Abstract
Since congenital cytomegalovirus (CMV) infection causes late-onset sequelae, the identification of CMV-infected newborns is important. For this purpose, we established a simple real-time PCR assay using a filter disk. Combined with the collection of urine using filter papers placed in the diaper, this assay can make CMV screening more feasible and cost-effective.
Congenital cytomegalovirus (CMV) infection occurs in 0.2 to 1% of all births. In addition to infants with symptomatic cases, a proportion of asymptomatically infected newborns face a significant risk of late-onset sequelae (8, 9, 12, 16, 17, 22, 26). Since the early identification of congenitally infected newborns may lead to early intervention and antiviral treatment options (14), a simple and inexpensive assay for CMV detection is required to implement screening programs for congenital CMV infection (19).
In spite of the progress in PCR-based assays, the need for DNA purification from body fluid specimens as well as the labor, costs, and other problems associated with specimen collection, transportation, and storage have limited the convenience of these assays. Although a small volume of urine can be used directly for PCR, inhibitors in urine reduce PCR efficiency (6, 7, 13, 23). Robotic systems that may simplify the DNA purification process (20) are not affordable for every facility. The use of filter papers can resolve the problems associated with collection and storage, as exemplified by the use of dried blood spots (10). Washing filter papers has reduced the amount of inhibitors, which in turn has allowed for conventional PCR on the filter disk (25). Alternatively, DNA samples have been extracted and/or purified from specimens on filters and used for PCR (3, 5, 18, 27). Although dried blood spots can be used for CMV screening (2), the assay may not detect some cases of congenital infection, as blood specimens contain smaller amounts of CMV than urine specimens (4, 11; N. Inoue and S. Koyano, unpublished results).
In this study, we developed a real-time PCR assay using urine specimens on filter disks as a template for the reaction and demonstrated the assay's technological potential for screening for congenital CMV infection.
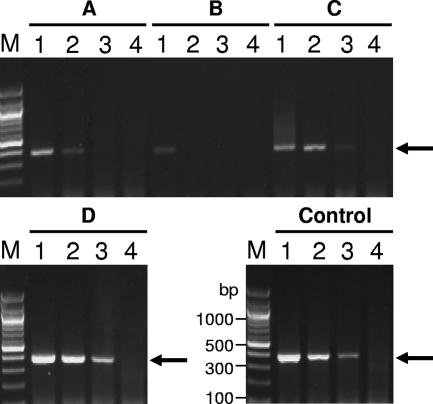
First, we selected a filter paper on which PCRs could proceed efficiently (Fig. 1). Commercially available filter papers were spiked with dilutions of purified CMV, and filter disks obtained from the filters were added directly to PCR mixtures. Since Isocode filters allowed more efficient amplification than the others, these filters were used for the following experiments.
FIG. 1.
Selection of filter paper. Filter disks of 3 mm in diameter (A to D) and a solution (Control) containing 5 × 105 (lane 1), 5 × 104 (lane 2), 5 × 103 (lane 3), and 0 (lane 4) genome copies of purified CMV (Towne strain) were added to reaction mixtures for the conventional PCR assay. Numbers of CMV genome copies were determined by the real-time PCR assay described previously (17). Filter papers compared were as follows: FTA Micro card (Whatman, United Kingdom) (A), Generation Capture card (Gentra Systems Inc., Minneapolis, MN) (B), plain filter for blood collection (Advantec, Japan) (C), and Isocode (Schleicher & Schuell, Germany) (D). After PCR amplification, the PCR products were analyzed by gel electrophoresis. Arrows indicate the expected PCR products. Lane M, 1-kb DNA ladder marker.
Next, we found that only instruments equipped with a photomultiplier-tube scanning system (e.g., Stratagene MX3500P) could be used for real-time PCR assays with filter disks in the reaction mixture, due to the fact that instruments using a charge-coupled device camera (e.g., ABI7700) were adversely affected by nonspecific signals from the disks. The optimized real-time PCR conditions were as follows. Fifty microliters of reaction mixture contained 1× Brilliant quantitative PCR master mix (Stratagene), 5 μg of bovine serum albumin, 100 ng of salmon sperm DNA, 0.2 μM primers, 0.125 μM TaqMan probe, and a 3-mm-diameter filter disk. The primers, probe, and standards were described previously (17). The cycle conditions were one cycle of 2 min at 50°C and 15 min at 95°C followed by cycles of 15 s at 95°C, 30 s at 58°C, and 30 s at 72°C.
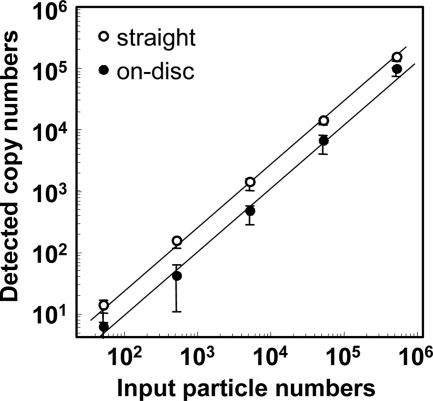
CMV diluted in CMV-negative urine was either added directly to the reaction mixture or applied to filter disks that were then used as a template in the PCR. Both templates provided a linear dose dependency in the real-time PCR assay (Fig. 2). The ratios of the detected copy number to the input genome copy number at each dilution were averaged, and the result was defined as the detection efficiency. Detection efficiencies for specimens added directly and specimens applied to disks were 25 and 11% of input, respectively. With the detection limit for standard DNA as a cutoff value (5 copies/reaction mixture), 50 CMV genome copies on a disk were enough to generate a positive signal.
FIG. 2.
Efficiency of CMV detection by real-time PCR with a filter disk in the reaction mixture. Purified CMV samples were diluted in CMV-negative urine. Three microliters of the dilutions containing 5 × 105 to 5 × 101 CMV genome copies per reaction mixture was added directly to 50 μl of the PCR mixture (“straight”). Three microliters of the dilutions was also applied to filter disks of 3 mm in diameter. The disks were rinsed with water and then added directly to a reaction plate for real-time PCR (“on-disc”). The x axis indicates the quantity of CMV virions added to each reaction mixture, and the y axis indicates CMV copy numbers measured by the real-time PCR. The experiment was done in triplicate.
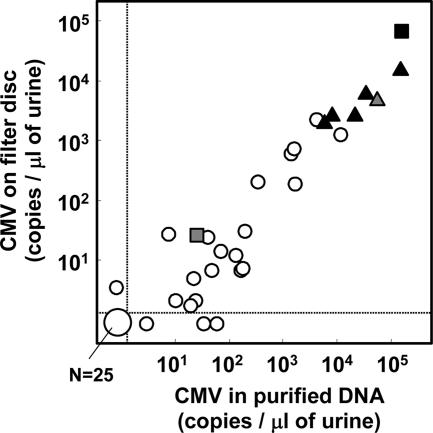
To examine the practical applicability of the assay, 55 urine specimens from 34 newborns and infants were collected. Eight of the specimens corresponded to six congenital cases. Five of these congenitally infected subjects were asymptomatic at birth. Their congenital infections were identified by the detection of CMV-specific immunoglobulin M and confirmed by PCR using dried umbilical cords (15). The collection and use of the specimens was approved by the Ethical Committee on Human Subjects, and informed consent was obtained from each parent(s). The three following forms of templates obtained from the same original volumes of urine were compared by PCR: (i) DNA purified from urine by using the QIAamp viral RNA kit (QIAGEN), (ii) straight urine specimens, and (iii) urine specimens on filter disks. There were good correlations among the CMV copy numbers measured in the assays using different forms of templates (correlation coefficients between each pair of the assays were 0.89 to 0.94) (Fig. 3; data not shown). With 5 copies in a reaction mixture as the cutoff value, 96% of the specimens negative in the assay using purified DNA were negative in the filter-based assay and 90% of the specimens positive in the assay using purified DNA were positive in the filter-based assay. Importantly, the CMV copy numbers in urine specimens from all six infants with congenial infections were >1,000-fold higher than the detection limit.
FIG. 3.
Comparison of real-time PCR assays using clinical urine specimens. CMV viral loads in 55 urine specimens were measured by the real-time PCR assays using DNA samples (equivalent to 3 μl of urine specimens) purified from urine specimens and using filter disks containing 3 μl of urine specimens. The obtained results were expressed as copy numbers per microliter of original urine specimen. For each specimen, the CMV copy numbers obtained in the assay using DNA purified from urine specimens (x axis) and those obtained in the assay using the filter (y axis) are plotted. Specimens from five asymptomatic newborns with congenital CMV infection obtained at birth (n = 5) and 1 month later (n = 1) are indicated with closed and hatched triangles, respectively. Specimens from a symptomatic newborn with congenital CMV infection obtained at birth and 1 month later, after treatment with ganciclovir, are indicated with closed and hatched squares. Other specimens are indicated with open circles. Dashed lines indicate the cutoff values.
By adding CMV-spiked urine on filter papers inserted underneath the inside surfaces of baby diapers, we confirmed that the expected amounts of CMV were detected on those filters. Finally, from 100 apparently healthy newborns and 1 infant with congenital CMV infection, urine specimens were collected onto filter papers placed in diapers. The detection of CMV on the filters was performed blindly, and only the filter from the infant with the congenital infection yielded a positive signal in the filter-based assay.
We found that >60% of CMV DNA on filter disks was eluted into a solution by incubation at 95°C for 30 min. DNA recovery allowed for the confirmation of the filter-based PCR results and genotyping of the positive specimens.
Saliva specimens are as useful as urine specimens for congenital CMV screening (1, 24). It is noteworthy that our filter-based real-time PCR was applicable for saliva specimens also (data not shown).
The advantages of the filter-based real-time PCR were as follows: (i) minimum specimen handling; (ii) no liquid specimen phase before PCR, which eliminates the potential for PCR contamination; and (iii) sensitive and quantitative measurements. The stability of DNA in filters has been demonstrated previously (21). The assay cost, including the costs for filters, reagents, and disposables and the wages of a technician, totaled <$8 per specimen. Combined with urine collection from diapers, our filter-based assay can be most suitable for screening programs for CMV infections in newborns. Because small sample volumes on filter disks might introduce unexpected errors in clinical settings, a pilot screening program to verify the assay performance is under way.
Acknowledgments
We thank Phil Pellett and John A. Stewart (deceased) for their intellectual input.
This work was supported by a Grant for Child Health and Development from the Ministry of Health, Labor and Welfare, Japan (N.I.), and by a Grant for the Research on Health Sciences Focusing on Drug Innovation program from the Japanese Human Science Foundation (N.I.).
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Balcarek, K. B., W. Warren, R. J. Smith, M. D. Lyon, and R. F. Pass. 1993. Neonatal screening for congenital cytomegalovirus infection by detection of virus in saliva. J. Infect. Dis. 167:1433-1436. [DOI] [PubMed] [Google Scholar]
- 2.Barbi, M., S. Binda, V. Primache, S. Caroppo, P. Didò, P. Guidotti, C. Corbetta, and D. Melotti. 2000. Cytomegalovirus DNA detection in Guthrie cards: a powerful tool for diagnosing congenital infection. J. Clin. Virol. 17:159-165. [DOI] [PubMed] [Google Scholar]
- 3.Barbi, M., S. Binda, V. Primache, C. Luraschi, and C. Corbetta. 1996. Diagnosis of congenital cytomegalovirus infection by detection of viral DNA in dried blood spots. Clin. Diagn. Virol. 6:27-32. [DOI] [PubMed] [Google Scholar]
- 4.Boppana, S. B., K. B. Fowler, R. F. Pass, L. B. Rivera, R. D. Bradford, F. D. Lakeman, and W. J. Britt. 2005. Congenital cytomegalovirus infection: association between virus burden in infancy and hearing loss. J. Pediatr. 146:817-823. [DOI] [PubMed] [Google Scholar]
- 5.Cassol, S., T. Salas, M. Arella, P. Neumann, M. T. Schechter, and M. O'Shaughnessy. 1991. Use of dried blood spot specimens in the detection of human immunodeficiency virus type 1 by the polymerase chain reaction. J. Clin. Microbiol. 29:667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, D., M. Wang, W. C. Ou, R. T. Tsai, C. Y. Fung, and Y. J. Hwang. 1996. A simple method for detecting human polyomavirus DNA in urine by the polymerase chain reaction. J. Virol. Methods 58:131-136. [DOI] [PubMed] [Google Scholar]
- 7.Demmler, G. J., G. J. Buffone, C. M. Schimbor, and R. A. May. 1988. Detection of cytomegalovirus in urine from newborns by using polymerase chain reaction DNA amplification. J. Infect. Dis. 158:1177-1184. [DOI] [PubMed] [Google Scholar]
- 8.Fowler, K. B., A. J. Dahle, S. B. Boppana, and R. F. Pass. 1999. Newborn hearing screening: will children with hearing loss caused by congenital cytomegalovirus infection be missed? J. Pediatr. 135:60-64. [DOI] [PubMed] [Google Scholar]
- 9.Fowler, K. B., F. P. McCollister, A. J. Dahle, S. Boppana, W. J. Britt, and R. F. Pass. 1997. Progressive and fluctuating sensorineural hearing loss in children with asymptomatic congenital cytomegalovirus infection. J. Pediatr. 130:624-630. [DOI] [PubMed] [Google Scholar]
- 10.Guthrie, R., and A. Susi. 1963. A simple phenylalanine method for detecting phenylketonuria in large populations of newborn infants. Pediatrics 32:338-343. [PubMed] [Google Scholar]
- 11.Halwachs-Baumann, G., B. Genser, S. Pailer, H. Engele, H. Rosegger, A. Schalk, H. H. Kessler, and M. Truschnig-Wilders. 2002. Human cytomegalovirus load in various body fluids of congenitally infected newborns. J. Clin. Virol. 25(Suppl. 3):S81-S87. [DOI] [PubMed] [Google Scholar]
- 12.Hicks, T., K. Fowler, M. Richardson, A. Dahle, L. Adams, and R. Pass. 1993. Congenital cytomegalovirus infection and neonatal auditory screening. J. Pediatr. 123:779-782. [DOI] [PubMed] [Google Scholar]
- 13.Khan, G., H. O. Kangro, P. J. Coates, and R. B. Heath. 1991. Inhibitory effects of urine on the polymerase chain reaction for cytomegalovirus DNA. J. Clin. Pathol. 44:360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimberlin, D. W., C. Y. Lin, P. J. Sánchez, G. J. Demmler, W. Dankner, M. Shelton, R. F. Jacobs, W. Vaudry, R. F. Pass, J. M. Kiell, S. J. Soong, and R. J. Whitley. 2003. Effect of ganciclovir therapy on hearing in symptomatic congenital cytomegalovirus disease involving the central nervous system: a randomized, controlled trial. J. Pediatr. 143:16-25. [DOI] [PubMed] [Google Scholar]
- 15.Koyano, S., A. Araki, Y. Hirano, K. Fujieda, T. Suzutani, K. Yagyu, K. Murono, and N. Inoue. 2004. Retrospective diagnosis of congenital cytomegalovirus infection using dried umbilical cords. Pediatr. Infect. Dis. J. 23:481-482. [DOI] [PubMed] [Google Scholar]
- 16.Kumar, M. L., G. A. Nankervis, I. B. Jacobs, C. B. Ernhart, C. E. Glasson, P. M. McMillan, and E. Gold. 1984. Congenital and postnatally acquired cytomegalovirus infections: long-term follow-up. J. Pediatr. 104:674-679. [DOI] [PubMed] [Google Scholar]
- 17.Ogawa, H., T. Suzutani, Y. Baba, S. Koyano, N. Nozawa, K. Ishibashi, K. Fujieda, N. Inoue, and K. Omori. 2007. Etiology of severe sensorineural hearing loss in children: independent impact of congenital cytomegalovirus infection and GJB2 mutations. J. Infect. Dis. 195:782-788. [DOI] [PubMed] [Google Scholar]
- 18.Pitcovski, J., E. Shmueli, S. Krispel, and N. Levi. 1999. Storage of viruses on filter paper for genetic analysis. J. Virol. Methods 83:21-26. [DOI] [PubMed] [Google Scholar]
- 19.Revello, M. G., and G. Gerna. 2002. Diagnosis and management of human cytomegalovirus infection in the mother, fetus, and newborn infant. Clin. Microbiol. Rev. 15:680-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang, Y. W., S. E. Sefers, H. Li, D. J. Kohn, and G. W. Procop. 2005. Comparative evaluation of three commercial systems for nucleic acid extraction from urine specimens. J. Clin. Microbiol. 43:4830-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uttayamakul, S., S. Likanonsakul, R. Sunthornkachit, K. Kuntiranont, S. Louisirirotchanakul, A. Chaovavanich, V. Thiamchai, S. Tanprasertsuk, and R. Sutthent. 2005. Usage of dried blood spots for molecular diagnosis and monitoring HIV-1 infection. J. Virol. Methods 128:128-134. [DOI] [PubMed] [Google Scholar]
- 22.Williamson, W. D., G. J. Demmler, A. K. Percy, and F. I. Catlin. 1992. Progressive hearing loss in infants with asymptomatic congenital cytomegalovirus infection. Pediatrics 90:862-866. [PubMed] [Google Scholar]
- 23.Yamaguchi, Y., T. Hironaka, M. Kajiwara, E. Tateno, H. Kita, and K. Hirai. 1992. Increased sensitivity for detection of human cytomegalovirus in urine by removal of inhibitors for the polymerase chain reaction. J. Virol. Methods 37:209-218. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto, A. Y., M. M. Mussi-Pinhata, L. J. Marin, R. M. Brito, P. F. C. Oliveira, and T. B. Coelho. 2006. Is saliva as reliable as urine for detection of cytomegalovirus DNA for neonatal screening of congenital CMV infection? J. Clin. Virol. 36:228-230. [DOI] [PubMed] [Google Scholar]
- 25.Yourno, J., and J. Conroy. 1992. A novel polymerase chain reaction method for detection of human immunodeficiency virus in dried blood spots on filter paper. J. Clin. Microbiol. 30:2887-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yow, M. D., D. W. Williamson, L. J. Leeds, P. Thompson, R. M. Woodward, B. F. Walmus, J. W. Lester, H. R. Six, and P. D. Griffiths. 1988. Epidemiologic characteristics of cytomegalovirus infection in mothers and their infants. Am. J. Obstet. Gynecol. 158:1189-1195. [DOI] [PubMed] [Google Scholar]
- 27.Zerr, D. M., M. L. Huang, L. Corey, M. Erickson, H. L. Parker, and L. M. Frenkel. 2000. Sensitive method for detection of human herpesviruses 6 and 7 in saliva collected in field studies. J. Clin. Microbiol. 38:1981-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]