of a patient undergoing peritoneal dialysis. Bacterial identification included electron microscopy and 16S rDNA sequencing. To our knowledge, this is the first report of human Caulobacter infection. Special growth requirements suggest that Caulobacter spp. may be overlooked in the clinical microbiology laboratory.
CASE REPORT
A 64-year-old man with chronic renal insufficiency undergoing intermittent peritoneal dialysis was admitted to the hospital in August 2005 because of peritonitis (abdominal cramps and cloudy dialysis fluid). The patient had been undergoing intermittent peritoneal dialysis since February 2005 because of progressive renal insufficiency due to arteriosclerosis. His medical history also included hypertension and smoking. On examination, the patient was relatively asymptomatic except for the abdominal cramps. He was not febrile (37.1°C); however, he had received acetaminophen before admission. The white blood cell count showed neutrophilic leukocytosis (14.3 × 109/liter). The C-reactive protein level was less than 5 mg/liter but rose to 52 mg/liter the following day. Empirical antibiotic treatment with intraperitoneal gentamicin and vancomycin was initiated.
Two sets of blood culture bottles with peritoneal fluid (40 ml, total), each set including an aerobic and an anaerobic bottle (BACTEC Plus aerobic/F and anaerobic/F, respectively), were obtained from the patient before antibiotics were administered. The two sets were obtained a few minutes apart. The blood culture bottles were incubated in a BACTEC 9240 automated instrument (Becton Dickinson Diagnostic Instrument Systems, Franklin Lakes, NJ). Growth was observed in the two aerobic bottles after 4 days of incubation. Gram staining revealed a gram-negative curved rod (Fig. 1A). Material was subcultured on 5% horse blood agar, bromthymol-blue lactose agar (a selective medium for Enterobacteriaceae), and Danish blood agar, used for antibiotic susceptibility testing (Statens Serum Institut [SSI] Diagnostica, Hillerød, Denmark) and incubated at 36°C in an aerobic atmosphere. After 2 days, small, convex, smooth white to pale gray colonies were seen only on the Danish blood agar. The isolate was also subcultured on 10% horse blood agar, chocolate agar, simple agar, and Mueller-Hinton agar (SSI Diagnostica), but no growth was observed after 48 h. Subsequent subculturing for further examination was performed on Danish blood agar. The isolate was non-acid fast. Polar motility was observed in a wet mount preparation. A test for oxidase was positive in 4 to 5 s. The isolate was also peroxidase positive, but the catalase test was negative.
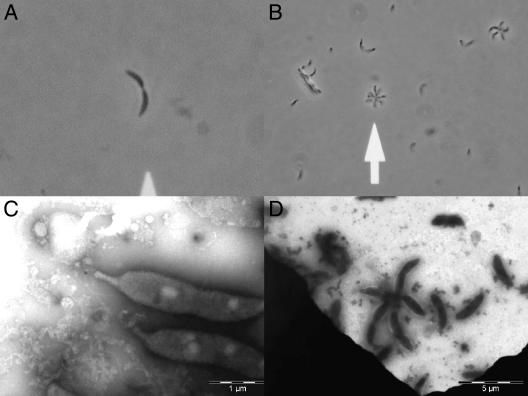
FIG. 1.
Caulobacter case isolate (SSI4214). Wet mount (A and B). (A) Dividing Caulobacter and (B) rosette formation. Electron microscopy, negative staining (C and D). (C) Dividing Caulobacter, stalk to the left, and (D) rosette formation, typical five-pointed star (the Caulobacter pixie).
Antibiotic susceptibility of the isolate was determined by standard disk diffusion with Neo-Sensitabs (Rosco Diagnostica, Taastrup, Denmark). This was supplemented by an E-test (AB Biodisk, Solna, Sweden) for specific antibiotics (Table 1). The isolate was classified as resistant, intermediate, or susceptible by the growth inhibition zones, according to the guidelines of the disk and the E-test manufacturer. The isolate was found to be sensitive to meropenem, imipenem, erythromycin (disk only), aminoglycosides, tetracycline, chloramphenicol, and sulfadiazine. Surprisingly, the isolate was also sensitive to rifampin, linezolid, and vancomycin (disk only) but not to clindamycin (Table 1). The isolate was resistant to almost all penicillins and cephalosporins. However, sensitivity to amdinocillin and intermediate susceptibilities to amoxicillin-clavulanic acid and to cefotaxime were noted, although the amdinocillin minimal inhibitory concentration according to E-testing was high (Table 1). The isolate was also resistant to nalidixic acid, ciprofloxacin, moxifloxacin, polymyxin, and trimethoprim.
TABLE 1.
Antibiotic susceptibility of the Caulobacter case isolate (SSI4214) according to E-testing
| Antibiotic | MIC (μg/ml) | E-test interpretationa |
|---|---|---|
| Benzylpenicillin | >32 | R |
| Ampicillin | >256 | R |
| Amoxicillin-clavulanic acid | 16 | I |
| Piperacillin-tazobactam | >256 | R |
| Aztreonam | >256 | R |
| Amdinocillin | 24 | R |
| Cefoxitin | >256 | S |
| Cefpodoxime | >256 | R |
| Cefuroxime | >256 | R |
| Ceftazidime | 32 | R |
| Cefotaxime | 16 | I |
| Ceftriaxone | >32 | R |
| Meropenem | 2 | S |
| Imipenem | 3 | S |
| Erythromycin | 4 | I |
| Gentamicin | 0.5 | S |
| Streptomycin | 3 | S |
| Netilmicin | 0.5 | S |
| Tobramycin | 0.38 | S |
| Nalidixic acid | >256 | R |
| Ciprofloxacin | >32 | R |
| Moxifloxacin | 16 | R |
| Polymyxin | 384 | R |
| Tetracycline | 0.094 | S |
| Sulfadiazine | 0.094 | S |
| Vancomycin | 24 | R |
| Clindamycin | 48 | R |
| Linezolid | 1.5 | S |
| Rifampin | 1 | S |
S, susceptible; I, intermediate; R, resistant.
The patient completed a 21-day course of intraperitoneal gentamicin and made an uneventful recovery. The white blood cell count and the C-reactive protein level were normalized at day 7.
Biochemical identification with API ID 32 GN and VITEK 2 GN systems (bioMérieux, Marcy l'Etoile, France) was performed. The two systems identified the isolates as Brevundimonas vesicularis (probability, 90.6%) and Sphingomonas paucimobilis (probability, 97.24), respectively. However, the distinct curved (crescent) shape and the color of the colonies were not in accordance with either Brevundimonas vesicularis or Sphingomonas paucimobilis, which are both usually straight rods with yellow colonies.
The cellular fatty acid profile was determined by gas chromatography (6890 series gas chromatograph; Agilent Technologies, Palo Alto, CA), with a culture grown for 2 days on casein soy agar (SSI Diagnostica). The results of the cellular fatty acid analysis were as follows: major (>10%) C16:1ω7c, 12.26%; C16:0, 25.13%; C18:1ω7c, 33.93%; C18:1ω7c11CH3, 14.63%; minor (<10%) C12:0, 1.27%; C12:1 3OH, 0.83%; C12:0 3OH, 0.93%; C14:0, 2.33%; C15:0, 1.03%; C14:0 2OH, 0.92%; C18:0, 1.10%; C17:0 ISO 3OH, 1.93%; C19:0 CYCLO ω8c, 3.70%. The profile was analyzed with Sherlock microbial identification system software (MIDI, Newark, DE), but there were no significant matches in the Sherlock library, in which the highest match had an unacceptable similarity index.
To identify the isolate further, sequencing of the 16S rDNA gene was performed. Genomic DNA was extracted from culture material on Danish blood agar, using a QIAamp DNA mini-kit (QIAGEN, Hilden, Germany) according to the manufacturer's specifications. PCR-mediated amplification of the 16S ribosomal DNA (rDNA) gene was performed with primers BSF-8 (5′-AGAGTTTGATCCTGGCTCAG-3′) and BSR-1407 (5′-GACGGGCGGTGTGTRC-3′), including controls (Escherichia coli DNA as a positive control and distilled water as a negative control). A Quantitect SYBR Green kit (QIAGEN) was used for real-time PCR, as described previously (4). PCR conditions were as follows: 95°C for 15 min, followed by 40 cycles at 94°C for 30 s, 63°C for 30 s, and 72°C for 30 s. The PCR product was purified for later DNA sequencing using spin columns (Microcon YM-100 filter units; Millipore, Billerica, MA). Four primers were used as sequencing primers, as follows: BSF-8 and BSR-1407 and BSF-815 (5′-AGGATTAGATACCCTGGTAGTCC-3′) and BSR-838 (5′-GGACTACCAGGGTATCTAATCCT-3′), to obtain an overlap of alignment of the sequences. Both DNA strands of the amplicons were sequenced on an ABI PRISM 3100 Avant genetic analyzer (Applied Biosystems, Foster City, CA) using a BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems). Sequencing data were edited using SeqScape Software (Applied Biosystems), and the data (1,274-bp sequence) were then compared using the BLAST search engine for sequences deposited in the NCBI database. Evaluations for the number of identities (a percentage) for the best match and the next-best match were performed. The sequence similarity (16S rDNA gene) with the highest number of identities (1,254/1,254 bp [100%]), was obtained with Caulobacter sp. strain FWC41 (GenBank accession no. AJ227775). The next best match was with Caulobacter crescentus strain CB15 (number of identities, 1,268/1,274 [99.5%]; GenBank accession numbers AE005673, AE006011, and AE005930).
Finally, the morphological characteristics of the present Caulobacter case isolate were confirmed by electron microscopy (Fig. 1C and D).
The genus Caulobacter was defined by Henrici and Johnson in 1935, based on microscopic findings of organisms attached to slides which had been suspended in a freshwater lake (similar bacteria had also been described in 1890 by Loeffler) (2, 3, 8). Caulobacter spp. are characterized by their asymmetric cell division and stalk. Caulobacter spp. are motile and have a single polar flagellum. The flagellum is lost, and a stalk is developed (Caulobacter, stalked rod). When Caulobacter divides, the younger cell emerging from the opposite pole bears a single flagellum at the free pole (Fig. 1C). In the laboratory, a rosette formation is often observed in wet mounts (Fig. 1B and D). Caulobacter spp. are aerobic, and the optimal temperature for growth for most Caulobacter spp. is 25°C to 30°C (ranging from 10°C to 35°C). NaCl usually inhibits growth, and typically, no growth is seen with NaCl concentrations above 2% (wt/vol) (2). Special requirements for growth are often needed (riboflavin, cyanocobalamin, biotin, and unidentified growth factors from peptone) (8, 9). Caulobacter can be isolated from distilled, tap, and commercially bottled drinking water; river, canal, and pond water; and sewage (activated sludge during secondary treatment) (3, 6, 9). Caulobacter vibrioides has also been cultured from water used for hemodialysis (5). The genus is very well characterized, based on 16S rDNA sequencing, including several species which have been placed among the alphaproteobacterial class (7, 9).
The diagnosis of the present case isolate (SSI4214) based on 16S rDNA sequencing is supported by the cellular fatty acid profile (1). The profile corresponds to the pattern C according to Abraham et al. (2) and classifies the case isolate as a non-group IV Caulobacter sp. with 16S rDNA pattern III (7, 9). Also, the antibiotic susceptibility of the case isolate is in accordance with the Caulobacter genus (6). Resistance to polymyxin and sensitivity to chloramphenicol, tetracycline, erythromycin, tobramycin, and streptomycin are characteristics similar to those of Caulobacter sp. strain FWC41, which was isolated from activated sludge at a secondary treatment facility in Canada. An isolate with a similar 16S rDNA sequence has been obtained from an industrial production site in Sweden (CCUG number 45663; Culture Collection, University of Göteborg, Sweden).
To our knowledge, this is the first human case of Caulobacter infection. The final diagnosis was made several months after the patient had been discharged from the hospital. Unfortunately, the patient died of other causes before any information about possible exposure to freshwater except for showers, e.g., bathtubs, swimming pools, freshwater lakes, or public baths, could be obtained. The possibility that the case isolate was a contaminant was considered; however, the fact that the case isolate was recovered from two separate sets of blood culture bottles as the only isolate does not support this theory. Also, Caulobacter spp. are not a part of the normal microbial flora of the skin. Another possible source of infection might have been the dialysis bags used for peritoneal dialysis. As Caulobacter can be recovered from many freshwater facilities, the source of infection in this case remains unknown (3, 9).
The special requirements for growth, the low temperatures, and the NaCl intolerance suggest that several species of Caulobacter may be overlooked or misidentified by standard methods in the clinical microbiology laboratory. In this case, the isolate was misidentified as Brevundimonas vesicularis and Sphingomonas paucimobilis by automated identification systems, as Caulobacter spp. are not in the API or the VITEK database. The suspicion of Caulobacter is based on the primary microscopic findings (crescent shape and rosette formation). Subsequently, the final diagnosis should be confirmed by 16S rDNA sequencing, if possible. However, the facts that Caulobacter infection-induced peritonitis is probably rare and that the pathogenic potential of Caulobacter is low, combined with a high degree of sensitivity to aminoglycosides, imply that a systematic detection effort of Caulobacter spp. is not imperative. However, for a patient with recurrent culture-negative peritonitis, Caulobacter may be considered.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Abraham, W.-R., H. Meyer, S. Lindholst, M. Vancanneyt, and J. Smit. 1997. Phospho- and sulfolipids as biomarkers of Caulobacter sensu lato, Brevundimonas and Hyphomonas. Syst. Appl. Microbiol. 20:522-539. [Google Scholar]
- 2.Abraham, W.-R., C. Strompl, H. Meyer, S. Lindholst, E. R. Moore, R. Christ, M. Vancanneyt, B. J. Tindall, A. Bennasar, J. Smit, and M. Tesar. 1999. Phylogeny and polyphasic taxonomy of Caulobacter species. Proposal of Maricaulis gen. nov. with Maricaulis maris (Poindexter) comb. nov. as the type species, and emended description of the genera Brevundimonas and Caulobacter. Int. J. Syst. Bacteriol. 49:1053-1073. [DOI] [PubMed] [Google Scholar]
- 3.Bowers, L. E., R. H. Weaver, E. A. Grula, and O. F. Edewards. 1954. Studies on a strain of Caulobacter from water. I. Isolation and identification as Caulobacter vibrioides Henrici and Johnson with emended description. J. Bacteriol. 68:194-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, J. J., K. Andresen, T. Justesen, and M. Kemp. 2005. Ribosomal DNA sequencing: experiences from use in the Danish National Reference Laboratory for Identification of Bacteria. APMIS 113:621-628. [DOI] [PubMed] [Google Scholar]
- 5.Gomila, M., J. Gasco, A. Busquets, J. Gil, R. Bernabeu, J. M. Buades, and J. Lalucat. 2005. Identification of culturable bacteria present in haemodialysis water and fluid. FEMS Microbiol. Ecol. 52:101-114. [DOI] [PubMed] [Google Scholar]
- 6.MacRae, J. D., and J. Smit. 1991. Characterization of caulobacters isolated from wastewater treatment systems. Appl. Environ. Microbiol. 57:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poindexter, J. S. 2005. Genus I. Caulobacter, p. 287-303. In J. G. Holt, N. R. Krieg, P. H. Sneath, J. T. Staley, and S. T. Williams (ed.), Bergey's manual of determinative bacteriology, 10th ed. Williams & Wilkins Co., Baltimore, MD.