Abstract
Hepatitis C virus (HCV) isolates have been classified into six main genotypes. Genotyping methods, and especially the widely used line probe assay (LiPA), are frequently based on the 5′-untranslated region (5′UTR). However, this region is not appropriate for discriminating HCV strains at the subtype level and for distinguishing many genotype 6 samples from genotype 1. We investigated the capacity of a novel LiPA (Versant HCV Genotype 2.0 assay) based on the simultaneous detection of 5′UTR and Core regions for genotypes 1 and 6 to provide correct HCV genotypes (characterized with a phylogenetic analysis) in a set of HCV strains mainly encountered in Western countries. The improvement was assessed by comparing the results to those obtained with the previous version of the assay. Of the 135 tested samples, 64.7% were concordant for genotype group and subtype with sequencing reference results using the Versant HCV Genotype 2.0 assay versus 37.5% with the previous version. The yield was mainly related to a better characterization of genotype 1, since the accuracy, tested in 62 genotype 1 samples, increased from 45.2% with the first version to 96.8% with the new one. However, this new version necessitates a specific PCR and could no longer be used after 5′UTR PCR used for current HCV infection diagnosis. Moreover, the information provided by 5′UTR hybridization is not reliable for correctly identifying the diversity within genotypes 2 and 4. Thus, the Versant HCV Genotype 2.0 assay remains a useful tool for clinical practice when only the discrimination between major HCV genotypes is necessary.
Hepatitis C virus (HCV) infection is one of the most common causes of chronic liver disease, with a risk of evolution toward cirrhosis and hepatocellular carcinoma (36). In order to avoid the occurrence of such severe complications, efficient antiviral treatments have been developed (16), but it has been shown that their efficacy is largely influenced by several biological parameters, such as the virus genotype. It has been demonstrated that genotypes 1 and 4 are more resistant than genotypes 2 and 3 to the current pegylated alpha interferon and ribavirin combination therapy (17). For this reason, in association with the determination of viral load and different host's related markers, HCV genotyping is used to predict the response to antiviral therapy (12, 26, 33) and to optimize the duration of treatment (3, 34). Furthermore, HCV genotyping is an essential tool for epidemiological studies (4, 24, 25, 32) and for tracing a source of contamination (1, 6, 15, 18, 23, 31). For clinical concerns, the determination of the genetic group is sufficient, whereas the subtype designation is crucial for epidemiological and transmission investigations.
Analysis of the HCV genome has demonstrated a high degree of genetic heterogeneity. HCV isolates have been classified into six main genotypes, and most of them have been divided into several subtypes (38). Since the sequencing of the entire HCV genome has not yet been performed, genotyping methods focused on several segments of the genome have been developed. Many of them, based on the 5′-untranslated coding region (5′UTR), are widely used since the 5′UTR is one of the most conserved and best-characterized regions of the HCV genome. In addition, the possibility of using PCR products generated by the diagnostic assays, which are often based on this region, is an attractive and practical option for genotyping assays. However, the 5′UTR has been shown to be inappropriate to discriminate HCV strains, especially at the subtype level (5, 8, 14). The main failures of 5′UTR-based genotyping that have been described concern (i) the misclassification of 1a genotype frequently identified as 1b (6, 8, 21); (ii) the lack of subtyping genotypes 2, 3, and 4 related to the diversity within these genetic groups, which could not be correctly distinguished from each other when only the 5′UTR is analyzed because they are not divergent enough in this region (38); and (iii) the occasional misclassification of genotype 6 as genotype 1 (9, 44) due to the identity of the genotype 6 5′UTR to that of genotype 1a or 1b (27, 37, 47, 48).
To compensate for these failures, alternative genomic regions have been proposed for the genotyping (7, 11, 21, 28, 35, 44). Recently, a new version (Versant HCV Genotype 2.0; Bayer Health Care, Eragny, France) of a currently commercially available genotyping assay (Versant HCV Genotype assay; Bayer Health Care) based on the reverse hybridization of a 5′UTR segment (42) has been developed by the addition of Core sequence information, in order to improve the accuracy of HCV subtype classification for genotypes 1 and 6. A recent study performed in Asia indicated that this novel line probe assay (LiPA) allows a more accurate discrimination of genotype 6 from genotype 1 (30). The aim of our study was to investigate the ability of this new assay to provide correct HCV genotypes in a subset of HCV subtypes mainly encountered in Western countries (especially genotypes 1 to 4) and to assess the improvement by comparing the results with those obtained with the previous version of the assay.
MATERIALS AND METHODS
Panel.
The panel was made up of 136 retrospective samples collected from HCV-infected blood donors and selected as a subset of HCV subtypes mainly encountered in clinical practice. All samples had been previously genotyped with INNO-LiPA HCV 1.0 and by a method based on NS5b region sequence analysis (some of them, especially genotype 4 samples, were also genotyped by analyzing a 363-base fragment in the region of the E1 envelope [positions 648 to 1041]) (5). HCV viral loads were determined when necessary with Amplicor HCV monitor version 2.0 (Roche, Meylan, France). The characteristics of these samples are given in Table 1.
TABLE 1.
Comparison of HCV genotyping results obtained with the NS5b sequencing method, the Versant HCV Genotype assay, and the Versant HCV Genotype 2.0 assay for 136 samples
| Genotype(s) detected by NS5b sequencing (no. of samples) | Versant HCV Genotype assay result (no. of samples)a
|
Genotype(s) detected by the Versant HCV Genotype 2.0 assay | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 or 1a/1b | 1a | 1b | NIe | 2 | 2b | 2a/2c | 3a | 4 | 4c/4d | 4f | 5a | 6 | 1b | ||
| 1a (41) | 23 | 10 | 8 | 0 | 1a | ||||||||||
| 1b (21) | 1 | 0 | 0 | 0 | 1a | ||||||||||
| 1 | 0 | 17 | 1 | 1b | |||||||||||
| 0 | 0 | 1 | 0 | NAd | |||||||||||
| 2a (5) | 0 | 0 | 5 | 2a or 2c | |||||||||||
| 2b (6) | 5 | 1 | 0 | 2b | |||||||||||
| 2c (2) | 0 | 0 | 1 | 2 | |||||||||||
| 0 | 0 | 1 | 2a or 2c | ||||||||||||
| Two othersb (17) | 4 | 0 | 4 | 2 | |||||||||||
| 0 | 0 | 9 | 2a or 2c | ||||||||||||
| 3a (15) | 14 | 3a | |||||||||||||
| 1 | NA | ||||||||||||||
| 4a (6) | 2 | 3 | 0 | 4 or 4a/4c/4d | |||||||||||
| 1 | 0 | 0 | 4h | ||||||||||||
| 4d (8) | 2 | 0 | 0 | 4 | |||||||||||
| 0 | 6 | 0 | 4a/4c/4d | ||||||||||||
| 4f (1) | 0 | 0 | 1 | 4f | |||||||||||
| Four othersc (7) | 2 | 1 | 0 | 4 | |||||||||||
| 0 | 3 | 0 | 4a/4c/4d | ||||||||||||
| 0 | 1 | 0 | 4e | ||||||||||||
| 5a (5) | 5 | 5a | |||||||||||||
| 6a (2) | 1 | 0 | 6 | ||||||||||||
| 1 | 0 | 6a or 6b | |||||||||||||
The correct results obtained with INNO-LiPA 1.0 are in italics, and those obtained with INNO-LiPA 2.0 are indicated in boldface.
Genotypes 2i (n = 5), 2k (n = 5), 2l (n = 4), and 2 undesignated (n = 3).
Genotypes 4h (n = 2) and 4 undesignated (n = 5).
NA, not amplified.
NI, not interpretable.
Sequencing methods.
Subtyping was performed by amplification and sequencing directly with amplification primers of a 339-bp segment in the NS5b region (positions 8002 to 8340 with reference to the open reading frame of strain D10750) (5). Using the CLUSTAL W 1.8 software package, the nucleotide-sequences of HCV strains were aligned with a reference panel of sequences representative of each subtype retrieved from the GenBank database (46). Pairwise distances were generated by using the p-distance algorithm of the program MEGA3 (20). Phylogenetic analysis was performed by using the neighbor-joining method for tree drawing. The reliability of phylogenetic classification was evaluated by a 1,000-cycle bootstrap test.
INNO LiPA HCV assays.
The Versant HCV Genotype assay (INNO-LiPA HCV 1.0) was performed according to the manufacturer's instructions except for the amplification step, which was obtained with the Amplicor HCV (Roche Molecular Systems, Meylan, France) assay. The procedure used here was described elsewhere (43).
A Versant HCV Genotype 2.0 assay (INNO-LiPA HCV 2.0) was performed according to the manufacturer's instructions. RNA was isolated from 140 μl of plasma by using QIAamp viral RNA (QIAgen, Hilden, Germany). Extracted RNA was resuspended in 60 μl of buffer. Reverse transcription-PCR was performed in a single tube: 20 μl of extracted RNA was added to 30 μl of PCR master mix containing two pairs of biotinylated synthetic oligonucleotides (5′UTR and Core region) in buffer with deoxynucleoside triphosphate/dUTP mix, MgCl2, RNAsin, and enzymes (Sensiscript and Omniscript reverse transcriptase, HotStarTaq polymerase, and uracil-N-glycosylase). The reverse transcription was performed at 50°C for 30 min, followed by an initial PCR activation step of 95°C for 15 min. Then, 40 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 15 s were carried out, with a final extension at 72°C for 2 min on an Applied Biosystems thermal cycler model GeneAmp PCR System 9700. Two distinct biotinylated DNA fragments of 240 and 270 bp representing the 5′UTR and Core HCV region, respectively, were produced. After denaturation, the biotinylated DNA-PCR product was hybridized to immobilized oligonucleotide probes. The probes which are bound to a nitrocellulose strip by poly(T) tail are specific for the 5′UTR region and the Core region of different HCV genotypes. Each strip contains three control lines (conjugate, amplification from 5′UTR, and Core region) and 22 DNA probe lines (19 for the 5′UTR and 3 for the Core region) with specific sequences for HCV genotypes 1 to 6. After the hybridization step, the unhybridized PCR product was washed from the strip, and alkaline phosphatase-labeled streptavidin (conjugate) was bound to the biotinylated hybrid. BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium chromogen (substrate) reacts with the streptavidin-alkaline phosphate complex, forming a purple-brown precipitate, resulting in a visible banding pattern on the strip. AutoLiPA 2.0 (Innogenetics, Zwijndrecht, Belgium) was used to carry out hybridization of the DNA PCR product and the developing color step.
Samples that gave noninterpretable results or that were not successfully amplified were tested once. Samples with discrepancies in the results between the INNO-LiPA HCV 2.0 and the NS5b sequencing method were tested once with the two methods and sometimes with a commercial 5′UTR-based sequencing method (Trugene HCV; Bayer Healthcare, Eragny, France).
Interpretation of the results.
The results of genotypes and subtypes obtained by NS5b sequencing were considered the reference genotypes. Each HCV genotype obtained with INNO-LiPA HCV 2.0, according to the interpretation chart provided by the manufacturer (two independent readings by two persons), was compared to the NS5b sequencing result and to the genotype obtained with INNO-LiPA HCV 1.0. The result was considered correct when both the correct genotype and the correct subtype were identified. An incomplete result was defined as an exact genotype result with an unidentified subtype or with an absence of discrimination between two subtypes. A correct genotype associated with a wrong subtype defined a misclassification. Accuracy was defined as the percentage of correct results (right genotype and right subtype).
For genotype 1 samples, the results obtained with INNO-LiPA HCV 2.0 were interpreted with or without Core information.
RESULTS
Table 1 gives the results obtained in the 136 samples studied by the NS5b sequencing method and the two INNO-LiPA HCV assays.
Among the 41 genotype 1a samples, 10 (23.8%) were correctly genotyped, 23 (57.7%) were incompletely genotyped, and 8 (19%) were misclassified (as 1b) with INNO-LiPA HCV 1.0, whereas 100% were classified as 1a with INNO-LiPA HCV 2.0. Of the 21 genotype 1b samples, 18 (85.7%) were correctly genotyped, 2 (9.5%) were incompletely genotyped, and 1 (4.8%) provided an uninterpretable result with INNO-LiPA HCV 1.0, whereas 19 samples (90.4%) were correctly identified, 1 (4.8%) was misclassified as 1a, and 1 (4.8%) was not amplified (viral load at 400 IU/ml) with INNO-LiPA HCV 2.0. The overall accuracies for the 62 genotype 1 samples were 45.2% and 96.8% for INNO-LiPA HCV 1.0 and INNO-LiPA HCV 2.0, respectively. By interpreting the results obtained in all genotype 1 samples with INNO-LiPA 2.0 according to the amplified region, one independently of the other, 40 (64.5%) would have been correctly genotyped (24 of 41 genotype 1a and 16 of 21 genotype 1b) by taking into account 5′UTR alone, whereas 60 (96.8%; 41 of 41 genotype 1a and 19 of 21 genotype 1b) were correctly genotyped by the additional information on the Core region.
Of the 30 genotype 2 samples, 1 (genotype 2b) was correctly classified with INNO-LiPA HCV 1.0, 16 were incompletely classified as 2a/2c (5 were 2a, and 2 were 2c) or as 2 with an assigned subtype (5 were 2b, and 4 were other genotype 2 samples), and 13 were misclassified as 2a/2c. INNO-LiPA HCV 2.0 provided 6 correct results (all were genotypes 2b), 15 incomplete results (5 were 2a, 2 were 2c, and 8 were other genotype 2 samples), and 9 misclassifications (2 genotypes other than 2a, 2b, or 2c). Core region amplification failed for 15 of 30 (50%) of the genotype 2 samples.
All of the 15 genotype 3a samples were correctly identified by INNO-LiPA HCV 1.0, whereas one sample (with a viral load of 7,000 IU/ml) could not be classified by INNO-LiPA HCV 2.0 due to the absence of amplification in the 5′UTR region. This sample was confirmed to be a 3a genotype when retested by NS5b sequencing genotyping and the 5′UTR-based sequencing Trugene method.
Only 1 (4.5%) of the 22 genotype 4 samples was correctly identified by the two INNO-LiPA assays. This sample was the only genotype 4f represented in the panel. Thirty (59.1%) were incompletely identified, and eight (36.4%) were misclassified with INNO-LiPA HCV 1.0, while sixteen (72.3%) were incomplete results and five (22.7%) were misclassifications with INNO-LiPA HCV 2.0.
The five genotype 5a samples were classified as 5a by both INNO-LiPA assays.
Of the two genotype 6 samples, one was determined to be genotype 1b, and the other was correctly identified as genotype 6 with INNO-LiPA HCV 1.0, whereas INNO-LiPA HCV 2.0 provided correct results for both samples.
Considering the whole panel, the overall rate of concordance (correct genotype and correct subtype) was 37.5% (51 samples) for INNO-LiPA HCV 1.0 versus 64.7% (88 samples) for INNO-LiPA HCV 2.0. The improved accuracy observed with INNO-LiPA 2.0 compared to the previous version of the assay concerned 39 samples: 31 (79.5%) were genotype 1a that were wrongly classified as 1b with INNO-LiPA HCV 1.0 (Fig. 1), 2 were genotype 1b, 5 were genotype 2b unassigned 2 with INNO-LiPA HCV 1.0, and 1 was genotype 6 misclassified as 1b with the previous version of the assay. On the other hand, one 1b sample and one 3a sample correctly classified by INNO-LiPA HCV 1.0 failed to be amplified with the INNO-LiPA HCV 2.0 PCR procedure. The percentages of incomplete results (undistinguishable or not identified subtype) were 39.7% (54 samples) for INNO-LiPA HCV 1.0 and 22.8% (31 samples) for INNO-LiPA HCV 2.0. Misclassifications were observed for 22.1% of the tested samples (30 samples) for INNO-LiPA HCV 1.0 and 11% (15 samples) for INNO-LiPA HCV 2.0. Finally, the amplification step failed for one and two samples for INNO-LiPA HCV 1.0 and INNO-LiPA HCV 2.0, respectively.
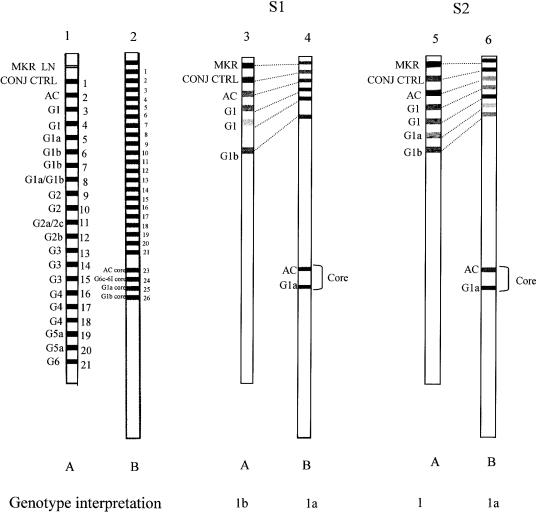
FIG. 1.
Comparison of INNO-LiPA patterns obtained with the Versant HCV Genotype assay (A) and the Versant HCV Genotype 2.0 assay (B). Type-specific line numbers with corresponding interpreted genotypes of 5′UTR (the same for both assays) and Core region (specific for the Versant HCV Genotype 2.0 assay) are indicated at the left side (strips 1 and 2). Patterns of the Versant HCV Genotype assay (strip 3 and 5) and the Versant HCV Genotype 2.0 assay (strips 4 and 6) obtained from two genotype 1a samples (S1 and S2) are presented. The genotype interpretations are indicated at the bottom of each strip. Dashed lines represent the correspondence between 5′UTR type-specific lines of both assays. MKR, marker line; CONJ CTRL, conjugate control; AC, amplification control.
DISCUSSION
For the purpose of treatment management, current 5′UTR-based genotyping assays are acceptably accurate since they have been shown to present more than 95% concordance with genotypes identified by nucleotide sequencing (13, 22, 40, 45, 50). However, several studies demonstrated that 5′UTR region analysis is not appropriate for definitive genotype identification and for the identification of subtypes (7, 21, 44). Moreover, although not frequent but probably increasing due to the hybrid generation in multiply exposed individuals, the recombination forms (10, 19, 29) limit the accuracy of genotyping assays when only a segment of the genome is analyzed.
In order to improve the genotype determination of genotype 1 and 6 samples, the widely used reverse hybridization assay for HCV genotyping based on 5′UTR was recently modified by the addition of Core probes. Our study showed an improvement in the accuracy of the results obtained with the new strip since 64.7% of tested samples were concordant for genotype group and subtype with sequencing reference results versus 37.5% with the previous version of the assay. Obviously, the improved accuracy is mainly related to the better characterization of subtypes 1, since the accuracy increased from 45.2% with the first version to 96.8% with the new version of the assay. The misclassification of genotype 1a as 1b or the contrary has been extensively described on the basis of the sequence polymorphism at position −99 of the genome, frequently used to differentiate genotypes 1 (2, 7, 8, 14, 41, 44, 49). Thus, we confirmed the benefit of the inclusion of the Core region in the assay, since only 58.5% of genotype 1a would have been correctly characterized by using 5′UTR information alone, whereas 100% of them have been genotyped 1a with Core information. One of the two genotype 1 samples, missed by INNO-LiPA HCV 2.0, was a sample with a low viral load probably situated under the limit of the detection of the PCR used in the INNO-LiPA 2.0 procedure. Interestingly, the second sample classified as 1a was confirmed as 1b by the NS5b sequencing method and as 1a by 5′UTR-based sequencing assay due to the presence of a nucleotide A at position −99. This result is consistent with other reports (8, 14, 49), which reported the same phenomenon. These findings could be explained by a mixed infection of subtypes 1a and 1b, by an A/G polymorphism that may exist at nucleotide −99 in some HCV isolates, or by an infection with a recombinant form 1a/1b. Except for this particular sample, which necessitates further molecular investigations, the results obtained in other genotype 1 samples included in the present study emphasize the usefulness of the inclusion of Core region for differentiating subtypes 1a and 1b. This improvement has a particular impact in Western countries, where the genotype 1 is widely distributed (38).
The other benefit of INNO-LiPA HCV 2.0 is its ability to correctly classify genotype 6. The two genotype 6 samples included here were correctly identified, whereas one of them was genotyped as 1b with INNO-LiPA HCV 1.0. The number of studied genotype 6 specimens was insufficient to evaluate the capacity of the new version of the assay to provide an accurate classification of this genotype. Nevertheless, a recent published study including several genotype 6 strains from Southeast Asia showed the improved performance of INNO-LiPA HCV 2.0 in the correct characterization of this genotype (30). Indeed, of the 33 genotype 6 samples included in the present study, only 12 (36.3%) were genotyped as 6 (all were 6a) with INNO-LiPA HCV 1 versus 32 (97%) (1 sample was nontypeable) with INNO-LiPA HCV 2.0.
The 5′UTR is not heterogeneous enough for use in the determination of HCV subtypes in genotypes 2 and 4 due to the high degree of diversity in these groups (39). This is illustrated by the lack of precision in determining subtypes with both INNO-LiPA HCV assays. However, INNO-LiPA 2.0 correctly identified the six genotypes 2b included in the panel. Surprisingly, INNO-LiPA HCV 1.0 missed five of them (genotype 2 with an unassigned subtype), although the interpretation pattern is the same for the two assays and despite the fact that the Core region is not taken into account for genotype 2 classification. However, when we performed once more the INNO-LiPA HCV 1.0 with the PCR procedure recommended by the manufacturer instead of Amplicor HCV PCR (data not shown), all of them were correctly subtyped as 2b, demonstrating that the dedicated PCR leads to more accurate results. The use of the specific protocol for amplification according to the manufacturer's instructions could increase the overall concordance between the two assays for subtype identification. However, for practical reasons and because the use of amplified products from the Roche Amplicor HCV assay is also recommended by the manufacturer, this procedure is extensively adopted and corresponds to the main routine practice that was evaluated here.
Genotypes 3 and 5 did not raise the issue of identification. However, a larger number of these genotypes should be tested to give more information. Interestingly, one sample failed to be amplified twice with the specific 5′UTR primers of the INNO-LiPA HCV 2.0 assay, whereas the amplification with Core primers succeeded. Moreover, 5′UTR and NS5b sequencing methods successfully identified the sample as genotype 3a (data not shown). The viral load (7,000 IU/ml) could not be at the origin of the lack of PCR. Thus, a mismatch of primers with this particular sample is one of the hypotheses to explain this false-negative result.
In conclusion, hybridization typing based on genotyping methods represent an attractive genotyping option compared to sequencing methods. Incontestably, INNO-LiPA HCV 2.0 demonstrates better performance than INNO-LiPA 1.0, especially for the subtyping of genotype 1 samples and the characterization of genotype 6 (30), due to the addition of Core motifs, which provide a useful complement of information. However, this new version necessitates a specific PCR and could no longer be used after 5′UTR PCR used for current HCV infection diagnosis. Moreover, the 5′UTR information provided by this assay is not reliable for correctly identifying the diversity within genetic groups as seen in genotypes 2 and 4.
Acknowledgments
The NS5b sequencing method was used for genotype determination in a study supported by grant 2003-09 from the Conseil Scientifique de l'Etablissement Français du Sang.
Footnotes
Published ahead of print on 24 January 2007.
REFERENCES
- 1.Ackerman, Z., E. Ackerman, and O. Paltiel. 2000. Intrafamilial transmission of hepatitis C virus: a systematic review. J. Viral Hepat. 7:93-103. [DOI] [PubMed] [Google Scholar]
- 2.Andonov, A., and R. K. Chaudhary. 1995. Subtyping of hepatitis C virus isolates by a line probe assay using hybridization. J. Clin. Microbiol. 33:254-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anonymous. 2002. Consensus conference treatment of hepatitis C virus. Gastroenterol. Clin. Biol. 26:B312-B320. [PubMed] [Google Scholar]
- 4.Bourliere, M., J. M. Barberin, M. Rotily, V. Guagliardo, I. Portal, L. Lecomte, S. Benali, C. Boustiere, H. Perrier, M. Jullien, G. Lambot, R. Loyer, O. LeBars, R. Daniel, H. Khiri, and P. Halfon. 2002. Epidemiological changes in hepatitis C virus genotypes in France: evidence in intravenous drug users. J. Viral Hepat. 9:62-70. [DOI] [PubMed] [Google Scholar]
- 5.Cantaloube, J., H. Venault, J. Zappitelli, P. Gallian, M. Touinssi, H. Attoui, P. Biagini, X. de Lamballerie, and P. de Micco. 2000. Molecular analysis of HCV type 1 to 5 envelope gene: application to investigations of posttransfusion transmission of HCV. Transfusion 40:712-717. [DOI] [PubMed] [Google Scholar]
- 6.Cantaloube, J. F., P. Gallian, H. Attoui, P. Biagini, P. De Micco, and X. De Lamballerie. 2001. Erroneous HCV genotype assignment by a hybridization typing assay in a case of posttransfusion HCV infection. Transfusion 41:429-430. [DOI] [PubMed] [Google Scholar]
- 7.Cantaloube, J. F., S. Laperche, P. Gallian, F. Bouchardeau, X. de Lamballerie, and P. de Micco. 2006. Analysis of the 5′ noncoding region versus the NS5b region in genotyping hepatitis C virus isolates from blood donors in France. J. Clin. Microbiol. 44:2051-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Z., and K. E. Weck. 2002. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 40:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chinchai, T., J. Labout, S. Noppornpanth, A. Theamboonlers, B. L. Haagmans, A. D. Osterhaus, and Y. Poovorawan. 2003. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J. Virol. Methods 109:195-201. [DOI] [PubMed] [Google Scholar]
- 10.Colina, R., D. Casane, S. Vasquez, L. Garcia-Aguirre, A. Chunga, H. Romero, B. Khan, and J. Cristina. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31-37. [DOI] [PubMed] [Google Scholar]
- 11.Corbet, S., J. Bukh, A. Heinsen, and A. Fomsgaard. 2003. Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J. Clin. Microbiol. 41:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, G. L., and J. Y. Lau. 1997. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology 26:122S-127S. [DOI] [PubMed] [Google Scholar]
- 13.Gault, E., P. Soussan, Y. Morice, L. Sanders, A. Berrada, B. Rogers, and P. Deny. 2003. Evaluation of a new serotyping assay for detection of anti-hepatitis C virus type-specific antibodies in serum samples. J. Clin. Microbiol. 41:2084-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germer, J. J., P. N. Rys, J. N. Thorvilson, and D. H. Persing. 1999. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor HCV test. J. Clin. Microbiol. 37:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halfon, P., P. Trimoulet, M. Bourliere, H. Khiri, V. de Ledinghen, P. Couzigou, J. M. Feryn, P. Alcaraz, C. Renou, H. J. Fleury, and D. Ouzan. 2001. Hepatitis C virus genotyping based on 5′ noncoding sequence analysis (Trugene). J. Clin. Microbiol. 39:1771-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayashi, N., and T. Takehara. 2006. Antiviral therapy for chronic hepatitis C: past, present, and future. J. Gastroenterol. 41:17-27. [DOI] [PubMed] [Google Scholar]
- 17.Hnatyszyn, H. J. 2005. Chronic hepatitis C and genotyping: the clinical significance of determining HCV genotypes. Antivir. Ther. 10:1-11. [PubMed] [Google Scholar]
- 18.Izopet, J., C. Pasquier, K. Sandres, J. Puel, and L. Rostaing. 1999. Molecular evidence for nosocomial transmission of hepatitis C virus in a French hemodialysis unit. J. Med. Virol. 58:139-144. [DOI] [PubMed] [Google Scholar]
- 19.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 21.Laperche, S., F. Lunel, J. Izopet, S. Alain, P. Deny, G. Duverlie, C. Gaudy, J. M. Pawlotsky, J. C. Plantier, B. Pozzetto, V. Thibault, F. Tosetti, and J. J. Lefrere. 2005. Comparison of hepatitis C virus NS5b and 5′ noncoding gene sequencing methods in a multicenter study. J. Clin. Microbiol. 43:733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau, J. Y., M. Mizokami, J. A. Kolberg, G. L. Davis, L. E. Prescott, T. Ohno, R. P. Perrillo, K. L. Lindsay, R. G. Gish, K. P. Qian, et al. 1995. Application of six hepatitis C virus genotyping systems to sera from chronic hepatitis C patients in the United States. J. Infect. Dis. 171:281-289. [DOI] [PubMed] [Google Scholar]
- 23.Le Pogam, S., D. Le Chapois, R. Christen, F. Dubois, F. Barin, and A. Goudeau. 1998. Hepatitis C in a hemodialysis unit: molecular evidence for nosocomial transmission. J. Clin. Microbiol. 36:3040-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martial, J., Y. Morice, S. Abel, A. Cabie, C. Rat, F. Lombard, A. Edouard, S. Pierre-Louis, P. Garsaud, O. Bera, R. Chout, E. Gordien, P. Deny, and R. Cesaire. 2004. Hepatitis C virus (HCV) genotypes in the Caribbean island of Martinique: evidence for a large radiation of HCV-2 and for a recent introduction from Europe of HCV-4. J. Clin. Microbiol. 42:784-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinot-Peignoux, M., F. Roudot-Thoraval, I. Mendel, J. Coste, J. Izopet, G. Duverlie, C. Payan, J. M. Pawlotsky, C. Defer, M. Bogard, V. Gerolami, P. Halfon, Y. Buisson, B. Fouqueray, P. Loiseau, J. Lamoril, J. J. Lefrere, and P. Marcellin. 1999. Hepatitis C virus genotypes in France: relationship with epidemiology, pathogenicity, and response to interferon therapy. J. Viral. Hepat. 6:435-443. [DOI] [PubMed] [Google Scholar]
- 26.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 27.Mellor, J., E. A. Walsh, L. E. Prescott, L. M. Jarvis, F. Davidson, P. L. Yap, and P. Simmonds. 1996. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core-based genotyping assay. J. Clin. Microbiol. 34:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nolte, F. S., A. M. Green, K. R. Fiebelkorn, A. M. Caliendo, C. Sturchio, A. Grunwald, and M. Healy. 2003. Clinical evaluation of two methods for genotyping hepatitis C virus based on analysis of the 5′ noncoding region. J. Clin. Microbiol. 41:1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noppornpanth, S., T. X. Lien, Y. Poovorawan, S. L. Smits, A. D. Osterhaus, and B. L. Haagmans. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 80:7569-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noppornpanth, S., E. Sablon, K. De Nys, T. X. Lien, J. Brouwer, M. Van Brussel, S. L. Smits, Y. Poovorawan, A. D. Osterhaus, and B. L. Haagmans. 2006. Genotyping hepatitis C viruses from South East Asia using a novel line probe assay that simultaneously detects 5′UTR and core regions. J. Clin. Microbiol. 44:3969-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norder, H., A. Bergstrom, I. Uhnoo, J. Alden, L. Weiss, J. Czajkowski, and L. Magnius. 1998. Confirmation of nosocomial transmission of hepatitis C virus by phylogenetic analysis of the NS5-B region. J. Clin. Microbiol. 36:3066-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawlotsky, J. M., L. Tsakiris, F. Roudot-Thoraval, C. Pellet, L. Stuyver, J. Duval, and D. Dhumeaux. 1995. Relationship between hepatitis C virus genotypes and sources of infection in patients with chronic hepatitis C. J. Infect. Dis. 171:1607-1610. [DOI] [PubMed] [Google Scholar]
- 33.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, J. Albrecht, et al. 1998. Randomised trial of interferon alpha2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 34.Poynard, T., J. McHutchison, Z. Goodman, M. H. Ling, J. Albrecht, et al. 2000. Is an “a la carte” combination interferon alfa-2b plus ribavirin regimen possible for the first line treatment in patients with chronic hepatitis C? Hepatology 31:211-218. [DOI] [PubMed] [Google Scholar]
- 35.Sandres-Saune, K., P. Deny, C. Pasquier, V. Thibaut, G. Duverlie, and J. Izopet. 2003. Determining hepatitis C genotype by analyzing the sequence of the NS5b region. J. Virol. Methods 109:187-193. [DOI] [PubMed] [Google Scholar]
- 36.Seeff, L. B. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35-S46. [DOI] [PubMed] [Google Scholar]
- 37.Simmonds, P. 2001. The origin and evolution of hepatitis viruses in humans. J. Gen. Virol. 82:693-712. [DOI] [PubMed] [Google Scholar]
- 38.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. T. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 39.Simmonds, P., J. Mellor, T. Sakuldamrongpanich, C. Nuchaprayoon, S. Tanprasert, E. C. Holmes, and D. B. Smith. 1996. Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J. Gen. Virol. 77(Pt. 12):3013-3024. [DOI] [PubMed] [Google Scholar]
- 40.Simmonds, P., K. A. Rose, S. Graham, S. W. Chan, F. McOmish, B. C. Dow, E. A. Follett, P. L. Yap, and H. Marsden. 1993. Mapping of serotype-specific, immunodominant epitopes in the NS-4 region of hepatitis C virus (HCV): use of type-specific peptides to serologically differentiate infections with HCV types 1, 2, and 3. J. Clin. Microbiol. 31:1493-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, D. B., J. Mellor, L. M. Jarvis, F. Davidson, J. Kolberg, M. Urdea, P. L. Yap, P. Simmonds, et al. 1995. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J. Gen. Virol. 76(Pt. 7):1749-1761. [DOI] [PubMed] [Google Scholar]
- 42.Stuyver, L., A. Wyseur, W. van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation line probe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stuyver, L., A. Wyseur, W. van Arnhem, F. Lunel, P. Laurent-Puig, J. M. Pawlotsky, B. Kleter, L. Bassit, J. Nkengasong, L. J. van Doorn, et al. 1995. Hepatitis C virus genotyping by means of 5′-UR/core line probe assays and molecular analysis of untypeable samples. Virus Res. 38:137-157. [DOI] [PubMed] [Google Scholar]
- 44.Tamalet, C., P. Colson, H. Tissot-Dupont, M. Henry, C. Tourres, N. Tivoli, D. Botta, I. Ravaux, I. Poizot-Martin, and N. Yahi. 2003. Genomic and phylogenetic analysis of hepatitis C virus isolates: a survey of 535 strains circulating in southern France. J. Med. Virol. 71:391-398. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka, T., K. Tsukiyama-Kohara, K. Yamaguchi, S. Yagi, S. Tanaka, A. Hasegawa, Y. Ohta, N. Hattori, and M. Kohara. 1994. Significance of specific antibody assay for genotyping of hepatitis C virus. Hepatology 19:1347-1353. [PubMed] [Google Scholar]
- 46.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tokita, H., H. Okamoto, P. Luengrojanakul, K. Vareesangthip, T. Chainuvati, H. Iizuka, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d), and ninth (9b, 9c) major genetic groups. J. Gen. Virol. 76(Pt. 9):2329-2335. [DOI] [PubMed] [Google Scholar]
- 48.Tokita, H., H. Okamoto, F. Tsuda, P. Song, S. Nakata, T. Chosa, H. Iizuka, S. Mishiro, Y. Miyakawa, and M. Mayumi. 1994. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc. Natl. Acad. Sci. USA 91:11022-11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeuzem, S., B. Ruster, J. H. Lee, T. Stripf, and W. K. Roth. 1995. Evaluation of a reverse hybridization assay for genotyping of hepatitis C virus. J. Hepatol. 23:654-661. [DOI] [PubMed] [Google Scholar]
- 50.Zheng, X., M. Pang, A. Chan, A. Roberto, D. Warner, and B. Yen-Lieberman. 2003. Direct comparison of hepatitis C virus genotypes tested by INNO-LiPA HCV II and TRUGENE HCV genotyping methods. J. Clin. Virol. 28:214-216. [DOI] [PubMed] [Google Scholar]