Abstract
Cryptococcus neoformans is a fungal pathogen that causes life-threatening infections primarily in immunocompromised hosts. Based on the genetic characteristics and serologic properties of capsular polysaccharides, three varieties and five serotypes have been defined: C. neoformans var. neoformans (serotype D), C. neoformans var. grubii (serotype A), hybrid serotype AD, and C. neoformans var. gattii (serotypes B and C). Epidemiologic features, such as geographic distribution and ecologic niche, and clinical characteristics have been shown to be associated with serotypes. At the present time, serotyping is based on agglutination tests with either commercial or “homemade” antisera or on immunofluorescence assays using a monoclonal antibody directed against the capsule polysaccharide. In this paper, we describe two molecular methods (PCR-restriction enzyme analysis and length polymorphism analysis) for C. neoformans serotype identification. Both are based on the sequence characteristics of a fragment of the CAP59 gene required for capsule biosynthesis. Testing of 72 C. neoformans strains including representatives of the five serotypes demonstrated the reliability of these methods.
Cryptococcus neoformans is a basidiomycetous encapsulated yeast that causes life-threatening infections, mainly in immunocompromised hosts and particularly in human immunodeficiency virus-infected patients (1, 13). Based on the genetic characteristics and serologic properties of capsular polysaccharides (CPS) (9), three varieties and four nonhybrid serotypes have been defined, namely, C. neoformans var. neoformans (serotype D), C. neoformans var. grubii (serotype A), and C. neoformans var. gattii (serotypes B and C) (4). Serotypes correspond to different sexual teleomorphs, namely, Filobasidiella neoformans and Filobasidiella bacillispora for serotypes A and D and serotypes B and C, respectively. Moreover, hybrid strains that most likely correspond to either diploid or aneuploid organisms have been characterized as serotype AD strains (11, 14, 17, 20). Some epidemiologic properties, such as geographic distribution, have been associated with serotypes, as serotypes A, D, and AD are found worldwide while serotypes B and C are restricted mainly to tropical and subtropical regions (1). While serotype A and D strains are usually isolated from pigeon droppings and cause disease mainly in immunocompromised hosts, serotype B and C strains are more commonly isolated from eucalyptus trees and more often infect hosts with normal immune status, causing cryptococcoma (1). The need for prolonged treatment and surgery and sequelae including impaired visual acuity and a higher mortality rate have also been found to be more frequently associated with serotype B strains than with serotype A strains (2, 18).
Serotyping is presently performed by using agglutination with commercial (Crypto Check kit; Iatron Labs, Tokyo, Japan) or “homemade” antisera components (9) or a combination of canavanine-glycine-bromothymol (CGB) blue agar diagnostic medium and a direct immunofluorescence assay using E1 monoclonal antibody directed against CPS (3). These methods are widely used but may be either inapplicable or unreliable in the case of noncapsulated or nontypeable isolates, respectively (8, 16). Moreover, cases of misidentification have already been reported (3, 19). In the present paper, we report two molecular methods, based on the sequence characteristics of a fragment of the CAP59 gene required for capsule biosynthesis (5). Both methods allow for the rapid and reliable identification of all C. neoformans serotypes.
MATERIALS AND METHODS
Strains.
Seventy-two C. neoformans strains, including 33 reference strains, representing the five serotypes and one Cryptococcus uniguttulatus strain were tested (Table 1 ). The identification of all strains was confirmed by using the ID32C auxanogram panel (BioMérieux, Marcy l'Etoile, France). Strains were serotyped by agglutination with homemade antisera based on the procedure described by Ikeda et al. (9) or with a commercial kit (Crypto Check kit; Iatron Labs, Tokyo, Japan). Some strains have been serotyped by the Centre National de Référence Mycologie et Antifongiques at the Pasteur Institute of Paris by combining CGB diagnostic medium with a direct immunofluorescence assay using E1 monoclonal antibody specific for CPS, as described by Dromer et al. (3). Isolates were stored at −80°C and were grown on yeast-peptone-glucose agar plates for 2 days at 30°C before testing.
TABLE 1.
C. neoformans strains tested in this study and results of immunoserotyping and molecular identification of the serotype
| Strain no. | Strain name | Isolationa
|
Immunologic serotyping
|
Molecular identification
|
|||
|---|---|---|---|---|---|---|---|
| Origin | Site | Methodb | Result | REA patternc | Fragment size(s) (bp)d | ||
| 1 | H99 | United States | CSF | Commercial kit | A | A | 394 |
| 2 | IHEM 11752 | Congo | CSF | Commercial kit | A | A | 400 |
| 3 | IHEM 11753 | Congo | CSF | Commercial kit | A | A | 394 |
| 4 | IHEM 11756 | Congo | CSF | Commercial kit | A | A | 394 |
| 5 | IHEM 11757 | Congo | CSF | Commercial kit | A | A | 394 |
| 6 | IHEM 11876 | Congo | CSF | Commercial kit | A | A | 394 |
| 7 | IHEM 11877 | Congo | CSF | Commercial kit | A | A | 394 |
| 8 | IHEM 11880 | Congo | CSF | Commercial kit | A | A | 394 |
| 9 | IHEM 11882 | Congo | CSF | Commercial kit | A | A | 394 |
| 10 | Tou | Mali | CSF | Homemade Ab | A | A | 394 |
| 11 | Fat | Congo | CSF | Homemade Ab | A | A | 400 |
| 12 | coc 119 | Cochin, India | CSF | Homemade Ab | A | A | 394 |
| 13 | 333 | Gabon | CSF | Homemade Ab | A | A | 394 |
| 14 | 2598 P | Ivory Coast | CSF | Homemade Ab | A | A | 394 |
| 15 | g 57497 | Gabon | CSF | Homemade Ab | A | A | 394 |
| 16 | Oug 1 | Uganda | CSF | Homemade Ab | A | A | 394 |
| 17 | PP38 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 18 | RV66254 | Rwanda | CSF | Homemade Ab | A | A | 394 |
| 19 | 3/768 | Congo | CSF | Homemade Ab | A | A | 394 |
| 20 | Lim | France | CSF | Homemade Ab | D | A | 394 |
| 21 | C3 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 22 | C6 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 23 | C7 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 24 | Oug 7 | Uganda | CSF | Homemade Ab | A | A | 394 |
| 25 | PP29 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 26 | RV68535 | Rwanda | CSF | Homemade Ab | A | A | 394 |
| 27 | RV68528 | Rwanda | CSF | Homemade Ab | A | A | 394 |
| 28 | Tnn Ci | France | CSF | Homemade Ab | A | A | 394 |
| 29 | g 59546 | Gabon | CSF | Homemade Ab | A | A | 394 |
| 30 | RV66254 | Rwanda | CSF | Homemade Ab | A | A | 394 |
| 31 | C57 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 32 | C68 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 33 | Oug 3 | Uganda | CSF | Homemade Ab | AD | A | 394 |
| 34 | Oug 9 | Uganda | CSF | Homemade Ab | A | A | 394 |
| 35 | PP43 | Cambodia | CSF | Homemade Ab | A | A | 394 |
| 36 | 241 | Gabon | CSF | Homemade Ab | A | A | 394 |
| 37 | Tnn Mb | France | Blood | mAb IF | A | A | 394 |
| 38 | Tnn Da | France | Blood | mAb IF | A | A | 394 |
| 39 | Tnn Mo | France | CSF | mAb IF | A | A | 394 |
| 40 | Bel | Belgium | CSF | Commercial kit | A | A | 394 |
| 41 | IHEM 4157 | Belgium | NS | Commercial kit | AD | AD | 382, 394 |
| 42 | IHEM 13877 | Belgium | CSF | Commercial kit | AD | AD | 382, 394 |
| 43 | IHEM 14440 | Belgium | CSF | Commercial kit | AD | AD | 382, 394 |
| 44 | IHEM 16079 | Belgium | Voice prosthesis | Commercial kit | AD | AD | 382, 394 |
| 45 | IHEM 16513 | Peru | CSF | Commercial kit | AD | AD | 382, 394 |
| 46 | IHEM 17861 | Peru | CSF | Commercial kit | AD | AD | 382, 394 |
| 47 | RV 66423 | Rwanda | CSF | Homemade Ab | A | D | 382 |
| 48 | JEC21 | NS | NS | Commercial kit | D | D | 382 |
| 49 | B 3501 A | NS | NS | Commercial kit | D | D | 382 |
| 50 | NIH D52 | United States | CSF | Commercial kit | D | D | 382 |
| 51 | IHEM 15038 | Peru | NS | Commercial kit | D | D | 382 |
| 52 | IHEM 18620 | Belgium | Skin | Commercial kit | D | D | 382 |
| 53 | IHEM 16711 | Belgium | Skin | Commercial kit | D | D | 382 |
| 54 | IHEM 4158 | Italy | NS | Commercial kit | D | D | 382 |
| 55 | AKD 30 | Ivory Coast | CSF | Homemade Ab | A | D | 380 |
| 56 | T 6073 | France | CSF | Homemade Ab | D | D | 382 |
| 57 | 2815 | Cambodia | CSF | Homemade Ab | A | D | 382 |
| 58 | Tnn Re | France | BAL fluid | mAb IF | D | D | 382 |
| 59 | JLP34 | France | CSF | mAb IF | D | D | 382 |
| 60 | JLP23 | France | Urine | mAb IF | D | D | 382 |
| 61 | IHEM 11758 | Congo | CSF | Commercial kit | D | D | 380 |
| 62 | C9 | Cambodia | CSF | Homemade Ab | A | D | 382 |
| 63 | IHEM 14934 | Brazil | NS | Commercial kit | B | B | 373 |
| 64 | IHEM 4161e | United States | Sputum | Commercial kit | B | B | 373 |
| 65 | IHEM 11796e | Zaire | CSF | Commercial kit | B | B | 373 |
| 66 | IHEM 4170 | Malaysia | NS | Commercial kit | B | B | 373 |
| 67 | IHEM 4164 | China | CSF | Commercial kit | B | B | 371 |
| 68 | WM 276 | Australia | Environment | Commercial kit | B | B | 371 |
| 69 | VanR265 | Canada | BWF | Commercial kit | B | B | 380 |
| 70 | RV68522 | Rwanda | CSF | Homemade Ab | D | B | 371 |
| 71 | IHEM 10079 | NS | CSF | Commercial kit | C | C | 366 |
| 72 | IHEM 4159e | United States | CSF | Commercial kit | C | C | 366 |
NS, not specified; CSF, cerebrospinal fluid; BAL, bronchoalveolar lavage; BWF, bronchial wash fluid; Congo, Democratic Republic of the Congo.
The commercial kit used was the Crypto Check kit (Iatron Labs, Tokyo, Japan). Homemade Ab, agglutination with homemade antisera (9); mAb IF, immunofluorescence with monoclonal antibody specific for CPS (3).
REA pattern deduced from in silico analysis (Table 2).
PCR fragments are supposed to be 8 bases longer than the sequences due to a 7-base tail added to the reverse primer used to promote the addition of an extra A to the amplified fragment.
Type strain.
CAP59 gene amplification.
Sequences of CAP59 genes from the four nonhybrid serotypes of C. neoformans were retrieved from the GenBank database and the Cryptococcus neoformans Serotype B Sequencing Project of the Broad Institute of Harvard and the Massachusetts Institute of Technology (http://www.broad.mit.edu). Sequences were then aligned using ClustalX (version 1.83), and two primers (CH-Cap 59F, 5′-CCTTGCCGAAGTTCGAAACG, and CH-Cap 59R, 5′-AATCGGTGGTTGGATTCAGTGT) were designed from the conserved regions of the CAP59 genes. DNA was extracted using a rapid method based on thermal shock and the chelation of components other than nucleic acids by using a resin suspension, as previously described (7). The PCR was carried out with a 50-μl reaction volume containing 1× PCR buffer, 1.6 μM (each) primers (Eurogentec, Liege, Belgium), 0.2 mM (each) deoxynucleoside triphosphates (equimolar concentrations of dATP, dCTP, dGTP, and dTTP), and 0.5 U of Taq polymerase (New England BioLabs Inc.). A touchdown amplification program was performed as follows: 7 min at 94°C; three cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C; three cycles of 30 s at 94°C, 30 s at 58°C, and 30 s at 72°C; three cycles of 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C; 28 cycles of 30 s at 52°C and 30 s at 72°C; and a final 15-min extension at 72°C.
Restriction enzyme analysis (REA).
Restriction maps of unique sites obtained using DNA Strider version 1.4x-4e (15) were used to select restriction enzymes allowing for the identification of the various serotypes (Table 2). In a first-line identification, PCR products were subjected to separate restriction procedures with BsmFI and HpaII according to the instructions of the manufacturer (New England BioLabs Inc.). Digested fragments were separated by electrophoresis in agarose gel (3% in Tris-borate-EDTA buffer) stained with ethidium bromide (0.5 μg/ml) at 90 V for 3 h. PCR products with patterns compatible with the B or C serotype were further digested with the AgeI enzyme (New England BioLabs Inc.).
TABLE 2.
Expected sizes (bp) of CAP59 gene fragments from different serotypes before and after restriction with BsmFI, HpaII, and AgeI enzymes
| Assay or fragment | Expected size(s) (bp) for serotypea:
|
|||
|---|---|---|---|---|
| A | B | C | D | |
| Native amplicon | 387 | 373 | 359 | 375 |
| REA with BsmFI | 206, 191 | NRS | NRS | NRS |
| REA with HpaII | NRS | 270, 101 | 256, 101 | 211, 162 |
| REA with AgeI | NRS | 269, 100 | NRS | NRS |
NRS, no restriction site; serotype A, strain H99 (GenBank accession number AF337639); serotype B, strain R265 (http://www.broad.mit.edu/annotation/genome/cryptococcus_neoformans_b/Assembly.html); serotype C, strain IHEM 4159 (sequence obtained in the present study); serotype D, JEC21 strain (GenBank accession number AE017341).
Fragment length polymorphism analysis.
For fragment length analysis, a similar amplification protocol was performed but with a fluorolabeled (6-carboxyfluorescein dye; Applied Biosystems, Courtaboeuf, France) forward primer. A 7-base tail used to promote the addition of an extra A to the amplified fragment was added to the reverse primer in order to facilitate sizing. Each PCR product was run with an internal molecular marker (GenScan HD400Rox, ABI) for size determination on a DNA sequencer (ABI 3100; Applied Biosystems).
In the case of conflicting results, a second serotyping determination was performed with the Crypto Check commercial kit (Iatron Labs, Tokyo, Japan).
Based on fragment length results, we selected certain PCR products to be sequenced (Table 3). Fragments of a serotype AD strain (IHEM 13877) not digested by either BsmFI or HpaII and corresponding to the D or A allele, respectively, were gel purified (Gel Band purification kit; Amersham Biosciences, Freiburg, Germany) before sequencing. Direct sequencing was performed using a Big Dye terminator protocol as recommended by the manufacturer (Applied Biosystems). Double-strand sequencing was performed using the same primers as those used for PCR amplification. After manual correction, sequences were compared using the BLASTN software on the GenBank database limited to the organism C. neoformans.
TABLE 3.
Comparison of the partial sequence of the CAP59 gene with sequences in the GenBank database by using BLASTN software
| Strain | Serotype | Fragment length (bases) | Results (best hit) | Accession no. |
|---|---|---|---|---|
| IHEM 11752 | A | 392 | Best score obtained when aligned with 2 fragments from serotype A strain M9253 (accession no. AB066120) | EF392812 |
| Partial 5′ end: 99% identity over a 203-nucleotide alignment | ||||
| Partial 3′ end: 97% identity over a 131-nucleotide alignment | ||||
| Better score for partial 3′ end obtained with sequence from serotype A strain H99: 98% identity over a 173-nucleotide alignment | ||||
| IHEM 13877 | AD | 387 (A allelea) | 99% identity over a 382-nucleotide alignment with sequence from serotype A strain H99 (accession no. AF337639) | EF392816 |
| 375 (D alleleb) | 98% identity over a 375-nucleotide alignment with sequence from serotype D strain JEC21 (accession no. AF337639) | EF392813 | ||
| IHEM 11796 | B | 373 | 99% identity over a 324-nucleotide alignment with sequence from serotype B strain M9244 (accession no. AB066134) | EF392816 |
| IHEM 4159 | C | 359 | 99% identity over a 312-nucleotide alignment with sequence from serotype C strain M9247 (accession no. AB066126)c | EF392815 |
No restricted fragment after digestion with HpaII.
No restricted fragment after digestion with BsmFI.
Complete sequence of the fragment not available in the GenBank database.
Nucleotide sequence accession number.
The sequence of IHEM 4159 obtained in the present study has been deposited in GenBank with accession number EF392815.
RESULTS
All tested C. neoformans strains but not the C. uniguttulatus strain gave positive amplification.
PCR-REA.
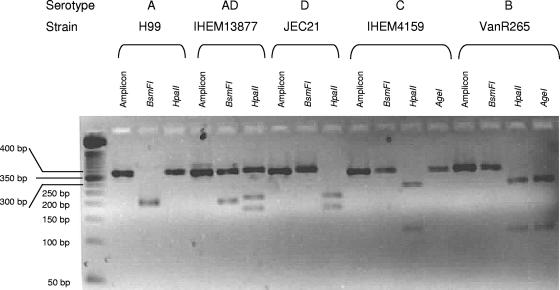
As expected from in silico analysis, enzymatic restriction with BsmFI and HpaII gave three different patterns for serotypes A, D, and B and C (Fig. 1). Serotype AD strains exhibited a mixed A and D restriction profile. All strains were tested, and no profile variation among strains of a given serotype was observed. Serotype D and serotypes B and C, while having similar profiles (unique cutting site for HpaII and no cutting site for BsmFI), can be easily differentiated based on the size difference of the digested fragments. Serotypes B and C can be further differentiated by AgeI digestion and gel electrophoresis. Two fragments corresponding to serotype B strains (one cutting site) were detected, while serotype C amplicons remained unchanged (no cutting site) (Fig. 1).
FIG. 1.
Agarose gel (3%) electrophoresis of native amplicons of the CAP59 gene fragments and restriction profiles of C. neoformans serotypes.
Length polymorphism analysis.
Because in silico analysis predicted length polymorphism corresponding to serotypes in the amplified fragments (Table 2), we designed a fluorescent fragment length analysis method using a DNA sequencer. Seven different alleles accounting for eight different genotypes were detected (Table 1). Fragments amplified from the strains of serotypes A, B, C, and D used as a reference for in silico analysis were sized with the predicted length plus or minus 1 base, which is the instrument resolution provided by the manufacturer.
A small degree of intraserotype polymorphism was seen: serotype A strains exhibited an allele of either 394 bp (n = 38) or 400 bp (n = 2); serotype D strains had an allele of either 380 bp (n = 2) or 382 bp (n = 14); and serotype B strains had an allele of 371 bp (n = 3), 373 bp (n = 4), or 380 bp (n = 1), while the two serotype C strains tested had an allele of 366 bp (Table 1). The six serotype AD strains exhibited two alleles, of 382 and 394 bp. Fragments with sizes different from the predominant size for a given serotype were sequenced. Table 3 summarizes the results of the BLASTN comparison of these sequences with sequences in the GenBank database. In all cases, the results of the sequence comparison agreed with the molecular identifications. Sequence alignment showed polymorphism among and within serotypes, mainly in the intronic part the fragments, located between nucleotide positions 200 and 280 (data not shown).
Analysis of discrepancies between conventional and molecular serotype identifications.
In a first analysis, we found seven cases of discrepancies in identifications (strains 20, 33, 47, 55, 57, 62, and 64). These discrepancies were seen only for strains for which the serotype had been determined by a homemade agglutination method. For these strains, a new serotype determination was performed using the Crypto Check kit, which agreed with the PCR-REA and length polymorphism identifications in every case.
DISCUSSION
Serotyping has been useful for the description of the epidemiologic and clinical characteristics associated with C. neoformans. More recently, it has played a major role in the investigation of the endemic infection on Vancouver Island (Canada) (10). Serotyping has no diagnostic value, but differences in treatments and outcomes corresponding to serotypes have been reported (2, 18). Thus, considering that the Crypto Check kit (Iatron, Japan) is no longer available and in view of the relative complexity of obtaining monoclonal antibodies, new techniques need to be developed for the typing of C. neoformans strains.
Both methods developed in this work allow the reliable identification of the five serotypes of C. neoformans by testing a unique genomic region. Our assays based on molecular analysis of a fragment of the CAP59 gene appeared to be sensitive and specific, providing interpretable results for all C. neoformans strains tested but no amplification for a C. uniguttulatus strain.
PCR-REA is an easy-to-use approach and requires only common molecular materials. Its value for distinguishing among fungal species such as Malassezia spp. has already been emphasized (6). As restriction sites were chosen within conserved regions of the fragments, all tested strains exhibited the restriction pattern predicted by in silico analysis. A flow chart for serotype identification by combining separate digestions with BsmFI and HpaII, followed by subsequent digestion with AgeI when the pattern suggests a B or C serotype, can therefore be proposed (Table 2). Serotype AD strains exhibited a mixed A and D restriction profile. Our results were in accordance with those of previous studies suggesting that AD serotype strains are diploid or aneuploid and possess, at least for a number of genes including the CAP59 gene, both serotype A-like and serotype D-like alleles (14, 17, 19, 20). This indication was further confirmed by a sequence analysis performed with the purified fragments (Table 3).
Length polymorphism analysis showed different lengths for different serotypes, with the exception of two serotype D strains (no. 55 and 61) and a serotype B strain (no. 69), which all exhibited a 380-bp allele. Note that these particular cases concerned only strains with alleles of the minority size within the serotype considered. These difficult cases can be solved by identifying the serotype by using either REA with AgeI (only serotype B is restricted) or the results of culture on CGB blue agar, on which serotype B but not serotype D strains produce a color change in the medium (12). Fragment length polymorphism may be easier to perform but requires a DNA sequencer, which is now more frequently available in teaching hospitals in industrialized countries. Following PCR amplification, results can be obtained within 1 h and are entirely objective.
Overall, results from both molecular methods fit well with serotype identifications obtained with immunologic procedures, but the homemade technique is probably less reliable. Both methods presented in this study appear to be reliable alternatives to immunologic methods.
Acknowledgments
We are grateful to J. Heitman and C. Arndt (Duke University Medical Center, Durham, NC); M.A. Viviani (Istituto di Igiene e Medicina Preventiva, Milano, Italy); M. Kombila (Département de Parasitologie-Mycologie-Médecine Tropicale, FMSS, Libreville, Gabon); D. Swinne (Institut Scientifique de Santé Publique-Mycologie, Brussels, Belgium); K. Adou-Bryn (Laboratoire de Parasitologie-Mycologie, UFR des Sciences Médicales, Abidjan, Ivory Coast); Y. Buisson (IMTSSA-Le Pharo, Marseille, France); and A. Paugam, A. Datry, and M. Develoux (Laboratoires de Parasitologie-Mycologie, Hôpital Pitié-Salpêtrière, Hôpital Tenon, and Hôpital Cochin, APHP, Paris, France) for the generous gift of some of the strains tested in this study. We are also indebted to the CNRMA (Centre National de Référence Mycologie et Antifongiques) directed by F. Dromer for serotyping some clinical isolates and to the Cryptococcus neoformans Serotype B Sequencing Project of the Broad Institute of Harvard and the Massachusetts Institute of Technology (http://www.broad.mit.edu) for making available C. neoformans serotype B sequences.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Casadevall, A., and J. R. Perfect. 1998. Cryptococcus neoformans. ASM Press, Washington, DC.
- 2.Chen, Y. C., S. C. Chang, C. C. Shih, C. C. Hung, K. T. Luhbd, Y. S. Pan, and W. C. Hsieh. 2000. Clinical features and in vitro susceptibilities of two varieties of Cryptococcus neoformans in Taiwan. Diagn. Microbiol. Infect. Dis. 36:175-183. [DOI] [PubMed] [Google Scholar]
- 3.Dromer, F., E. Gueho, O. Ronin, and B. Dupont. 1993. Serotyping of Cryptococcus neoformans by using a monoclonal antibody specific for capsular polysaccharide. J. Clin. Microbiol. 31:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzot, S. P., B. C. Fries, W. Cleare, and A. Casadevall. 1998. Genetic relationship between Cryptococcus neoformans var. neoformans strains of serotypes A and D. J. Clin. Microbiol. 36:2200-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia-Rivera, J., Y. C. Chang, K. J. Kwon-Chung, and A. Casadevall. 2004. Cryptococcus neoformans CAP59 (or Cap59p) is involved in the extracellular trafficking of capsular glucuronoxylomannan. Eukaryot. Cell 3:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillot, J., M. Deville, M. Berthelemy, F. Provost, and E. Gueho. 2000. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett. Appl. Microbiol. 31:400-403. [DOI] [PubMed] [Google Scholar]
- 7.Hennequin, C., E. Abachin, F. Symoens, V. Lavarde, G. Reboux, N. Nolard, and P. Berche. 1999. Identification of Fusarium species involved in human infections by 28S rRNA gene sequencing. J. Clin. Microbiol. 37:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horta, J. A., C. C. Staats, A. K. Casali, A. M. Ribeiro, I. S. Schrank, A. Schrank, and M. H. Vainstein. 2002. Epidemiological aspects of clinical and environmental Cryptococcus neoformans isolates in the Brazilian state Rio Grande do Sul. Med. Mycol. 40:565-571. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda, R., T. Shinoda, Y. Fukazawa, and L. Kaufman. 1982. Antigenic characterization of Cryptococcus neoformans serotypes and its application to serotyping of clinical isolates. J. Clin. Microbiol. 16:22-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd, S. E., F. Hagen, R. L. Tscharke, M. Huynh, K. H. Bartlett, M. Fyfe, L. Macdougall, T. Boekhout, K. J. Kwon-Chung, and W. Meyer. 2004. A rare genotype of Cryptococcus gattii caused the cryptococcosis outbreak on Vancouver Island (British Columbia, Canada). Proc. Natl. Acad. Sci. USA 101:17258-17263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon-Chung, K. J., J. E. Bennett, and J. C. Rhodes. 1982. Taxonomic studies on Filobasidiella species and their anamorphs. Antonie Leeuwenhoek 48:25-38. [DOI] [PubMed] [Google Scholar]
- 12.Kwon-Chung, K. J., I. Polacheck, and J. E. Bennett. 1982. Improved diagnostic medium for separation of Cryptococcus neoformans var. neoformans (serotypes A and D) and Cryptococcus neoformans var. gattii (serotypes B and C). J. Clin. Microbiol. 15:535-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon-Chung, K. J., T. C. Sorrell, F. Dromer, E. Fung, and S. M. Levitz. 2000. Cryptococcosis: clinical and biological aspects. Med. Mycol. 38(Suppl. 1):205-213. [PubMed] [Google Scholar]
- 14.Lengeler, K. B., G. M. Cox, and J. Heitman. 2001. Serotype AD strains of Cryptococcus neoformans are diploid or aneuploid and are heterozygous at the mating-type locus. Infect. Immun. 69:115-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marck, C. 1988. ‘DNA Strider': a ′C’ program for the fast analysis of DNA and protein sequences on the Apple Macintosh family of computers. Nucleic Acids Res. 16:1829-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moyrand, F., B. Klaproth, U. Himmelreich, F. Dromer, and G. Janbon. 2002. Isolation and characterization of capsule structure mutant strains of Cryptococcus neoformans. Mol. Microbiol. 45:837-849. [DOI] [PubMed] [Google Scholar]
- 17.Okabayashi, K., R. Kano, Y. Nakamura, S. Watanabe, and A. Hasegawa. 2006. Capsule-associated genes of serotypes of Cryptococcus neoformans, especially serotype AD. Med. Mycol. 44:127-132. [DOI] [PubMed] [Google Scholar]
- 18.Speed, B., and D. Dunt. 1995. Clinical and host differences between infections with the two varieties of Cryptococcus neoformans. Clin. Infect. Dis. 21:28-36. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka, R., Y. Imanishi, and K. Nishimura. 2003. Difference in FKS1 gene sequences between serotypes A and D of Cryptococcus neoformans. J. Clin. Microbiol. 41:4457-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu, J., G. Luo, R. J. Vilgalys, M. E. Brandt, and T. G. Mitchell. 2002. Multiple origins of hybrid strains of Cryptococcus neoformans with serotype AD. Microbiology 148:203-212. [DOI] [PubMed] [Google Scholar]