Abstract
We prospectively evaluated a new PCR-enzyme-linked immunosorbent assay kit (Onychodiag; BioAdvance, France) for the diagnosis of dermatophytic onychomycosis by testing nail samples from 438 patients with suspected onychomycosis and from 108 healthy controls in three independent laboratories. In two laboratories, samples were collected by trained mycologists as close as possible to the lesions (proximal samples). In one laboratory, samples were collected by other physicians. All samples were processed by conventional mycological techniques and by Onychodiag, blindly to the mycological results. An additional distal sample, collected by clipping the nail plate, was obtained from 75 patients and tested with Onychodiag alone. In patients with culture-proven dermatophytic onychomycosis, the sensitivity of Onychodiag was 83.6% (87.9% including the gray zone) and ranged from 75 to 100% according to the laboratory and the sampling conditions. The specificity was 100% when healthy subjects were considered true negative controls. Onychodiag was positive on 68 patient samples that were sterile or yielded nondermatophyte species in culture. Based on the results of Onychodiag for mycologically proven positive samples and true-negative samples, these results were considered true positives, and the poor performance of mycology on these samples was attributed to inconvenient sampling conditions or to contaminants. When tested on distal samples, Onychodiag was positive in 49/53 (92%) cases of proven dermatophytic onychomycosis. Finally, with either proximal or distal samples, Onychodiag provided a diagnosis of dermatophytic onychomycosis within 24 to 48 h after sampling, and its sensitivity was close to that of mycological techniques applied to proximal samples.
Onychomycoses are very common infections responsible for about half of all nail dystrophies, and their incidence has been increasing worldwide (2, 6, 10). Dermatophytes account for about 90% cases of toenail onychomycosis and at least 50% of fingernail onychomycosis (5, 14). Identification of the fungal origin of the lesion is required to avoid misdiagnosis with nonfungal nail dystrophies and to justify the expense, long duration, and potential adverse effects of antifungal treatment (8, 15).
Laboratory diagnosis of onychomycosis currently relies on proper sampling of the nail and demonstration of hyphae by direct microscopic examination after treatment with KOH, followed by culture and species identification (1, 5, 22). Direct microscopy, although falsely negative in 5 to 15% of cases in ordinary practice (22), is an efficient screening technique, but it cannot always differentiate dermatophyte hyphae from mold hyphae. Culture requires up to 3 to 4 weeks to obtain typical macroscopic and microscopic features for specific dermatophyte identification and is also subject to a false-negative rate of about 30% (10). Appropriate sampling conditions are crucial for diagnosis. If the sample is not collected in proper conditions, i.e., as close as possible to the cuticle and to the advancing infected edge of the lesion, then both direct examination and culture are unreliable, with a high rate of false-negative results and also false-positive results due to fungal or bacterial contamination (1, 5, 7).
The last decade has seen significant advances in molecular methods for rapid and sensitive identification of dermatophytes. Most are based on real-time PCR, species-specific nested PCR, or a combination of PCR with restriction fragment length polymorphism or sequencing (3, 9, 11, 12, 13, 16, 18, 19). Menotti et al. (17) previously showed the excellent sensitivity and specificity of a PCR-restriction fragment length polymorphism assay for rapid diagnosis of dermatophytes and Scytalidium spp. on properly sampled clinical specimens, as well as the performance of this method on distal nail specimens that are improper for mycological examination. Based on these results and on the fact that dermatophytes account for most cases of fungal onychomycosis, a new PCR-enzyme-linked immunosorbent assay (ELISA) was recently developed and marketed as Onychodiag (Bio Advance, Bussy-Saint-Martin, France) for the diagnosis of dermatophyte onychomycosis.
We evaluated the performance of this test in three independent laboratories with different nail sampling practices and compared the results with those of conventional mycological techniques.
MATERIALS AND METHODS
Participating laboratories and sampling conditions.
Three university hospital laboratories participated in this study. Each laboratory has staff well trained in mycological diagnosis and routinely examines 10 to 30 dermatological samples per day.
In the Parasitology-Mycology laboratory of St-Louis hospital in Paris (laboratory A), nail samples were collected at the outpatient dermatology department of the hospital by well-trained laboratory staff.
The Institute of Parasitology of Strasbourg (laboratory B) received nail samples referred from laboratories or physicians in the surrounding area. Healthy control samples (CS) were collected from laboratory staff by a dermatologist of the Institute.
The Parasitology-Mycology laboratory of Michallon hospital in Grenoble (laboratory C) tested nail samples collected by well-trained laboratory biologists.
Clinical specimens.
In each center, various types of nail samples were collected prospectively from healthy controls and from patients with suspected onychomycosis.
Reference proximal samples (RPS) were collected by mycologists or other well-trained staff as recommended for optimally sensitive mycological diagnosis (5). Subungual debris were obtained from the infected nail bed as close as possible to the cuticle or to the advancing infected edge of the lesion. Material was also obtained from the underside of the nail bed. These samples were collected before treatment was prescribed.
“Distal samples” (DS) were collected in healthy controls and in some patients by clipping the free edge of the nail plate at the hyponychium. In patients, samples were collected by trained mycologists, just before collecting an RPS from the same dystrophic nail.
“Regular samples” (RegS) were referred to laboratory B with no information on the sampling conditions or site. These samples probably consisted of a mixture of RPS and DS.
CS were collected by clipping the nail plate of one healthy nail of volunteers with no nail dystrophy and no cutaneous fungal lesions. Sampling was performed at home or in the laboratories.
Mycological diagnosis.
In the three laboratories, nail samples were processed with the same mycological techniques. Part of the sample was examined microscopically after mounting in black chlorazol solution containing KOH, and the presence of hyphae was recorded. Part of the sample was cultured on both a Sabouraud chloramphenicol agar slant and a Sabouraud chloramphenicol plus cycloheximide agar slant (Bio-Rad, Marnes-la-Coquette, France) for fungal identification. Incubation for up to 3 to 4 weeks at 25 to 30°C is necessary for the growth and identification of all dermatophytes. The species of dermatophytes and the genus of other fungi were identified by their macroscopic and microscopic appearance after lactophenol cotton blue staining.
Part of the sample was kept at room temperature for molecular diagnosis.
Molecular diagnosis.
Nail pieces were cut into small fragments with a surgical blade before DNA extraction. DNA was extracted and purified using the High Pure PCR template preparation kit (Roche, Mannheim) and then processed with the Onychodiag kit in two steps, as recommended by the manufacturer.
The first step, consisting of PCR amplification of the fungal DNA, was performed in a 25-μl volume containing 5 μl of DNA template, 75 mM Tris-HCl, 20 mM (NH4)2SO4, 0.01% Tween 20, 1 mM MgCl2, 0.1 mM concentrations of each deoxynucleotide triphosphate (dNTP), 1 μl of each primer provided with the kit, and 1 U of Taq DNA polymerase (Eurogentec, Angers, France). Amplifications were performed using a TC-412 thermal cycler (Techne, Bioblock, Illkirch, France) in laboratory A and a Mastercycler 96 (Eppendorf, Le Pecq, France) in laboratory B, according to the manufacturer's instructions, with initial denaturation for 5 min at 94°C, 40 cycles of amplification (denaturation for 60 s at 94°C, annealing for 40 s at 60°C, and extension for 60 s at 72°C), a final extension step for 5 min at 72°C, and then cooling to 4°C.
In laboratory C, amplifications were performed using a PTC-100 thermal cycler (MJ Research, Inc., Watertown, MA) by following a protocol that agreed with Bio Advance's technical support. The amplification volume was increased from 25 to 50 μl, and the amounts of all other reagents were doubled. dNTP were replaced by a dNTP/dUTP mix (Eurogentec, Angers, France) consisting of 0.2 mM dATP, dGTP, and dCTP and 0.4 mM dUTP. Bovine serum albumin at a final concentration of 0.6% (wt/vol) and uracil-N-glycosylase (UNG) (Eurogentec, Angers, France) at a final concentration of 0.5 U/50 μl were also added. Finally, a drop of mineral oil was placed on the mix before amplification. The amplification conditions were as in laboratories A and B, including two additional steps before initial denaturation, namely, digestion with UNG for 10 min at 22°C and inactivation of UNG for 10 min at 94°C. The annealing temperature was reduced from 60°C to 55°C.
In the second step, amplified products were revealed with an ELISA as recommended by the manufacturer. Briefly, amplified products were mixed within dermatophyte-specific labeled probe and then subjected to chemical denaturation for 10 min at room temperature. Denatured products were transferred into a capture-probe-precoated microwell containing hybridization buffer and were incubated for 1 h at 37°C. After 6 washing steps with washing buffer, the diluted conjugate was added to each well and incubated for 1 h at 25°C. After 6 washing steps a colorimetric substrate was added to each well and incubated for 30 min at 25°C in the dark. The reaction was stopped with the stop solution.
Optical density values were recorded spectrophotometrically at 450 nm, using 620 nm as a reference wavelength. In these conditions, optical density (OD) values are <0.1 when the test is performed with human DNA or genomic DNA extracted from cultures of Scytalidium spp., Acremonium, Scopulariopsis, Fusarium, Aspergillus, Candida famata, Candida albicans, and Candida parapsilosis and >3.0 with DNA extracted from any species of dermatophyte (Bio Advance information).
The sensitivity of Onychodiag for the detection of dermatophyte DNA was examined in laboratory A by testing serial dilutions of a titrated suspension of plasmid (positive control provided with the kit) containing the target sequence.
Expression of results.
To be interpretable, each series of tests must include one negative and one positive control (both provided with the kit), the OD values of which should be <0.2 and >0.7, respectively. For clinical samples, results are expressed as the OD value obtained with the sample minus the OD value of the negative control. A calculated OD value of ≥0.5 is considered positive and indicates the presence of dermatophytic DNA in the sample. The test is considered negative with a calculated OD of <0.3 and borderline with an OD between 0.3 and 0.5 (gray zone). Moreover, an additional negative extraction control must be added for each DNA extraction series, and its calculated OD must be <0.3, showing the absence of cross-contamination.
Study design and analysis.
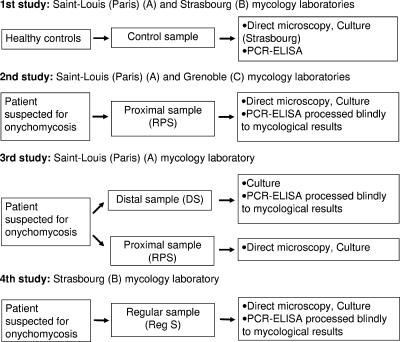
We blindly evaluated the performance of Onychodiag compared to mycological diagnosis, including direct microscopic examination (DE) and/or selective culture (Fig. 1).
FIG. 1.
Study design and participating laboratories assessing Onychodiag on healthy control samples (study 1), RPS (study 2), and DS compared to RPS (study 3) and Reg S (study 4).
For RPS, culture was used as the gold standard to calculate the sensitivity and specificity of Onychodiag, i.e., the number of positive Onychodiag tests/the number of positive dermatophyte cultures and the number of negative Onychodiag tests/the number of negative dermatophyte cultures, respectively.
In our experience, direct microscopy is totally unsuited to DS, and culture is positive in less than 20% of cases of clinically proven dermatophyte onychomycosis. These samples are also frequently contaminated by molds and bacteria (data not shown). As DS cannot be used as a reliable reference for the diagnosis of onychomycosis, a specific protocol was applied in laboratory A to assess the performance of Onychodiag on these samples. In 75 patients with suspected onychomycosis, two samples were successively collected from the same nail. The first, a DS, was tested with Onychodiag alone; the second, an RPS, was tested with Onychodiag and by mycological methods (DE and culture), the results of which were used as reference.
RegS were processed in the same way as RPS. CS were collected in laboratories A and B and were tested with Onychodiag alone in laboratory A and with Onychodiag and mycological methods in laboratory B.
These studies were conducted prospectively in the three laboratories between January 2004 and July 2005, and each laboratory was unaware of the results of the other laboratories until the end of the study.
RESULTS
Sensitivity of Onychodiag.
Testing of serial dilutions of the positive control showed positive OD values for samples containing at least 25 target copies, corresponding to less than 1 fg of target fungal DNA.
Clinical specimens.
A total of 546 clinical samples were studied in the three participating laboratories; they comprised 236 RPS (Table 1), 75 DS (Table 2), 127 RegS (Table 3), and 108 healthy CS.
TABLE 1.
Performance of Onychodiag relative to culture and DE alone or in combination for the diagnosis of dermatophyte onychomycosis on 236 RPS
| RPS mycology | Total no. of samples | No. of RPS with Onychodiag resulta:
|
||
|---|---|---|---|---|
| Positive | Gray zone | Negative | ||
| Dermatophyte culture | ||||
| Positive | 106 | 83 | 6 | 17 |
| Negativeb (no. sterile, no. nondermatophytes) | 130 (105, 25) | 30 (24, 6) | 12 (9, 3) | 88 (72, 16) |
| DE | ||||
| Positive | 151 | 103 | 13 | 35 |
| Negative | 85 | 10 | 5 | 70 |
| Combination of DE and culture | ||||
| Positive dermatophyte culture or positive DE | 151 | 103 | 13 | 35 |
| Negative dermatophyte culture and negative DEb | 85 | 10 | 5 | 70 |
Positive, OD ≥ 0.5; gray zone, 0.3 ≤ OD < 0.5; negative, OD < 0.3.
Including sterile cultures and cultures of nondermatophyte species.
TABLE 2.
Comparative performance of Onychodiag for diagnosis of dermatophyte onychomycosis on 75 DSa
| Corresponding RPS mycology | Total no. of samples | No. of DS with Onychodiag resultb:
|
||
|---|---|---|---|---|
| Positive | Gray zone | Negative | ||
| Dermatophyte culture | ||||
| Positive | 53 | 49 | 4 | 0 |
| Negativec (no. sterile, no. nondermatophytes) | 22 (11, 11) | 6 (3, 3) | 0 | 16 (8, 8) |
| DE | ||||
| Positive | 65 | 55 | 4 | 6 |
| Negative | 10 | 0 | 0 | 10 |
| Combination of DE and culture | ||||
| Positive dermatophyte culture or positive DE | 65 | 55 | 4 | 6 |
| Negative dermatophyte culture and negative DEc | 10 | 0 | 0 | 10 |
The mycology results are those for the corresponding RPS.
Positive, OD ≥ 0.5; gray zone, 0.3 ≤ OD < 0.5; negative, OD < 0.3.
Including sterile cultures and cultures of nondermatophyte species.
TABLE 3.
Performance of Onychodiag relative to culture and DE alone and in combination for the diagnosis of dermatophyte onychomycosis on 127 RegS
| RegS mycology | Total no. of samples | No. of RegS with Onychodiag resulta:
|
||
|---|---|---|---|---|
| Positive | Gray zone | Negative | ||
| Dermatophyte culture | ||||
| Positive | 34 | 34 | 0 | 0 |
| Negativeb (no. sterile, no. nondermatophytes) | 93 (48, 45) | 32 (14, 18) | 6 (4, 2) | 55 (30, 25) |
| DE | ||||
| Positive | 33 | 31 | 0 | 2 |
| Negative | 94 | 35 | 6 | 53 |
| Combination of DE and culture | ||||
| Positive dermatophyte culture or positive DE | 33 | 31 | 0 | 2 |
| Negative dermatophyte culture and negative DEb | 80 | 21 | 6 | 53 |
Positive, OD ≥ 0.5; gray zone, 0.3 ≤ OD < 0.5; negative, OD < 0.3.
Including sterile cultures and cultures of nondermatophyte species.
Healthy controls.
Among the 108 healthy control nails, 4/68 were positive by Onychodiag in laboratory A and 0/40 in laboratory B. A new sample was collected from the 4 positive patients, who were asked to carefully wash their hands and brush their nails before sampling. These four samples were negative by Onychodiag. Mycological culture was negative for 39/40 samples in laboratory B and was not performed in laboratory A. None of the controls who were positive by Onychodiag had visible lesions or a past history of fungal cutaneous or nail infection. None of these patients developed clinical lesions compatible with onychomycosis.
RPS.
A dermatophyte was isolated by culture in 106 of 236 RPS (Table 1). The proportion of sample cultures positive for dermatophytes was similar in laboratories A and C (65/136, 47.8% and 41/100, 41%, respectively). In both laboratories, the most frequently isolated dermatophyte species was Trichophyton rubrum (n = 94), followed by Trichophyton interdigitale (n = 11) and Microsporum gypseum (n = 1). Nondermatophyte fungi were identified in 25 cultures and comprised Candida (n = 8, including 2 Candida albicans cultures), Scopulariopsis (n = 6), Fusarium (n = 5), Acremonium (n = 3), Aspergillus (n = 3), Penicillium (n = 1), Scytalidium (n = 1), and other molds (n = 2); four of these 25 cultures contained two different species. One hundred five samples were sterile.
DE was positive for 151/236 RPS. All 106 samples that yielded a dermatophyte in culture were positive by direct examination. Among the remaining 45 samples, 26 were sterile and 19 grew nondermatophyte fungi.
Onychodiag was positive on 83 (78.3%) of the 106 samples that were dermatophyte positive in culture. The proportion of dermatophyte culture-positive samples that were also positive with Onychodiag was higher in laboratory C (82.9%) than in laboratory A (75.4%). The OD values for positive samples were within the same range in the two laboratories (0.54 to 3; mean, 1.52 ± 0.8). Among the 23 cases of culture-proven dermatophytic onychomycosis not diagnosed by Onychodiag, 6 were within the gray zone (3 T. interdigitale and 3 T. rubrum) and 17 were negative by Onychodiag (12 T. rubrum and 5 T. interdigitale).
Among the 130 samples that were sterile in culture (n = 105) or which grew a nondermatophyte fungus (n = 25), 30 were positive with Onychodiag, and 15 of these samples were also DE positive.
Onychodiag was positive for 103 of the 151 DE-positive samples. Of these, 83 corresponded to culture-proven dermatophytic onychomycosis, 15 were sterile, and 5 grew nondermatophyte fungi. Among the 48 samples which were DE positive but were negative or in the gray zone with Onychodiag, 23 were from patients with culture-proven dermatophyte infection (see above for details), 11 were sterile, and 14 grew other fungi. Among the 10 samples that were positive by Onychodiag and negative by DE, 9 were sterile and 1 grew another fungus. Similar performances were obtained when Onychodiag was compared to the combined numbers of samples with (i) positive dermatophyte culture and positive DE (i.e., proven or probable onychomycosis) and (ii) negative culture and negative DE (i.e., probable nonfungal lesion).
DS.
The results of Onychodiag on DS from 75 patients with suspected onychomycosis were compared to the results of mycological methods applied to an RPS from the same nail as a reference (Table 2). Culture yielded a dermatophyte in 53/75 of these RPS (51 T. rubrum and 2 T. interdigitale). Forty-nine of the corresponding DS were positive by Onychodiag (92.5%) and 4 were in the gray zone (7.5%). Twenty-two samples were culture-negative or yielded nondermatophyte species; with six samples, Onychodiag was positive, whereas the corresponding RPS was sterile (n = 3) or yielded nondermatophyte species (n = 3).
Onychodiag was positive on 55 of 65 DE-positive samples and gave gray zone or negative results for the other 10 samples. The four samples that were in the gray zone corresponded to culture-proven dermatophyte infection, and the 6 samples that were negative yielded nondermatophyte species in culture.
None of the 10 samples that were DE negative were positive by Onychodiag. As with the RPS, the performance of Onychodiag was similar compared to DE alone or to the combination of DE and culture.
RegS.
Among 127 RegS processed in laboratory B, 34 were culture positive for dermatophytes (T. rubrum in 27 cases Trichophyton mentagrophytes in 7 cases), and all 34 were positive by Onychodiag (Table 3).
Ninety-three samples were sterile (n = 48) or grew nondermatophyte fungi (n = 45). These fungi were Scopulariospis (n = 7), Candida (non-C. albicans species, n = 6), Trichosporon (n = 6), Penicillium (n = 4), Fusarium (n = 2), Acremonium (n = 2), Cryptococcus (n = 2), Rhodotorulla (n = 2), Aspergillus (n = 1), Aureobasidium (n = 1), Kloeckera (n = 1), Ulocladium (n = 1), Engyodontium (n = 1), and other molds (n = 23); 14 cultures contained two fungal species.
Onychodiag was positive for 32 of these 93 cases. Eighteen of these positive samples (of which 9 were also DE positive) grew nondermatophyte species, and 14 (of which 3 were DE positive) were sterile.
Microscopic examination was positive in 33 cases (20 corresponding to proven dermatophyte infections; 13 sterile or yielding a nondermatophyte in culture) and negative in 94 cases (14/20 proven dermatophyte infections, 45/48 sterile, and 35/45 growing a nondermatophyte species). Onychodiag was positive for 31/33 DE-positive samples and positive for 21/80 DE-negative samples, of which 14 grew a dermatophyte. These latter 14 cases account for the difference in the performance of Onychodiag relative to DE alone and to DE plus culture.
Finally, dermatophytic onychomycosis was diagnosed by culture in 140 patients (106 on RPS in laboratories A and C, and 34 on RegS in laboratory B). Onychodiag was positive for 117 cases (83.6%) or for 123 cases (87.9%) when gray zone samples were included.
DISCUSSION
In this multicenter study, we tested the performance of Onychodiag in various technical conditions and on different types of nail samples. Testing of healthy nails in laboratories A and B showed that Onychodiag was consistently negative when the sample was collected after careful washing and brushing of the nail. Physicians and biologists should follow this recommendation, as there is a small but significant risk of false positivity when the nails are not correctly washed before sampling.
In laboratories A and C, application of mycological methods and Onychodiag to RPS allowed us to calculate the sensitivity and specificity of Onychodiag on properly collected samples. In both laboratories, the rates of positive culture and direct examination were very close, probably because of similar sampling conditions. The sensitivity of Onychodiag on RPS from patients with mycologically proven dermatophytic onychomycosis was 78.3% (83.9% including the gray zone). The specificity was 76.9% when negative culture was used as a reference and 88.2% when negative microscopic examination alone was used as reference. Onychodiag was also positive in 30 other cases in which culture was negative or yielded a nondermatophyte fungus or a yeast. Twenty of these samples were positive by direct examination, suggesting that the patients were indeed infected. Given the lack of cross-reactivity of Onychodiag with other fungal DNAs, and the negative results for healthy control nails, these results are unlikely to be Onychodiag false positives.
The results obtained with Onychodiag on DS compared to culture of the corresponding RPS show that Onychodiag could simplify the diagnosis of onychomycosis in routine practice. Based on the results of mycological testing of the proximal sample from the same nail, the sensitivity of Onychodiag for DS was 92% (100% including the gray zone). Its specificity was 73% when culture was used as a reference and 100% when negative microscopic examination alone was used as a reference. Onychodiag was also positive in six other cases in which culture of the corresponding RPS was negative or yielded a nondermatophyte mold or a yeast. As discussed above for RPS, we consider these results true positives. Keeping in mind that DS are not suitable for mycological examination, because of the high rate of false-negative direct examinations and false-positive cultures due to environmental contaminants, this performance is worthy of note. It clearly shows that PCR can provide reliable diagnosis of dermatophytic onychomycosis on readily collected samples. It also guaranties good diagnostic performance when the sampling conditions are not known, which is the case for referral specimens.
The results obtained in laboratory B correspond to this situation. Nail samples were referred to this laboratory by other laboratories or physicians and probably consisted of a mixture of proximal samples and DS. This is supported by the fact that the rate of dermatophyte culture was much lower than in laboratories A and C (26.8% and 44.9%, respectively) and that nondermatophyte species were more frequently isolated in this laboratory (35.4% and 10.6%, respectively). Onychodiag confirmed the diagnosis in all mycologically proven cases of dermatophyte infection but was also positive in 32 other cases in which culture was negative or yielded a nondermatophyte mold or a yeast. As discussed above for proximal samples, we consider these results true positives, especially considering that all samples collected from healthy controls in the same laboratory were negative.
In the absence of a high-quality gold standard for the diagnosis of onychomycosis, it is difficult to estimate the sensitivity and specificity of Onychodiag on these samples.
Calculation of the sensitivity and specificity of Onychodiag based on the results of culture would yield high rates of false positives and false negatives, and the diagnosis of fungal onychomycosis is highly uncertain when a nondermatophytic species is isolated from such samples. Most of the isolates were nonpathogenic and were recovered from samples for which direct microscopic examination was negative (35/45 of cases), suggesting that most of them were contaminants (20).
Consequently, we calculated the specificity of Onychodiag for RegS based on the results for true-negative samples from healthy controls. Even with the sensitivity and specificity values calculated for DS (i.e., the worst sampling conditions), it can be considered that most RegS that were positive by Onychodiag and negative by culture were not false positives. Similarly, several samples that were negative by Onychodiag and that yielded nondermatophyte species in culture were probably not cases of true fungal onychomycosis, especially those with negative microscopic examination.
These difficulties in comparing a new technique to reference mycological methods that have their own limitations have already been underlined (3, 13). The poor sensitivity and specificity of direct examination (10) and the fact that culture is negative in a significant percentage of cases of true dermatophytic onychomycosis, despite positive direct microscopy, could account for the difference in sensitivity/specificity between PCR and mycology. Relatively high sensitivity is expected with PCR amplification (estimated detection limit of 25 target copies with Onychodiag), but high specificity is also necessary to avoid false-positive results. The specificity of Onychodiag was shown by the fact that all healthy control nails were negative when collected after hand washing and by the absence of cross-reactivity with genomic DNA of nondermatophytic fungi.
Finally, considering all the patients with mycologically proven dermatophytic onychomycosis (n = 140), the sensitivity of Onychodiag was 83.6% on average and ranged between 75 and 100% according to the laboratory. The reasons for the negative results with mycologically proven infections are unclear but could be related to different sampling conditions and/or sample sizes used for PCR in the three laboratories. No clear relationship was found between a negative PCR result and the dermatophyte species found in culture.
Less than 7% of the samples fell within the gray zone, in which dermatophyte infection cannot be proven nor ruled out. In such cases, a new nail sample should be collected by a trained mycologist in optimal conditions (i.e., a proximal sample) and should be tested by both Onychodiag and mycological methods (DE and culture).
Onychodiag was designed to detect any dermatophyte species, without species identification. Indeed, a positive PCR result indicating the presence of a dermatophyte in the nail provides the physician with sufficient evidence to start antifungal treatment. Species-level identification would require repeating the test on replicate wells with several specific probes, the cost of which might not be justified when T. rubrum accounts for most dermatophytic nail infections in Europe (4, 21).
In conclusion, this multicenter study shows the good performance of Onychodiag for the diagnosis of onychomycosis due to dermatophytes. Good results were obtained under various sampling conditions, including the use of DS that cannot be properly examined with mycological techniques.
A recent study from the European Onychomycosis Observatory showed that, in case of suspected onychomycosis, only 3.4% of general practitioners and 39.6% of dermatologists requested a sample (4), probably because of the difficulties of mycological diagnosis and the time required for culture. The use of Onychodiag could significantly increase these percentages and facilitate the routine diagnosis of onychomycosis, as samples (even DS) can easily be collected by the physician and sent to the laboratory for diagnosis. Any laboratory equipped with a PCR amplifier and an ELISA microplate spectrophotometer can test such nail samples with Onychodiag and provide the results within 48 h. Further studies are under way to determine the time required for Onychodiag to become negative following topical and/or oral treatment and to examine the value of this test for treatment evaluation.
Acknowledgments
We thank the clinicians who referred the patients and samples to the participating laboratories, the technicians for processing the samples, C. Lacroix and M. Feuilhade for their expert advice, and D. Young for reviewing the manuscript.
This study was supported by a grant from Bio Advance (Bussy-Saint-Martin, France).
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Baran, R., D. Chabasse, and M. Feuilhade de Chauvin. 2001. Onychomycosis, II: diagnostic approach. J. Mycol. Med. 11:5-13. [Google Scholar]
- 2.Chabasse, D. 2003. Can we evaluate the frequency of onychomycosis. Ann. Dermatol. Venerol. 130:1222-1230. [PubMed] [Google Scholar]
- 3.Dobrowolska, A., P. Staczek, A. Kaszuba, and M. Kozlowska. 2006. PCR-RFLP analysis of the dermatophytes isolated from patients in Central Poland. J. Dermatol. Sci. 42:71-74. [DOI] [PubMed] [Google Scholar]
- 4.Effendy, I., M. Lecha, M. Feuilhade de Chauvin, N. Di Chiacchio, and R. Baran. 2005. Epidemiology and clinical classification of onychomycosis. J. Eur. Acad. Dermatol. Venereol. 19(Suppl. 1):8-12. [DOI] [PubMed] [Google Scholar]
- 5.Elewski, B. E. 1998. Onychomycosis: pathogenesis, diagnosis, and management. Clin. Microbiol. Rev. 11:415-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faergemann, J., and R. Baran. 2003. Epidemiology, clinical presentation and diagnosis of onychomycosis. Br. J. Dermatol. 149:1-4. [DOI] [PubMed] [Google Scholar]
- 7.Feuilhade de Chauvin, M. 2005. New diagnostic techniques. J. Eur. Acad. Dermatol. Venereol. 19(Suppl. 1):20-24. [DOI] [PubMed] [Google Scholar]
- 8.Gupta, A. K., and N. H. Shear. 2000. An update, with a risk-benefit evaluation of the newer antifungal agents to treat onychomycosis. Drug Safety 22:33-52. [DOI] [PubMed] [Google Scholar]
- 9.Gutzmer, R., S. Mommert, U. Kuttler, T. Werfel, and A. Kapp. 2004. Rapid identification and differentiation of fungal DNA in dermatological specimens by LightCycler PCR. J. Med. Microbiol. 53(Pt 12):1207-1214. [DOI] [PubMed] [Google Scholar]
- 10.Hay, R. 2005. Literature review. Onychomycosis. J. Eur. Acad. Dermatol. Venereol. 19(Suppl. 1):1-7. [DOI] [PubMed] [Google Scholar]
- 11.Kamiya, A., A. Kikuchi, Y. Tomita, and T. Kanbe. 2004. PCR and PCR-RFLP techniques targeting the DNA topoisomerase II gene for rapid clinical diagnosis of the etiologic agent of dermatophytosis. J. Dermatol. Sci. 34:35-48. [DOI] [PubMed] [Google Scholar]
- 12.Kanbe, T., Y. Suzuki, A. Kamiya, T. Mochizuki, M. Fujihiro, and A. Kikuchi. 2003. PCR-based identification of common dermatophyte species using primer sets specific for the DNA topoisomerase II genes. J. Dermatol. Sci. 32:151-161. [DOI] [PubMed] [Google Scholar]
- 13.Kardjeva, V., R. Summerbell, T. Kantardjiev, D. Devliotou-Panagiotidou, E. Sotiriou, and Y. Graser. 2006. Forty-eight-hour diagnosis of onychomycosis with subtyping of Trichophyton rubrum strains. J. Clin. Microbiol. 44:1419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kemna, M. E., and B. E. Elewski. 1996. A U.S. epidemiologic survey of superficial fungal diseases. J. Am. Acad. Dermatol. 35:539-542. [DOI] [PubMed] [Google Scholar]
- 15.Lecha, M., I. Effendy, M. Feuilhade de Chauvin, N. Di Chiacchio, and R. Baran. 2005. Treatment option-development of consensus guidelines. J. Eur. Acad. Dermatol. Venereol. 19(Suppl. 1):25-33. [DOI] [PubMed] [Google Scholar]
- 16.Machouart-Dubach, M., C. Lacroix, M. Feuilhade De Chauvin, I. Le Gall, C. Giudicelli, F. Lorenzo, and F. Derouin. 2001. Rapid discrimination among dermatophytes, Scytalidium spp., and other fungi with a PCR-restriction fragment length polymorphism ribotyping method. J. Clin. Microbiol. 39:685-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Menotti, J., M. Machouart, M. Benderdouche, C. Cetre-Sossah, P. Morel, L. Dubertret, F. Derouin, M. Feuilhade De Chauvin, and C. Lacroix. 2004. Polymerase chain reaction for diagnosis of dermatophyte and Scytalidium spp. onychomycosis. Br. J. Dermatol. 151:518-519. [DOI] [PubMed] [Google Scholar]
- 18.Ninet, B., I. Jan, O. Bontems, B. Lechenne, O. Jousson, R. Panizzon, D. Lew, and M. Monod. 2003. Identification of dermatophyte species by 28S ribosomal DNA sequencing with a commercial kit. J. Clin. Microbiol. 41:826-830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin, J. H., J. H. Sung, S. J. Park, J. A. Kim, J. H. Lee, D. Y. Lee, E. S. Lee, and J. M. Yang. 2003. Species identification and strain differentiation of dermatophyte fungi using polymerase chain reaction amplification and restriction enzyme analysis. J. Am. Acad. Dermatol. 48:857-865. [DOI] [PubMed] [Google Scholar]
- 20.Summerbell, R. C., E. Cooper, U. Bunn, and F. Jamieson. 2005. Onychomycosis: a critical study of techniques and criteria for confirming the etiologic significance of non dermatophytes. Med. Mycol. 43:39-59. [DOI] [PubMed] [Google Scholar]
- 21.Tietz, H. J., V. Kunzelmann, and G. Schönian. 1995. Changes in fungal spectrum of dermatomycoses. Mycoses 38:33-39. [DOI] [PubMed] [Google Scholar]
- 22.Weitzman, I., and R. C. Summerbell. 1995. The dermatophytes. Clin. Microbiol. Rev. 8:240-259. [DOI] [PMC free article] [PubMed] [Google Scholar]