Abstract
Rickettsia are unique in inserting in-frame a number of palindromic sequences within protein coding regions. In this study, we extensively analyzed repeated sequences in the genome of Rickettsia conorii and examined their locations in regard to coding versus noncoding regions. We identified 656 interspersed repeated sequences classified into 10 distinct families. Of the 10 families, three palindromic sequence families showed clear cases of insertions into open reading frames (ORFs). The location of those in-frame insertions appears to be always compatible with the encoded protein three-dimensional (3-D) fold and function. We provide evidence for a progressive loss of the palindromic property over time after the insertions. This comprehensive study of Rickettsia repeats confirms and extends our previous observations and further indicates a significant role of selfish DNAs in the creation and modification of proteins.
Interspersed repeated DNA sequences are usually confined in the intergenic regions of bacterial genomes. However, Rickettsia appears to be a unique exception in this respect. In our previous work, we identified evolutionarily related sequences of 50 amino acid residues dispersed in protein-coding regions of Rickettsia conorii and other Rickettsia (Ogata et al. 2000). The peptide segments showed no sequence similarity to known protein domains. On the other hand, the corresponding nucleotide sequences (∼150 bases) showed imperfect palindromic (self-complementary) properties that resemble other bacterial intergenic repeats like IRU (Sharples and Lloyd 1990) and RSA (Bachellier et al. 1996). The repeats were designated as Rickettsia palindromic elements (RPEs). On the basis of the predicted locations of the inserts in the three-dimensional (3-D) folds of proteins, and on the observed transcript sizes, it is most likely that the repeat-derived peptide is expressed as part of the proteins encoded by those open reading frames (ORFs) (Ogata et al. 2000). Thus the RPE appears to have a unique capability to spread over the coding, as well as the noncoding, regions of the bacterial genome.
The completion of the genome sequence of R. conorii revealed a high density of repeated sequences in the genome (Ogata et al. 2001b). In this study, we systematically analyzed the repeat locations in regard to coding versus noncoding regions. Three different palindromic sequence families showed clear cases of insertions into ORFs. In addition, several palindromic sequences were identified within RNA coding genes. We also found that the palindromic property of the repeats has a tendency to be dimmed over time after their insertions in the genome.
RESULTS
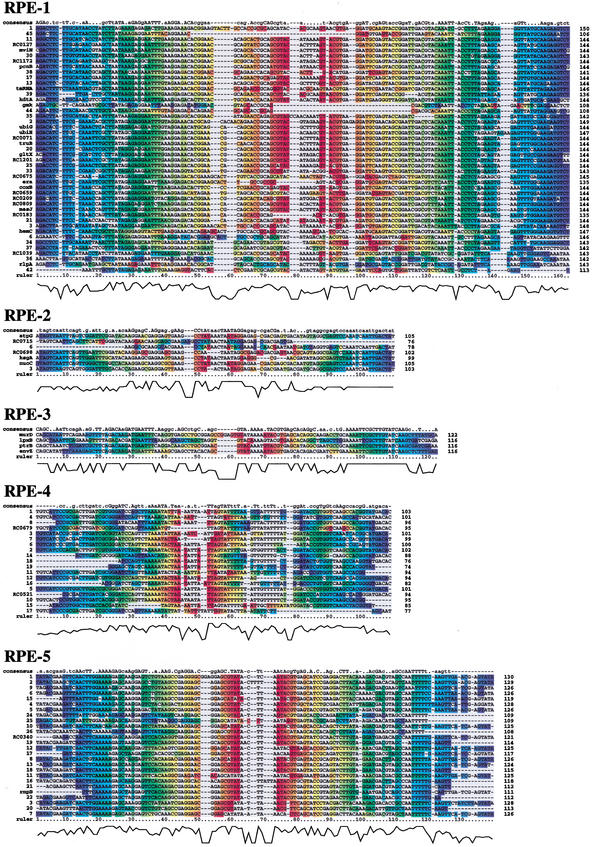
We identified 656 interspersed repeated DNA sequences in the genome of R. conorii. On the basis of sequence similarity, the repeated sequences were classified into 10 distinct families (Table 1). There is no significant sequence similarity between the various repeat families. Their copy numbers range from 5 to 223. Nucleotide sequence alignments of the repeated sequences are shown in Figure 1. A coloring scheme is used to help visualize the predicted RNA secondary structures in the alignments. Of the 10 families, eight showed palindromic sequences with consensus sizes from 95 to 149 bases. Stable RNA secondary structures were predicted for most of the sequences in those eight families. The predicted secondary structures showed hairpin-like forms or variants with additional branched stems. However, the precise base-pairing pattern in the structures varied across, as well as within, the repeat families. The eight palindromic sequence families were named RPE-1 to RPE-8. The previously reported 44 RPEs (Ogata et al. 2000; Ogata et al. 2001b) were classified into the RPE-1 family, for which 11 additional copies have been identified in this study. The two remaining families are composed of shorter repeats (25 bases and 27 bases) showing no stable predicted secondary structure. They were designated as Rickettsia repeat-1 (RR-1) and repeat-2 (RR-2).
Table 1.
Interspersed Palindromic Repeats and Short Repeats in Rickettsia conorii
| Name | Total number | Size (bp)a | RNA 2-D structure | G + C | Full length repeatb | Partial repeatb | Distribution in other species | Number in R. prowazekii | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Number | Within gene | Number | Within gene | |||||||
| Rickettsia palindromic elements (RPEs) | ||||||||||
| RPE-1 | 52 | 141 (25–150) | hairpin | 42% | 45 | 24c | 7 | 3 | Rickettsia spp. | 10 |
| RPE-2 | 7 | 105 (76–105) | hairpin | 43% | 7 | 5 | — | — | — | — |
| RPE-3 | 12 | 116 (28–122) | hairpin | 40% | 4 | 4 | 8 | 2 | Rickettsia spp. | 4e |
| RPE-4 | 94 | 95 (20–103) | hairpin | 43% | 19 | 2 | 75 | 11 | Rickettsia spp. | 15 |
| RPE-5 | 55 | 115 (22–130) | hairpin | 40% | 26 | 2d | 29 | 3 | Rickettsia spp. | 11f |
| RPE-6 | 168 | 136 (20–141) | clover leaf, | 39% | 23 | 1 | 145 | 26 | Rickettsia spp. | 3 |
| (RS3-like) | Y-shape | Neisseria spp. | ||||||||
| RPE-7 | 223 | 99 (20–136) | clover leaf, hairpin | 43% | 36 | 1 | 187 | 26 | Rickettsia spp. | 14 |
| RPE-8 | 31 | 149 (30–172) | hairpin | 22% | 15 | — | 16 | — | Rickettsia spp. | 17 |
| Rickettsia repeats (RRs) | ||||||||||
| RR-1 | 5 | 27 (19–27) | — | 49% | 5 | 1 | — | — | — | — |
| RR-2 | 9 | 25 (22–39) | — | 47% | 8 | — | 1 | — | — | — |
Sizes of the consensus sequences. The lengths for the smallest and the largest copies are shown in parentheses.
See text for the definition of the “full size” repeat and the “partial” repeats.
One of the repeats is within the tmRNA gene (ssrA) of R. conorii.
One of the repeats is within the M1 RNA gene (rnpB) of R. conorii.
Three copies are within the ORFs (RP037, RP012, RP707) of R. prowazekii.
One of the repeats is within the tmRNA gene of R. prowazekii.
Figure 1.
Nucleotide sequence alignments of the 10 repeat families identified in Rickettsia conorii. The coloring scheme is equivalent to the bracket representation of RNA secondary structure; the bases predicted to make base pairing are represented by the same color (instead of opening and closing brackets). The consensus sequences were derived from the aligned positions in which a single base is observed in at least 70% of the sequences (lower cases for 70%–90%; upper cases for ≥90% conservation). ClustalX alignment quality scores are shown at the bottom of the alignment.
Exhaustive BLAST searches against sequence databases revealed that all of the 10 families except the RPE-6 are specific to Rickettsia species or limited to R. conorii (Table 1). The RPE-6 repeat contains two directly oriented RS3 core motifs (ATTCCC-N8-GGGAAT) frequently found in Neisseria genomes (Haas and Meyer 1986). All the repeat families are relatively GC-rich (∼40% of GC) compared with the average GC content of the entire genome (32%), with the exception of the AT-rich RPE-8 (22%). Size variation within family is large for six families (RPE-3 to RPE-8). More than 50% of the identified repeats are “partial” repeats for those RPEs. Size variation is relatively small for the other families (RPE-1 and RPE-2; RR-1 and RR-2), which are mostly composed of “full-length” copies (see Methods).
The analysis of the whole 656 repeat locations revealed a large number of insertions within ORFs for seven RPEs (RPE-1 to RPE-7) and a single occurrence for RR-1 (Table 1). Those cases include full-length repeat insertions within “annotated” ORFs (ORFs with functions predicted by homology), as well as partial repeat insertions within ORFans (ORFs lacking similarity in other organisms). None of the insertions interrupts the reading frame of the host ORFs. Table 2 shows the 38 ORFs harboring the full-length repeats. Of the seven palindromic families found within ORFs, three families (RPE-1 to RPE-3) showed a number of full-length insertions into annotated ORFs. Most of those ORFs appear to be important for R. conorii, because they constitute parts of biological pathways or molecular complexes involving many other genes that are present in the R. conorii genome (Ogata et al. 2001b). However, there is no apparent functional relationship between the predicted functions of those altered ORFs.
Table 2.
Full Size Repeat Insertions within the Rickettsia conorii Genes
| Gene name | Function | Phylogenetic distributiona | Size (bp)b | Location of repeat insertionc | Structure datad |
|---|---|---|---|---|---|
| RPE-1 | |||||
| coxB (RC0555) | Cytochrome c oxidase polypeptide II | RPG--YOAE | 945 | 16..159 | 1OCC/Bos taurus |
| era (RC0158) | GTP-binding protein | RPG-SYO-E | 1017 | 10..144 | 1EGA/Escherichia coli |
| gltX (RC0966) | Glutamyl-tRNA synthetase | RPGCSYOAE | 1539 | 1006..1149 | 1GLN/Thermus thermophilus |
| gmk (RC1194) | Guanylate kinase | RPGC-YO-E | 687 | 19..126 | 1GKY/Saccharomyces cerevisiae |
| hemC (RC0706) | Porphobilinogen deaminase | RPGC-YOAE | 1053 | 772..918 | 1PDA/Escherichia coli |
| kdtA (RC0118) | 3-deoxy-D-manno-octulosonic-acid transferase | RP-C--O-- | 1392 | 147..290 | |
| mesJ (RC0067) | Cell cycle protein MesJ | RPGCSYO-- | 1434 | 625..768 | |
| mviN (RC0898) | Virulence factor MviN | RP-CSYO-- | 1665 | 6..149 | |
| pcnB (RC0015) | Poly(A)polymerase | RPGCSYO-E | 1308 | 120..263 | |
| rlpA (RC0537) | Rare lipoprotein A precursor | RP--SYO-- | 960 | 33..175 | |
| ssrA | tmRNA precursor | RPGCSYO-- | 474 | 78..223 | |
| truB (RC0665) | tRNA pseudouridine 55 synthase | RPGCSYOAE | 1035 | 784..927 | |
| ubiG (RC0965) | 3-demethylubiquinone-9 3-methyltransferase | RP------E | 867 | 148..291 | |
| uhiH (RC0848) | Ubiquinone biosynthesis protein | RP---Y--E | 1293 | 31..177 | |
| RC1039 | Split gene of mannose-1-phosphate guanylyltransferase | -P---YOA- | 627 | 9..151 | |
| RC0071 | Unknown function | RP---YO-E | 1218 | 985..1128 | |
| RC0127 | Unknown function | --------- | 222 | 42..185 | |
| RC0183 | Unknown function | --------- | 1158 | 706..840 | |
| RC0209 | Unknown function | R-------- | 279 | 22..165 | |
| RC0659 | Unknown function | R-------- | 582 | 31..174 | |
| RC0675 | Unknown function | --------- | 225 | 18..162 | |
| RC0809 | Unknown function | RPGCSYO-- | 735 | 349..492 | |
| RC1172 | Unknown function | --------- | 345 | 18..161 | |
| RC1201 | Unknown function | --------- | 240 | 100..243 | |
| RPE-2 | |||||
| atpG (RC1236) | ATP synthase γ chain | RPG--YO-E | 969 | 622..726 | 1H8E/Bos taurus |
| ksgA (RC1022) | Dimethyladenosine transferase | RPGCSYOAE | 945 | 370..468 | 1YUB/Streptococcus pneumoniae |
| nuoC (RC0483) | NADH dehydrogenase I chain C | RPG--YOAE | 726 | 199..303 | |
| RC0698 | Unknown function | R-------- | 1002 | 100..201 | |
| RC0715 | Unknown function | R-------- | 753 | 299..374 | |
| RPE-3 | |||||
| envZ (RC0592) | Osmolarity sensor protein EnvZ | RP------- | 1437 | 34..149 | |
| lpxB (RC0440) | Lipid-A-disaccharide synthase | RP-C-YO-- | 1338 | 1138..1253 | |
| murD (RC0560) | UDP-N-acetylmuramoylalanineD-glutamate ligase | RPGCSYO-- | 1500 | 796..917 | 1E0D/Escherichia coli |
| ptrB (RC0377) | Protease II | RPG------ | 2187 | 526..641 | 1QFM/Sus scrofa (pig) |
| RPE-4 | |||||
| RC0521 | Unknown function | --------- | 336 | 29..122 | |
| RC0679 | Unknown function | --------- | 1806 | 243..337 | |
| RPE-5 | |||||
| RC0340 | Unknown function | --------- | 171 | 45..165 | |
| rnpB | RNA subunit (M1 RNA) of ribonuclease P | RPGCSYOAE | 458 | 118..228 | |
| RPE-6 | |||||
| RC0614 | Unknown function | --------- | 387 | 198..334 | |
| RPE-7 | |||||
| RC1210 | Unknown function | --------- | 303 | 30..129 | |
| RR-1 | |||||
| RC1196 | Unknown function | --------- | 180 | 9..35 |
Abbreviations for the organism groups are as follows. R: Rickettsia (Rickettsia prowazekii); P: Proteobacteria (Escherichia coli K-12, Haemoplilus influenzae, Xylella fastidiosa, Vibrio cholerae, Pseudomonas aeruginosa, Buchnera sp., Neisseria meningitidis serogroup A and B, Helicobacter pylori 26695 and J99, Campylobacter jejuni); G: Gram positive bacteria (Bacillus subtilis, Bacillus halodurans, Mycoplasma genitalium, Mycoplasma pneumoniae, Ureaplasma urealyticum, Mycobacterium tuberculosis; C: Chlamydia (Chlamydia trachomatis, Chlamydia muridarum, Chlamydia pneumonia CWL029, AR39 and J138); S: Spirochete (Borrelia burgdorferi, Treponema pallidum); Y: Cyanobacteria (Synechocystis); O: Other bacteria (Deinococcus radiodurans, Aquifex aeolicus, Thermotoga maritima); A: Archaea (Methanococcus jannaschii, Methanobacterium thermoautotrophicum, Archaeoglobus fulgidus, Halobacterium sp., Thermoplasma acidophilum, Pyrococcus horikoshii; Pyrococcus abyssi, Aeropyrum pernix); E: Eukaryotes (Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila melanogaster). When there is no homolog within an organism group, ‘-’ replaces the organism abbreviation.
Gene size without stop codon.
The repeat location is indicated by the base position within the gene.
Protein Data Bank identifiers and the organism names for the available structure data.
By use of BLAST, we examined the occurrence of the homologous repeats in the Rickettsia prowazekii genome (Table 1). Interestingly, three copies of the RPE-3 were found within the ORFs of R. prowazekii: RP037 (putative O-sialoglycoprotein endopeptidase) and two ORFs of unknown functions (RP012 and RP707). Each of the three ORFs has a clear ortholog in R. conorii, which lacks the repeat insert. We previously reported nine cases of the RPE-1 within R. prowazekii ORFs (Ogata et al. 2000). In addition, a copy of an RS3-like element has been found in the N-terminal of the α subunit of DNA polymerase III (DnaE) in Rickettsia felis (Andersson and Andersson 1999). Thus, the insertions of palindromic sequences within ORFs seem to be a widespread phenomenon in different Rickettsia species.
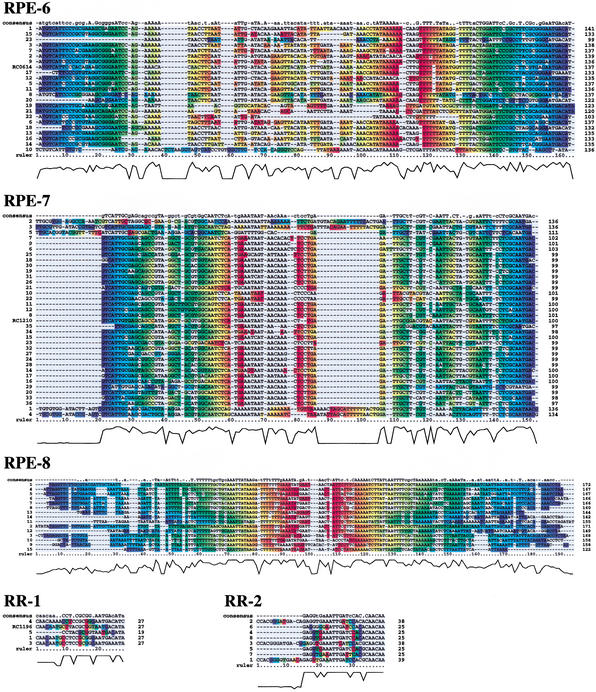
Multiple alignments of the peptide sequences derived from the RPE-1 to RPE-3 are shown in Figure 2. Each of the three RPEs shows distinct peptide sequences with unbiased amino acid compositions. The peptide sequences are well aligned within the family and correspond to the same reading frame. A remarkable feature of those RPEs is the capability of occupying any site, even in the middle, along the primary sequences of the ORFs (Table 2). For instance, an RPE-1 sequence is located in the middle part of the R. conorii MesJ protein. The predicted protein secondary structure is a central α-helix for the RPE-1 and the RPE-3. In contrast, extended conformations (β-strands) were predicted at both extremities of the peptide sequences derived from the RPE-2. Those three peptide families may thus show different 3-D folds.
Figure 2.
Multiple sequence alignments for the amino acid sequences translated from the palindromic sequences (RPE-1, RPE-2, and RPE-3). Amino acid residues are colored as follows: blue for F, W, and Y; yellow for C; orange for A, G, P, S, and T; green for I, L, M, and V; red for D, E, H, K, and R; purple for N and Q. The letters ‘H’ and ‘E’ in the first line of each alignment represent the predicted α-helices and β-strands, respectively. ClustalX alignment quality scores are shown at the bottom of the alignment.
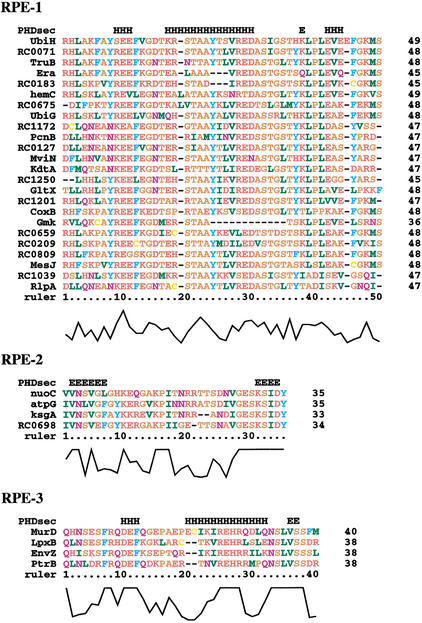
We then examined the insertion sites of the RPE-derived peptides with 3-D structure data for the homologs of the host proteins. Seven protein structures for RPE-1 insertions (Ogata et al. 2000), two for RPE-2 and two for RPE-3 (Table 2), were available for this analysis. In all cases, the insertion site corresponded to the solvent-exposed area of the proteins: mainly loops (nine cases) and occasionally short helices (one case) or beta strands (one case). Furthermore, none of the predicted insertions appeared to hinder known catalytic sites or protein/cofactor binding sites. Four cases corresponding to the ORFs with RPE-2 and RPE-3 insertions are shown in Figure 3.
Figure 3.
Predicted insertion sites of the RPE-2 (a, b) and RPE-3 (c, d). Green segments labeled by the green arrows in the reference structures indicate the insertion sites. (a) Bovine mitochondrial ATP synthase. γ chain is shown in red. (b) Streptococcus pneumoniae rRNA methyltransferase used as a reference for the R. conorii dimethyladonosine transferase. (c) E. coli UDP-N-acetylmuramoylalanine–D-glutamate ligase. (d) Pig prolyl oligopeptidase used as a reference for the R. conorii protease II. Seven cases corresponding to the predicted insertions sites for the RPE-1 have been previously reported (Ogata et al. 2000). The images are generated with MolScript (http://www.avatar.se/molscript/).
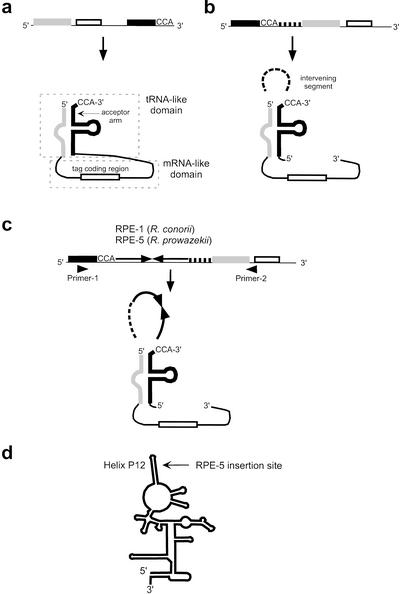
We identified several RPEs within RNA genes. The tmRNA coding genes (ssrA) of R. conorii and R. prowazekii harbor palindromic sequences of different families (an RPE-1 for R. conorii and an RPE-5 for R. prowazekii). tmRNA is an RNA molecule present in all known bacterial genomes. Its function is to rescue the ribosome stalled on an mRNA (Muto et al. 1998). tmRNA is composed of a tRNA-like domain and an mRNA-like domain (Fig. 4a). The structure of the tmRNA genes from α-proteobacteria (Keiler et al. 2000), together with the locations of the RPE-1 and RPE-5, are shown in Figure 4, b and c. The two insertions of the palindromic sequences within the tmRNA genes were both located right after the CCA-(3′) bases of the acceptor arm, where alanine is added by alanyl-tRNA synthetase. We examined the transcription status from the R. conorii tmRNA gene by reverse transcriptase-polymerase chain reaction (RT-PCR). The RT-PCR product showed the expected size with the repeat insert, demonstrating transcription of RPE-1 with the rest of the genes. Because there is no detectable sequence similarity between the RPE-1 and the RPE-5, those two insertions must have occurred independently in the different lineages of Rickettsia. Another case found in R. conorii is an RPE-1 within a ribozyme gene, rnpB, which encodes M1 RNA of the ribonuclease P. The insertion site corresponded to the P12 helix of the RNA secondary structure model of M1 RNA (Fig. 4d) (Brown et al. 1996). The P12 helix shows a highly variable sequence, and the helix is unlikely to involve functionally important tertiary interactions in vivo (Pomeranz Krummel and Altman 1999).
Figure 4.
RPEs found within the RNA genes. (a) The standard form of the tmRNA genes observed in most bacterial species. (b) The permutated form of the tmRNA gene observed in α-proteobacteria (Keiler et al. 2000). (c) Locations of the RPE-1 and RPE-5 inserted in the tmRNA genes of Rickettsia. Approximate locations of the primers are indicated by triangles. (d) The secondary structure model of the M1 RNA and the insertion site of the RPE-5 in R. conorii.
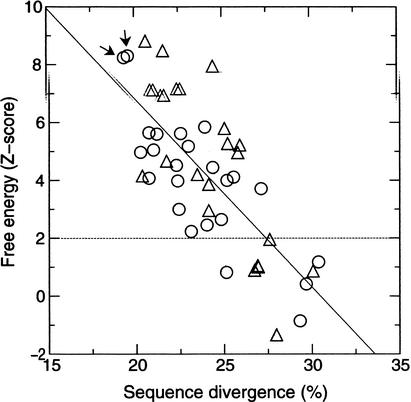
The palindromic property (hairpin-like secondary structure) of the RPEs is probably required for repeat insertion as suspected for IRU (Sharples and Lloyd 1990) and RSA (Bachellier et al. 1996). Alternatively, the hairpin structures might have an important function for Rickettsia. In the former case, the hairpin-like structure might lose its utility after insertions and could disappear over time. In the latter case, nucleotide secondary structures should be conserved despite sequence changes (as in ribosomal RNAs). To investigate the significance of the palindromic property of the RPEs, we computed the minimum free energy for every sequence of the RPE-1 to RPE-8 and obtained the relevant Z-score (see Methods). If we take Z-score > 2 (P-value < .0423) as a threshold, the energy values for 10/45 sequences (22%) of the RPE-1 failed to be significant. Such energy values below the threshold were also observed for the RPE-2 (4/7; 57%), the RPE-5 (6/20; 23%), and the RPE-8 (3/15; 20%). This result indicates that the palindromic properties of some repeats are unlikely to be constrained after their insertions. In Figure 5, the Z-score of the RPE-1 is plotted against the sequence divergence D, the average sequence difference against the other sequences of the RPE-1. The pair of most similar sequences, which might correspond to most recent inserts, showed very high Z-scores (Z = 8.30 and 8.24). The two sequences were identified in the truB gene and in RC0071. The 144 bases sequences are 94.4% identical with each other. There is also a global tendency for the better conserved sequences to show higher stabilities of the RNA secondary structures (the correlation coefficient is R = −0.74; P < .005). This decay of palindromic property indicates the absence of structural constraints on the repeats after their insertions. The lack of significant differences in the structural stability between the coding and the noncoding repeats also argues against a specific role of the palindromic structures at the transcription and translation levels.
Figure 5.
The minimum free energy of the predicted RNA secondary structures of the RPE-1 plotted against sequence divergence. The energy values are scaled into Z-score. Circles correspond to the repeats in coding regions. Triangles correspond to the repeats in noncoding regions. The arrows point to the two most similar sequences.
DISCUSSION
In this study, we identified 10 families of repeated sequences in the genome of R. conorii and examined their locations in regard to coding versus intergenic regions. Three palindromic sequence families (RPE-1, RPE-2, and RPE-3) showed clear cases of insertions within predicted coding regions, and eight families in total (RPE-1 to RPE-7 plus RR-1) showed insertions within coding regions including ORFans. Therefore, the surprising mechanism of repeat insertion within protein coding regions initially described for RPE-1 (Ogata et al. 2000) applies to many other repeat families in Rickettsia.
The analysis of the locations and the sequences of the repeat-derived peptides (RPE-1, RPE-2, and RPE-3) reinforced our previous observations (Ogata et al. 2000). First, they are inserted into ORFs with only one reading frame of six possibilities, as indicated by the aligned sequences of the repeat-derived peptides (Fig. 2). Second, there are no clear functional links between the ORFs harboring the repeats. Third, the insertion sites of the repeats vary along the primary sequence of the ORFs but always appear compatible with the preexisting protein folds.
This study revealed two additional aspects. First, the predicted protein secondary structures for the three RPEs (RPE-1 to RPE-3) correspond to two different conformations. α-Helices were predicted for the RPE-1 and RPE-3, whereas β-strands were predicted for the RPE-2. Thus, the two regular conformations in protein structure (α-helix and β-strand) could occur from repeat insertions. However, the possibility is not ruled out that those peptides are “neutral” and might adapt variable conformations in response to the surrounding structural environment at the insertions sites. Circular dichroism spectroscopy failed to show any property of the regular conformation for synthetic peptides (∼50 amino acids) corresponding to the RPE-1 (C. Abergel, unpublished data). Four repeat-containing proteins have been expressed in Escherichia coli (V. Monchois et al., in prep.), and experiments are in progress to determine the structural properties of the repeat-derived peptides within these proteins.
Second, we showed that some copies of the RPEs do not exhibit a significant palindromic structure. Because the pair of most similar RPE-1 sequences correspond to highly stable hairpins, it is plausible that this secondary structure is a feature of the original copies that are mobile within the genome. The structural property might then be lost after the insertion regardless of the site of the genome (coding or noncoding), as the initial repeat continued to diverge in both sequence and structure. However, the possibility is not ruled out that some of the repeats have been recruited for host cellular functions (Gilson et al. 1984; Gilson et al. 1986a; Gilson et al. 1986b; Sharples and Lloyd 1990) or recombination (Bi and Liu 1996; Oggioni and Claverys 1999; Shyamala et al. 1990; van der Ende et al. 1999), as already suggested for other bacterial repeats.
Proteins contain structurally flexible regions, usually corresponding to surface loops. Such loops are known to be tolerant of insertions of individual amino acids or peptides. For instance, insertions of peptides between 7 and 17 residues into a loop of the chymotrypsin inhibitor-2 (64 amino acids) have little effect on the stability and the folding rate (Ladurner and Fersht 1997). This physical flexibility parallels the evolutionary flexibility of protein sequences. Available sequence and structure data indicate a high preference of insertions and deletions within loops (Pascarella and Argos 1992). However, most (99%) of the accepted insertions and deletions are shorter than 10 amino acid residues. In contrast, the palindromic repeats described in this study could contribute to insertions up to ∼50 residues. Repeat insertion can freely occur within the 20% of R. conorii genome corresponding to the noncoding regions. If one accepts that surface loops account for a quarter of every protein sequence (Wootton 1994), another 20% of the genome (from the coding moiety) is available for additional repeat insertions. RPEs appear to invade both of the two genomic regions.
The mechanism by which the bacterial palindromic sequences of the size of RPEs spread within genomes is not known (Bachellier et al. 1999). However, the coincidence of the two insertion sites of the palindromic sequences in the tmRNA genes (ssrA) of Rickettsia is intriguing. The upstream sequences of those insertion sites are highly similar to bacteriophage attachment site (att) (Kirby et al. 1994). The homologous sites of the other tmRNA genes and tRNA genes have been known to harbor bacteriophages, retron phages, and pathogenicity islands in other bacteria (Billington et al. 1999; Haring et al. 1995; Inouye et al. 1991; Julio et al. 2000; Karaolis et al. 1998; Kirby et al. 1994; Pierson and Kahn 1987). Such retron phages and pathogenicity islands are supposed to be integrated by use of the integrases of phages. Some of the RPEs might have used a similar mechanism.
We have proposed that RPEs are selfish DNA elements that can break the barrier of genetic material between coding and noncoding sequences (Ogata et al. 2000). Recursive insertions of such selfish DNAs might provide the initial genetic material for rather “neutral” protein segments, which could then later evolve to create new functions (Dwyer 2001; Ogata et al. 2001a). The genomes of Rickettsia show by far the highest number of occurrence of such insertions, even if a few instances have been reported in other bacteria. For instance, a partial copy of RSA was found in the C-terminal of a hypothetical ORF of 99 amino acids in Salmonella typhimurium (Bachellier et al. 1996). Recently another case has been reported in Sinorhizobium meliloti. The DNA helicase II (UvrD) of this legume symbiont has a 47 amino acid residues insert encoded by a palindromic DNA sequence (motif C) (Capela et al. 2001). The ongoing accumulation of more bacterial genome sequences should lead to better appreciation of the importance of the phenomenon of repeat insertions in the origin and evolution of proteins.
METHODS
The genomic sequence and annotation data for R. conorii are available at RicBase (http://igs-server.cnrs-mrs.fr/RicBase) and NCBI GenBank (http://www.ncbi.nlm.nih.gov/; accession no. AE006914). Other complete genomes including those used in Table 2 were obtained from KEGG (Kanehisa and Goto 2000). Database searches were performed with the NCBI BLAST package (Altschul et al. 1997) against the complete genomes as well as the NCBI nonredundant sequence database.
Repeated DNA sequences of R. conorii were initially identified on the basis of the self-comparison of the genomic DNA by BLASTN (E-value < 10−4). The BLAST result was then analyzed to delineate the left and right edges of the repeated sequences with the repeat identification program Mocca (Notredame 2001). Some trivial repeats such as tRNAs or paralogous ORFs were removed from the dataset a posteriori. A complete list of the repeats described in this paper is available at RicBase.
The “full-length” repeats were defined as the sequences with lengths within 70% to 100% of the longest repeat of the family. The remaining shorter sequences were defined as “partial” repeats. The nucleotide sequence alignments and the consensus sequences in Figure 1 were constructed from the full-length repeats with T-Coffee (Notredame et al. 1998) and ClustalX (Thompson et al. 1997). Sequence divergence D used in Figure 5 was defined as the average sequence difference against the other sequences of the family. The definition of D does not take into account nucleotide secondary structures. In the computation of D, the positions with gaps in the pairwise alignments were omitted.
The minimum free energy and the corresponding RNA secondary structures were computed with the Vienna package at http://www.tbi.univie.ac.at/∼ivo/RNA/ (Hofacker et al. 1994). The minimum free energy value was then converted to Z-score. To compute Z-score, every sequence was randomly shuffled 30 times, from which the mean and the standard deviation values were computed. We used an approximation by the extreme value distribution (Gumbel 1958) to obtain the relevant P value. Protein secondary structures were predicted with PHDsec at http://dodo.cpmc.columbia.edu/predictprotein/ (Rost and Sander 1994).
The following protein structure data were obtained from the Protein Data Bank (http://www.rcsb.org/pdb/): Bovine mitochondrial ATP synthase F1 domain (1H8E); Streptococcus pneumoniae rRNA methyltransferase (1YUB); E. coli UDP-N-acetylmuramoylalanine–D-glutamate ligase (1E0D); and Pig prolyl oligopeptidase (1QFM). The rRNA methyltransferase was used as a reference for R. conorii dimethyladenosine transferase (KsgA); they belong to the rRNA adenine N-6-methyltransferase family. The prolyl oligopeptidase was used as a reference structure for R. conorii protease II (PtrB); they both belong to the prolyl oligopeptidase family.
The presence of the RPE-1 in the transcript from the tmRNA gene of R. conorii was assessed by RT-PCR by use of a primer pair, P1 (5′-TAA TTT AGA ATA GAG GTT GCG GAC T-3′) and P2 (5′-CGT TTG CGT TTC TTT GTT TT-3′), designed to be specific to the target gene. The expected size of the RT-PCR product was 311 bp including the RPE-1 (146 bp). For RNA extraction, a suspension of fresh R. conorii strain Malish (seven) was adjusted to 108/mL, and bacteria were separated from cells with a sucrose gradient. RNA extraction from bacteria was then performed with the RN easy Midi kit (Qiagen, Hilden, Germany) as recommended by the manufacturer. RT-PCR was performed on the resulting RNA with the One-step RT-PCR kit (Qiagen) following the manufacturer's instructions. Reverse transcription and amplification were performed in PTC-200 thermocyclers (MJ Research, Watertown, USA) with 40 PCR cycles and an annealing temperature of 50°C. RT-PCR products were run in 1% agarose gels, stained with ethidium bromide, and revealed on a UV box. Following the RT-PCR assay, we performed a PCR assay with the same primers on the RNA extract with the Elongase polymerase (Life Technologies, Cergy Pontoise, France). The PCR assay was negative, thus verifying the absence of contaminating DNA in the RNA. The RT-PCR product was sequenced with the same primers with the d-Rhodamine terminator cycle-sequencing ready reaction kit (PE Applied Biosystems, Les Ulis, France) and an ABI-PRISM 3100 automated DNA sequencer (PE Applied Biosystems), as recommended by the manufacturer. An RT-PCR product of 311 bp was obtained from the R. conorii RNA. The sequence of the RT-PCR product was 100% identical to the genomic sequence of the R. conorii tmRNA gene containing the RPE-1.
WEB SITE REFERENCES
http://dodo.cpmc.columbia.edu/predictprotein/; PHDsec.
http://igs-server.cnrs-mrs.fr/RicBase; RicBase.
http://www.avatar.se/molscript/; MolScript.
http://www.ncbi.nlm.nih.gov/; NCBI GenBank.
http://www.rcsb.org/pdb/; Protein Data Bank.
http://www.tbi.univie.ac.at/∼ivo/RNA/; Vienna package.
Acknowledgments
We thank Professors Philippe Derreumaux and Didier Raoult for helpful discussions.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Hiroyuki.Ogata@igs.cnrs-mrs.fr; FAX +33 4 91 16 45 49.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.227602. Article published online before print in April 2002.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson JO, Andersson SG. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- Bachellier S, Clement JM, Hofnung M. Short palindromic repetitive DNA elements in enterobacteria: A survey. Res Microbiol. 1999;150:627–639. doi: 10.1016/s0923-2508(99)00128-x. [DOI] [PubMed] [Google Scholar]
- Bachellier S, Gilson E, Hofnung M, Hill CW. Repeated sequences. In: Neidhardt FC, Curtiss III R, Ingraham JL, Lin ECC, Brooks Low K, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella. Washington D.C.: ASM Press; 1996. pp. 2012–2040. [Google Scholar]
- Bi X, Liu LF. DNA rearrangement mediated by inverted repeats. Proc Natl Acad Sci. 1996;93:819–823. doi: 10.1073/pnas.93.2.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington SJ, Huggins AS, Johanesen PA, Crellin PK, Cheung JK, Katz ME, Wright CL, Haring V, Rood JI. Complete nucleotide sequence of the 27-kilobase virulence related locus (vrl) of Dichelobacter nodosus: Evidence for extrachromosomal origin. Infect Immun. 1999;67:1277–1286. doi: 10.1128/iai.67.3.1277-1286.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Nolan JM, Haas ES, Rubio MA, Major F, Pace NR. Comparative analysis of ribonuclease P RNA using gene sequences from natural microbial populations reveals tertiary structural elements. Proc Natl Acad Sci. 1996;93:3001–3006. doi: 10.1073/pnas.93.7.3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capela D, Barloy-Hubler F, Gouzy J, Bothe G, Ampe F, Batut J, Boistard P, Becker A, Boutry M, Cadieu E, et al. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc Natl Acad Sci. 2001;98:9877–9882. doi: 10.1073/pnas.161294398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DS. Selfish DNA and the origin of genes. Science. 2001;291:252–253. doi: 10.1126/science.291.5502.252. [DOI] [PubMed] [Google Scholar]
- Gilson E, Clement JM, Brutlag D, Hofnung M. A family of dispersed repetitive extragenic palindromic DNA sequences in E. coli. EMBO J. 1984;3:1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson E, Perrin D, Clement JM, Szmelcman S, Dassa E, Hofnung M. Palindromic units from E. coli as binding sites for a chromoid-associated protein. FEBS Lett. 1986a;206:323–328. doi: 10.1016/0014-5793(86)81005-5. [DOI] [PubMed] [Google Scholar]
- Gilson E, Rousset JP, Clement JM, Hofnung M. A subfamily of E. coli palindromic units implicated in transcription termination? Ann Inst Pasteur Microbiol. 1986b;137B:259–270. doi: 10.1016/s0769-2609(86)80116-8. [DOI] [PubMed] [Google Scholar]
- Gumbel EJ. Statistics of extremes. New York, NY: Columbia University Press; 1958. [Google Scholar]
- Haas R, Meyer TF. The repertoire of silent pilus genes in Neisseria gonorrhoeae: Evidence for gene conversion. Cell. 1986;44:107–115. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- Haring V, Billington SJ, Wright CL, Huggins AS, Katz ME, Rood JI. Delineation of the virulence-related locus (vrl) of Dichelobacter nodosus. Microbiology. 1995;141:2081–2089. doi: 10.1099/13500872-141-9-2081. [DOI] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Sebastian Bonhoeffer L, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatsh Chem. 1994;125:167–188. [Google Scholar]
- Inouye S, Sunshine MG, Six EW, Inouye M. Retronphage phi R73: An E. coli phage that contains a retroelement and integrates into a tRNA gene. Science. 1991;252:969–971. doi: 10.1126/science.1709758. [DOI] [PubMed] [Google Scholar]
- Julio SM, Heithoff DM, Mahan MJ. ssrA (tmRNA) plays a role in Salmonella enterica serovar Typhimurium pathogenesis. J Bacteriol. 2000;182:1558–1563. doi: 10.1128/jb.182.6.1558-1563.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaolis DK, Johnson JA, Bailey CC, Boedeker EC, Kaper JB, Reeves PR. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci. 1998;95:3134–3139. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler KC, Shapiro L, Williams KP. tmRNAs that encode proteolysis-inducing tags are found in all known bacterial genomes: A two-piece tmRNA functions in Caulobacter. Proc Natl Acad Sci. 2000;97:7778–7783. doi: 10.1073/pnas.97.14.7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby JE, Trempy JE, Gottesman S. Excision of a P4-like cryptic prophage leads to Alp protease expression in Escherichia coli. J Bacteriol. 1994;176:2068–2081. doi: 10.1128/jb.176.7.2068-2081.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladurner AG, Fersht AR. Glutamine, alanine or glycine repeats inserted into the loop of a protein have minimal effects on stability and folding rates. J Mol Biol. 1997;273:330–337. doi: 10.1006/jmbi.1997.1304. [DOI] [PubMed] [Google Scholar]
- Muto A, Ushida C, Himeno H. A bacterial RNA that functions as both a tRNA and an mRNA. Trends Biochem Sci. 1998;23:25–29. doi: 10.1016/s0968-0004(97)01159-6. [DOI] [PubMed] [Google Scholar]
- Notredame C. Mocca: Semi-automatic method for domain hunting. Bioinformatics. 2001;17:373–374. doi: 10.1093/bioinformatics/17.4.373. [DOI] [PubMed] [Google Scholar]
- Notredame C, Holm L, Higgins DG. COFFEE: An objective function for multiple sequence alignments. Bioinformatics. 1998;14:407–422. doi: 10.1093/bioinformatics/14.5.407. [DOI] [PubMed] [Google Scholar]
- Ogata H, Audic S, Barbe V, Artiguenave F, Fournier P-E, Raoult D, Claverie J-M. Selfish DNA in protein-coding genes of Rickettsia. Science. 2000;290:347–350. doi: 10.1126/science.290.5490.347. [DOI] [PubMed] [Google Scholar]
- Ogata H, Audic S, Claverie J-M. Selfish DNA and the origin of genes. Science. 2001a;291:252–253. doi: 10.1126/science.291.5502.252. [DOI] [PubMed] [Google Scholar]
- Ogata H, Audic S, Renesto-Audiffren P, Fournier P-E, Barbe V, Samson D, Roux V, Cossart P, Weissenbach J, Claverie J-M, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001b;293:2093–2098. doi: 10.1126/science.1061471. [DOI] [PubMed] [Google Scholar]
- Oggioni MR, Claverys JP. Repeated extragenic sequences in prokaryotic genomes: A proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology. 1999;145:2647–2653. doi: 10.1099/00221287-145-10-2647. [DOI] [PubMed] [Google Scholar]
- Pascarella S, Argos P. Analysis of insertions/deletions in protein structures. J Mol Biol. 1992;224:461–471. doi: 10.1016/0022-2836(92)91008-d. [DOI] [PubMed] [Google Scholar]
- Pierson LS, 3rd, Kahn ML. Integration of satellite bacteriophage P4 in Escherichia coli. DNA sequences of the phage and host regions involved in site-specific recombination. J Mol Biol. 1987;196:487–496. doi: 10.1016/0022-2836(87)90026-x. [DOI] [PubMed] [Google Scholar]
- Pomeranz Krummel DA, Altman S. Verification of phylogenetic predictions in vivo and the importance of the tetraloop motif in a catalytic RNA. Proc Natl Acad Sci. 1999;96:11200–11205. doi: 10.1073/pnas.96.20.11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- Sharples GJ, Lloyd RG. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990;18:6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamala V, Schneider E, Ames GF. Tandem chromosomal duplications: Role of REP sequences in the recombination event at the join-point. EMBO J. 1990;9:939–946. doi: 10.1002/j.1460-2075.1990.tb08192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ende A, Hopman CT, Dankert J. Deletion of porA by recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect Immun. 1999;67:2928–2934. doi: 10.1128/iai.67.6.2928-2934.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton JC. Sequences with ‘unusual’ amino acid compositions. Curr Opin Struct Biol. 1994;4:413–421. [Google Scholar]