Abstract
Nucleotide sequence analysis of the NS5B region was performed to identify genotypes of 8,479 hepatitis C virus (HCV) RNA-positive patient samples collected in the Canadian province of Quebec. Genotypes could be determined for 97.3% of patients. Genotypes 1 to 6 were found in 59.4, 9.0, 25.7, 3.6, 0.6, and 1.8% of patients, respectively. Two isolates did not classify within the six genotypes. The subtype 1 distribution was 76.7% 1a, 22.6% 1b, and 0.7% others, while the subtype 2 distribution was 31.8% 2a, 47.6% 2b, 10.9% 2c, 4.1% 2i, and 5.6% others. Subtype 3a accounted for 99.1% of genotype 3 strains, while all genotype 5 samples were of subtype 5a. The subtype 4 distribution was 39.2% 4a, 15.4% 4k, 11.6% 4d, 10.2% 4r, and 23.6% others. The subtype 6 distribution was 40.4% 6e, 20.5% 6a, and 39.1% others. The 5′ untranslated region (5′UTR) sequences of subtype 6e were indistinguishable from those of genotype 1. All samples that did not classify within the established subtypes were also sequenced in C/E1 and 5′UTR. C/E1 phylogenetic reconstructions were analogous to those of NS5B. The sequences identified in this study allowed the provisional assignments of subtypes 1j, 1k, 2m, 2r, 3i, 4q, 6q, 6r, and 6s. Sixty-four (0.8%) isolates classifying within genotypes 1 to 6 could not be assigned to one of the recognized subtypes. Our results show that genotyping of HCV by nucleotide sequence analysis of NS5B is efficient, allows the accurate discrimination of subtypes, and is an effective tool for studying the molecular epidemiology of HCV.
Hepatitis C virus (HCV) was first identified in the late 1980s and is a major cause of liver disease throughout the world (40). It is a single-stranded, positive-polarity RNA virus classified in the genus Hepacivirus of the family Flaviviridae. The HCV genome encodes a single long polyprotein with the following gene order: 5′-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-3′ (22). The genomes of HCV variants display considerable sequence divergence and have been classified into six genotypes. Genotypes 1, 2, 3, 4, and 6 can further be subdivided into a series of subtypes. HCV genotypes display different geographic distributions worldwide (43).
Typing of HCV isolates is of specific clinical interest, as the genotype is a marker of the likelihood of a response to pegylated interferon-ribavirin combination therapy and serves as a guideline for the duration of therapy (50). Determination of the HCV genotype is now part of medical practice for pretreatment patient management. Genotyping is also useful for investigating outbreaks of infections and for understanding the epidemiology and virological features of this virus. Several methods targeting different regions of the HCV genome have been used for assessing genotypes. The most accurate method is to sequence an appropriate coding region that is divergent enough to allow the discrimination of types and subtypes (32, 36). The three most studied regions are the core, E1, and NS5B (4).
The 5′ untranslated region (5′UTR) is the region of choice for qualitative and quantitative HCV RNA detection due to its high level of conservation and sensitivity. For this reason, it has most often been used by clinical laboratories for routine genotyping of HCV. However, due to this high level of conservation, the 5′UTR is limited in its ability to discriminate genotype 6 from genotype 1 and subtypes within genotypes 1, 2, 3, 4, and 6. Genotype 6 variants other than 6a and 6b show 5′UTR sequences identical or similar to those of type 1 and, as a consequence, cannot be differentiated (8, 25, 45). A large number of subtypes that often share the same 5′UTR sequence have been described (4, 6, 10, 42, 46). The inability to distinguish subtype 2c from subtype 2a in the 5′UTR is a good example of this phenomenon. Furthermore, some of the genotype-specific motifs that were initially identified in the 5′UTR are no longer found to be conserved. For example, the G residue at position 243 of the 5′UTR, originally considered to be representative of subtype 1b, is found to occur in a relative proportion of subtype 1a viruses (6, 7, 10, 14, 46).
The nomenclature of HCV genotypes provides a scientific opportunity to study the spread of the virus worldwide (43). The efforts put forward by the international HCV databases allowing for the retrieval of sequences by genotype and subtype designations have facilitated the study of HCV genetic variability and classification of viral strains (20). Recent developments in DNA sequencing technology such as the advent of automated DNA sequencers and computer programs for genome assembly have rendered sequencing more accessible for routine use in the clinical laboratory (16). Using this technology, we have applied direct sequencing analysis of amplicons derived from the NS5B region for HCV genotyping. Sequence analyses of C/E1 and 5′UTR were also performed on a subset of variants for comparison purposes. Here, we report the prevalence of HCV genotypes/subtypes in more than 8,000 patients with a precision unmatched compared to 5′UTR-based genotyping methods.
MATERIALS AND METHODS
Study population.
HCV genotyping in the Canadian province of Quebec is centralized at the Laboratoire de Santé Publique du Québec. Samples submitted between November 2001 and June 2006 were included in this study. Samples were submitted from hospital laboratories and public or private clinics. Specimens were either refrigerated and sent within 3 days of collection on ice packs or frozen and transported on dry ice. Samples not meeting these criteria or displaying RNA levels below 600 IU/ml (COBAS AMPLICOR HCV Monitor test, version 2.0; Roche Diagnostics, Branchburg, NJ) were not genotyped.
RNA extraction and RT-PCR.
Samples for RNA extraction were processed in batches of 24, including one positive and one negative control. Viral RNA was extracted from 140 μl of serum or EDTA-plasma using the QIAamp viral RNA kit and QIAvac 6S vacuum manifold (QIAGEN Inc., Mississauga, Ontario, Canada). One reverse transcription (RT)-PCR run consisted of two batches of RNA extractions (48 samples) processed simultaneously. RT-PCR was performed using a one-step RT-PCR kit (QIAGEN, Inc.) in a 50-μl reaction volume containing 20 μl of the extracted RNA, 1× QIAGEN OneStep RT-PCR buffer, 400 μM each dATP, dCTP, dGTP, and dUTP (Amersham Biosciences Inc., Baie d'Urfé, Quebec, Canada), 0.6 μM sense and antisense primers (Table 1), 2.0 μl QIAGEN OneStep RT-PCR enzyme mix, 10 U RNAguard RNase inhibitor (Amersham Biosciences Inc.), and 1 U heat-labile uracil-DNA glycosylase (Roche Diagnostics, Laval, Quebec, Canada). Samples were incubated at 20°C for 10 min, 50°C for 30 min, and 95°C for 15 min. DNA amplification was performed for 40 cycles each consisting of 55°C for 30 s, 72°C for 60 s, and 94°C for 15 s in a GeneAmp PCR System 9600 or 9700 (Applied Biosystems, Foster City, CA). The last cycle was followed by a 10-min extension step at 72°C.
TABLE 1.
Primers used for viral genome detection and sequencing
| Genomic region and primer | Polarity | Sequence | Positiona | Size of PCR product (bp) |
|---|---|---|---|---|
| NS5B | ||||
| DM100 | Antisense | 5′-TACCTVGTCATAGCCTCCGTGAA-3′ | 8616-8638 | 389 |
| DM101 | Sense | 5′-TTCTCRTATGAYACCCGCTGYTTTGA-3′ | 8250-8275 | |
| DM106 | Antisense | 5′-GGNGCYGAGTAYCTGGTCATGGC-3′ | 8625-8647 | 407 |
| DM107 | Sense | 5′-CCHATGGGGTTYTCCTAYGACACCAG-3′ | 8241-8266 | |
| DM104 | Antisense | 5′-TAYCTGGTCATAGCNTCCGTAAA-3′ | 8616-8638 | 389 |
| DM105 | Sense | 5′-TTCTCCTAYGACACCAGRTGYTTTGA-3′ | 8250-8275 | |
| DM87 | Antisense | 5′-CATAGCCTCCGTGAAGGCTCTC-3′ | 8609-8630 | |
| DM88 | Antisense | 5′-CATAGCCTCCGTGAAGACTCGT-3′ | 8609-8630 | |
| DM113 | Sense | 5′-GAYACCCGCTGYTTGACTC-3′ | 8259-8378 | |
| DM128 | Sense | 5′-TGAGGAGTCCATATACCAGGC-3′ | 8306-8326 | |
| DM129 | Sense | 5′-TGAGGAGTCAATCTACCAATG-3′ | 8306-8326 | |
| DM130 | Sense | 5′-ACTGAGAGCGAYATCCGTACG-3′ | 8286-8306 | |
| DM131 | Sense | 5′-ACTGAGARTGACATCCGTGTT-3′ | 8286-8306 | |
| DM132 | Antisense | 5′-GCCGCATACCAGCATTGTGGG-3′ | 8532-8552 | |
| DM133 | Antisense | 5′-TCCACACACCAGCATAACGGG-3′ | 8532-8552 | |
| DM134 | Antisense | 5′-CGTGAAGGCTCGCAGGCTCGC-3′ | 8601-8621 | |
| DM136 | Antisense | 5′-CTGGACCCCTGCACTTTCACA-3′ | 8571-8591 | |
| DM137 | Sense | 5′-AATGTTGTGACCTGGACCCCC-3′ | 8323-8343 | |
| DM138 | Antisense | 5′-ATCGACGCCATCACTCTCAGC-3′ | 8571-8591 | |
| DM139 | Sense | 5′-AATGTTGTAATCTTGAGCCGG-3′ | 8323-8343 | |
| C/E1 | ||||
| DM108 | Antisense | 5′-TTCATCATCATRTCCCANGCCAT-3′ | 1293-1315 | 473 |
| DM109 | Sense | 5′-AAYYTDCCCGGTTGCTCTTTYTCTAT-3′ | 843-868 | |
| DM110 | Antisense | 5′-GTRGGNGACCARTTCATCATCA-3′ | 1306-1327 | 494 |
| DM111 | Sense | 5′-GCAACAGGGAAYYTDCCYGGTTGCTC-3′ | 834-859 | |
| 5′UTR | ||||
| DM50 | Antisense | 5′-CTCGCAAGCACCCTATCAGG-3′ | 292-311 | 241 |
| DM51 | Sense | 5′-GAAAGCGTCTAGCCATGGCGTTAGT-3′ | 71-95 |
Relative to H77 (GenBank accession number AF009606).
DNA purification and sequencing.
Amplicons were purified by using the QIAquick 8 PCR purification kit and QIAvac 6S vacuum manifold (QIAGEN Inc.) and analyzed by ethidium bromide agarose gel electrophoresis. Samples showing a band of the appropriate size were further analyzed by DNA sequencing. Sequencing reactions were performed by using the ABI Prism Big Dye Terminator cycle sequencing ready reaction kit v2.0 with AmpliTaq DNA polymerase FS and electrophoresed on an ABI Prism 3100 genetic analyzer (Applied Biosystems). Amplicons were sequenced in both directions using the sense and antisense amplification primers except for NS5B-derived PCR products, which were generally sequenced using sense primer DM113 and antisense amplification primer DM100 (Table 1). PCR products from various mixed-genotype infections were directly sequenced using the following primer pairs: for 1a and 1b, DM130/DM87 (1a) and DM131/DM88 (1b); for 1a and 2a, DM130/DM134 (1a) and DM128/DM132 (2a); for 1a and 2b, DM129/DM134 (1a) and DM128/DM133 (2b); for 1a and 3a, DM137/DM136 (1a) and DM139/DM138 (3a); and for 1b and 2a, DM129/DM88 (1b) and DM128/DM87 (2a).
Base calling, sequence assembly, and editing.
The ABI sample files for the sense and antisense sequencing reactions were electronically transferred via an internal network to a Linux platform. Chromatograms were base called using Phred (version 0.000925.C) and assembled with gcPhrap (version 0.990329) (11, 12). Phrap output files were automatically returned to a dedicated PC running Windows. The Phrap assembly was viewed with Gap4 version 4.5, and the Phrap consensus sequence was manually edited with BioEdit (version 5.0.9) (15, 44). Sequence editing included the removal of primer sequences and the incorporation of International Union of Biochemistry degenerate base codes. After the removal of primer sequences, lengths of stored sequences were as follows: 340 bp for NS5B, positions 8276 to 8615; 424 to 436 bp for C/E1, positions 869 to 1292; and 196 to 199 bp for 5′UTR, positions 96 to 291 (19).
Genotype determination.
Genotypes were assigned following phylogenetic analysis. Sequences were aligned by using the ClustalX program (47). Phylogenetic trees were inferred by using the PHYLIP (versions 3.5 and 3.6) software package (13). Trees were constructed by using the programs DNADIST and NEIGHBOR. DNADIST was run using the F84 model of nucleotide substitution. Trees were displayed with the TreeView program (version 1.6.6) (35). For the neighbor-joining trees presented in Fig. 1, the estimate of the transition/transversion rate was determined by the program TREE-PUZZLE using the HKY model of substitution (39). The robustness of the reconstructed phylogenies was evaluated by bootstrap analysis. The program SEQBOOT was used to generate a data set of 1,000 sequence replicates. The consensus tree was derived by using the program CONSENSE. Variants identified prior to November 2001 and used in the reconstruction of the phylogenetic trees include QC2, QC6, QC12, QC13, QC26, QC29, QC30, QC56 to QC59, QC133, QC135, QC214, and QC215. Genotype 6c variant QC334, identified after June 2006, was used in the reconstruction of the genotype 6 phylogeny. The following sequences were also used for constructing trees shown in Fig. 1: HCV-1 (GenBank accession number M62321), HCV-BK (accession number M58335), HC-G9 (accession number D14853), NL29/HC1-N15 (accession number L38377), NL35/HC1-N16 (accession number L38378), CAM1078 (accession number L38361), FR2 (accession number L38371), 2152 (accession number AF271798), FRSSD98 (accession number AJ291279), FR16 (accession number L48495), FrSSD49 (accession number AJ291257), 98CM1427 (accession number AY257091), 98CM1383 (accession number AY257083), 98CM1565 (accession number AY257094), 98CM1521 (accession number AY257087), M5186N (accession number AY265431), HCV-J6 (accession number D00944), HCV-J8 (accession number D10988), BEBE1 (accession number D50409), NE92 (accession number L29634), JK020 (accession number D49760), NL50 (accession number L44602), JK081 (accession number D49769), NL33 (accession number L44601), FR13 (accession number L48492), VAT96 (accession number AB031663), BA045 (accession number D86529), BA047 (accession number D86530), FR15 (accession number L48494), FR4 (accession number L38373), accession number M5230N (accession number AY265442), MRS40 (accession number AF516029), P11.Don (accession number AF515978), Mart44 (accession number AY257462), 98CM1414 (accession number AY257088), A13160124C (accession number AY265422), M7881N (accession number AY685040), 98CM1581 (accession number AY257076), 98CM1453 (accession number AY257084), M5401N (accession number AY265443), M7649N (accession number AY685033), A07010128C (accession number AY265420), A07030139C (accession number AY265426), 98CM9854 (accession number AY257079), M4959N (accession number AY265437), M7841N (accession number AY685037), M5480N (accession number AY265444), 98CM9919 (accession number AY257080), M4831N (accession number AY265436), M7930N (accession number AY685043), NZL1 (accession number D17763), HCV-Tr (accession number D49374), NE048 (accession number D16612), NE274 (accession number D16620), NE145 (accession number D16618), NE125 (accession number D16614), IND1751 (accession number X91307), JK049 (accession number D63821), SOM1 (accession number AF216792/AF216786), ED43 (accession number Y11604), FRSSD120 (accession number AJ401097), Z1 (accession number L16677), GB358 (accession number L29606), D13 (accession number L16656), CAM600 (accession number L29589), GB809 (accession number L29629), CAMG22 (accession number L29595), GB549 (accession number L29620), GB538 (accession number L29606), CAR4/1205 (accession number L36439), B14 (accession number L39282), 2116 (accession number AF271881), 1797 (accession number AF271876), FRSSD158 (accession number AJ401099), 1359 (accession number AF271874), 2153 (accession number AF271882), EUHK2 (accession number Y12083), Th580 (accession number D84262), VN998 (accession number D30797), VN235 (accession number D84263), Th271 (accession number D37858), VN4 (accession number L38382), Th846 (accession number D37857), VN12 (accession number L38380), VN507 (accession number D87357), VN405 (accession number D84264), VN004 (accession number D84265), VN085 (accession number D87353), Th555 (accession number D37863), Th553 (accession number D37862), JK046 (accession number D63822), MYAN-6H (accession number AB103151), and MYAN-2D (accession number AB103140).
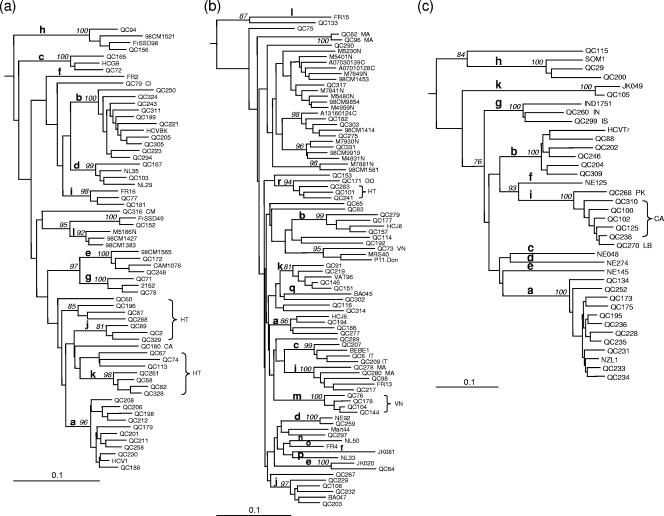
FIG. 1.
Neighbor-joining trees for NS5B sequences (340 bp) (positions 8276 to 8615) and C/E1 sequences (424 bp) (positions 869 to 1292). Phylogenies for genotype 1 NS5B (a), genotype 2 NS5B (b), genotype 3 C/E1 (c), genotype 4 C/E1 (d), and genotype 6 NS5B (e) sequences are shown. The phylogenetic tree for genotype 1 sequences was rooted using genotype 4a sequence ED43 as the outgroup, while the phylogenetic trees for sequences of genotypes 2, 3, 4, and 6 were rooted using genotype 1a sequence HCV-1 as the outgroup. The numbers at the nodes represent the percent bootstrap support for 1,000 replicates. Only values over 75% are shown. Bars at the base of the trees show the genetic divergence. Letters following isolate names indicate countries of origin of patients: BI, Burundi; CA, Canada; CD, Democratic Republic of Congo; CG, Congo; CI, Côte d'Ivoire; CM, Cameroon; DO, Dominican Republic; HT, Haiti; IN, India; IS, Indian subcontinent; IT, Italy; KH, Cambodia; LA, Laos; LB, Lebanon; MA, Morocco; PA, Pakistan; RW, Rwanda; VN, Vietnam.
Statistical analysis.
The proportion of males and females and mean age differences between groups were analyzed by the chi-square test and a Student's unpaired t test, respectively. P values of 0.01 were considered to be significant.
Variants.
Variants were named QC for their identification in the Canadian province of Quebec.
Nucleotide sequence accession numbers.
Sequences reported in this study have been submitted to GenBank and can be retrieved under accession numbers AY434105 to AY434158, AY706995 to AY707000, AY754610 to AY754639, AY894524 to AY894555, and EF115544 to EF116201.
RESULTS
HCV genome amplification.
Samples from 8,479 patients were included in this study. Primer pair DM100/DM101 (Table 1) proved to be efficient for amplifying genotypes 1 to 5. However, this primer pair failed to amplify genotype 6 sequences in approximately 50% of cases. A second primer pair, DM106/DM107, proved to be more efficient than primer pair DM100/DM101 in amplifying genotype 6 sequences. Consequently, samples from patients of Southeast Asian origin were simultaneously but separately amplified using both primer pairs. Still, approximately 5% of genotype 6 sequences failed to generate amplicons. These samples were further analyzed using a third primer set, DM104/DM105, which in general proved to be successful. Using this approach, 8,252 (97.3%) samples could be amplified and sequenced in NS5B.
All but 3 of the 227 samples that failed to amplify in NS5B could be amplified and sequenced in the 5′UTR using primer pair DM50/DM51. These three samples likely contained a PCR inhibitor, as they could be successfully amplified and sequenced following a 10-fold dilution in HCV RNA-negative plasma. All three were of genotype 1a. Finally, a subset of variants including those failing to classify within existing genotypes and subtypes were amplified and sequenced in the C and E1 regions and the 5′UTR using primer pair DM108/DM109 or DM110/DM111 and primer pair DM50/DM51, respectively (Table 1).
HCV genotypes derived from NS5B.
Among the 8,252 patients for whom NS5B sequences were obtained, 15 were shown to be infected by more than one genotype/subtype (see below). In the remaining 8,237 patients, genotypes 1 to 6 occurred in 59.4, 9.0, 25.7, 3.6, 0.6, and 1.8% of the patients, respectively (Table 2). Two patients were infected with an HCV variant that failed to classify within the existing six genotypes (see below). Nine novel subtypes were identified: 1j, 1k, 2m, 2r, 3i, 4q, 6q, 6r, and 6s. Sixty-four (0.8%) samples classifying within genotypes 1 to 6 could not be assigned to a specific subtype (Table 2).
TABLE 2.
Prevalence of HCV genotypes for 8,237 patients determined by sequence analysis of the NS5B region
| Subtype | Prevalence for genotype:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1
|
2
|
3
|
4
|
5
|
6
|
NGa
|
||||||||
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| Total | 4,889 | 59.4 | 741 | 9.00 | 2113 | 25.7 | 293 | 3.56 | 53 | 0.64 | 146 | 1.77 | 2 | 0.02 |
| a | 3,748 | 76.7 | 236 | 31.8 | 2095 | 99.1 | 115 | 39.2 | 53 | 100 | 30 | 20.5 | ||
| b | 1,105 | 22.6 | 353 | 47.6 | 7 | 0.33 | 1 | 0.34 | 0 | |||||
| c | 2 | 0.04 | 81 | 10.9 | 0 | 10 | 3.41 | 0 | ||||||
| d | 6 | 0.12 | 1 | 0.13 | 0 | 34 | 11.6 | 0 | ||||||
| e | 2 | 0.04 | 1 | 0.13 | 0 | 1 | 0.34 | 59 | 40.4 | |||||
| f | 0 | 0 | 0 | 2 | 0.68 | 1 | 0.68 | |||||||
| g | 5 | 0.10 | 0 | 2 | 0.09 | 1 | 0.34 | 0 | ||||||
| h | 2 | 0.04 | 0 | 1 | 0.05 | 6 | 2.05 | 2 | 1.37 | |||||
| i | 2 | 0.04 | 30 | 4.05 | 7 | 0.28 | 0 | 0 | ||||||
| j | 2 | 0.04 | 4 | 0.54 | 0 | 0 | ||||||||
| k | 4 | 0.08 | 6 | 0.81 | 1 | 0.05 | 45 | 15.4 | 0 | |||||
| l | 0 | 0 | 13 | 4.44 | 2 | 1.37 | ||||||||
| m | 4 | 0.54 | 5 | 1.71 | 0 | |||||||||
| n | 0 | 4 | 1.37 | 0 | ||||||||||
| o | 0 | 4 | 1.37 | 10 | 6.85 | |||||||||
| p | 0 | 1 | 0.34 | 7 | 4.79 | |||||||||
| q | 0 | 6 | 2.05 | 6 | 4.11 | |||||||||
| r | 4 | 0.54 | 30 | 10.2 | 8 | 5.48 | ||||||||
| s | 3 | 2.05 | ||||||||||||
| t | 2 | 0.68 | ||||||||||||
| UCb | 11 | 0.22 | 21 | 2.83 | 1 | 0.05 | 13 | 4.44 | 18 | 12.3 | ||||
NG, novel genotype.
UC, unclassified.
Genotype 1 variants.
Genotype 1a was the most prevalent, occurring in 45.5% of all patients (Table 2). Subtypes 1a and 1b accounted for 99.3% of genotype 1 isolates. Subtype 1b-infected patients showed an underrepresentation of males and were older than subtype 1a-infected patients (P < 0.001) (Table 3). Variants belonging to subtypes 1c to 1e, and 1g to 1k were also identified (Fig. 1a). Patients infected with newly recognized subtypes 1j and 1k originated from Haiti (43). 1j and 1k variants did not display specific 5′UTR sequence motifs that could distinguish them from subtype 1a or 1b. Eleven variants did not group with previously recognized subtypes. QC196, QC87, and QC288 were related to each other, grouping on a separate branch, and were found in patients of Haitian origin. This cluster was supported by a high bootstrap value in both NS5B and C/E1 (tree not shown). However, in C/E1, the genetic distance between these three isolates was higher than that observed between isolates of other subtypes (data not shown). QC67, QC74, and QC113 were also related to each other, clustering on a separate branch, and were also found in patients originating from Haiti. This cluster was supported by a high bootstrap value in C/E1 only. Finally, QC60, QC79, QC180, and QC316 were distinct from each other and from other previously described strains, while QC152 grouped with FrSSD49 (26). QC60, QC79, QC180, and QC316 were found in patients originating from Haiti, Côte d'Ivoire, Canada, and Cameroon, respectively. The NS5B tree topology was found to be analogous in C/E1.
TABLE 3.
Sex ratio and mean age values of patients among the various HCV genotypes
| Genotype | No. of patients | Sex ratioa | Age (yr) (mean ± SD)
|
||
|---|---|---|---|---|---|
| All patients | Men | Women | |||
| All | 8,237 | 2.2 | 44.3 ± 11.4 | 44.0 ± 10.3 | 45.0 ± 13.7 |
| 1 all | 4,889 | 2.4 | 44.3 ± 11.0 | 44.1 ± 9.9 | 44.8 ± 13.4 |
| 1a | 3,748 | 2.7 | 42.4 ± 9.5 | 42.9 ± 8.8 | 41.1 ± 11.2 |
| 1b | 1,105 | 1.6 | 50.7 ± 13.0 | 49.2 ± 12.1 | 53.0 ± 14.1 |
| 2 all | 741 | 1.5 | 50.4 ± 13.2 | 48.6 ± 11.9 | 53.2 ± 14.4 |
| 2a | 236 | 1.1 | 52.8 ± 13.1 | 49.0 ± 11.4 | 57.0 ± 13.6 |
| 2b | 353 | 2.4 | 44.5 ± 10.4 | 44.3 ± 9.4 | 44.8 ± 12.4 |
| 2c | 81 | 1.1 | 62.4 ± 10.6 | 63.4 ± 9.5 | 61.3 ± 11.7 |
| 2i | 30 | 1.5 | 60.3 ± 13.1 | 58.4 ± 11.7 | 63.1 ± 14.4 |
| 3 all | 2,113 | 2.7 | 40.6 ± 9.5 | 41.3 ± 9.1 | 38.7 ± 10.3 |
| 3a | 2,095 | 2.7 | 40.5 ± 9.5 | 41.2 ± 9.1 | 38.7 ± 10.3 |
| 4 all | 293 | 1.3 | 49.2 ± 12.5 | 48.4 ± 11.8 | 50.2 ± 13.2 |
| 4a | 115 | 2.8 | 50.8 ± 12.9 | 49.1 ± 12.1 | 55.6 ± 14.1 |
| 4d | 34 | 1.6 | 42.5 ± 8.9 | 42.8 ± 8.8 | 42.1 ± 9.1 |
| 4k | 45 | 1.0 | 49.8 ± 13.7 | 50.2 ± 12.0 | 49.4 ± 15.1 |
| 4r | 30 | 0.5 | 50.6 ± 10.3 | 52.1 ± 12.0 | 49.9 ± 9.3 |
| 5 all (5a) | 53 | 0.7 | 60.4 ± 13.7 | 60.8 ± 16.0 | 60.1 ± 11.9 |
| 6 all | 146 | 1.5 | 51.9 ± 10.8 | 51.1 ± 10.1 | 53.1 ± 11.8 |
| 6a | 30 | 2.0 | 47.3 ± 12.3 | 46.9 ± 12.1 | 48.1 ± 12.5 |
| 6e | 59 | 1.1 | 53.3 ± 10.3 | 50.5 ± 9.4 | 56.5 ± 10.3 |
Male-to-female ratio.
Genotype 2 variants.
The most prevalent subtypes of genotype 2 were 2b (47.6%), 2a (31.8%), 2c (10.9%), and 2i (4.1%). Subtype 2b-infected patients showed an overrepresentation of males (P < 0.001) and were younger (P < 0.001) than other genotype 2-infected patients (Table 3). Variants belonging to subtypes 2d, 2e, 2j, 2k, and 2m were also identified (Fig. 1b). Patients infected by newly recognized subtype 2m were of Vietnamese origin (43). Subtype 2m variants did not display specific 5′UTR sequence motifs. Twenty-one variants were not assigned to a subtype. QC65, QC75, QC83, QC114, QC116, QC153, QC192, QC267, QC289, QC290, QC297, QC302, QC314, and QC317 were distinct from each other and from previously described variants. QC62 and QC96, both obtained from patients originating from Morocco, clustered together and showed a 12-nucleotide insertion within E1. QC73 was obtained from a patient of Vietnamese origin and clustered with MRS40 and P11.Don (5). QC182, QC275, and QC303 clustered with 98CM1414 and A13160124C, both collected in Cameroon. These isolates clustered closely to another group comprised of isolates 98CM9854, M4831N, and M7930N, also sampled in Cameroon (28, 30, 31), and of isolate QC331, obtained from a Canadian with an unknown risk factor. Since the relationship between both groups did not display strong bootstrap support, these isolates were not assigned a novel subtype. Finally, QC171, QC283, QC101, and QC241 clustered together, with strong bootstrap support in both NS5B and C/E1 (tree not shown). The latter three isolates were derived from patients born in Haiti, while QC171 was derived from a patient born in the Dominican Republic. These isolates were provisionally assigned to subtype 2r. QC153, although closely related to subtype 2r isolates, was not at the moment classified as such, since its relationship in NS5B was not supported by a high bootstrap value. Subtype 2r isolates and QC153 displayed a C polymorphism at position 204 in the 5′UTR not frequently observed in other genotype 2 variants. QC153 also displayed a three-nucleotide insertion within E1. The NS5B tree topology was found to be reproduced in C/E1.
Genotype 3 variants.
Subtype 3a was the second most prevalent genotype, occurring in 25.4% of all patients, and accounted for 99.1% of genotype 3 isolates (Table 2). The remaining genotype 3 isolates were comprised of subtypes 3b, 3g to 3i, and 3k and of variant QC115. A phylogenetic tree of C/E1 sequences is presented in Fig. 1c and shows the relationship between subtype 3g isolate IND1751 and isolates QC260 and QC299. QC260 and QC299 were obtained from patients originating from the Indian subcontinent. Seven subtype 3i variants were identified, five of which (QC100, QC102, QC125, QC238, and QC310) showed a 12-nucleotide insertion within E1, while the remaining two (QC268 and QC270) lacked this insertion. QC268 and QC270 were obtained from patients originating from Pakistan and Lebanon, while isolates displaying the 12-nucleotide insertion were obtained from patients of Canadian origin. The 5′UTR sequences of the seven subtype 3i isolates were identical except for QC238, which showed a one-nucleotide difference, and were most closely related to that of subtype 3f isolate NE125. The C/E1 tree topology was found to be reproduced in NS5B.
Genotype 4 variants.
The most prevalent subtypes of genotype 4 were 4a (39.2%), 4k (15.4%), 4d (11.6%), 4r (10.2%), and 4l (4.4%). Subtype 4a patients showed an overrepresentation of males (P < 0.001), while subtype 4r patients showed an overrepresentation of females (P = 0.01), in comparison to other subtype 4 patients (Table 3). Patients infected by subtype 4r were mostly of Central African origin. Patients infected by subtype 4d were younger than patients infected with other subtypes of genotype 4 (P < 0.001). Thirty-three, 58, and 38% of patients infected with subtypes 4a, 4d, and 4l, respectively, were of Canadian origin. Patients infected with subtypes 4b, 4c, 4e to 4h, 4m to 4q, and 4t were also identified. A phylogenetic tree of genotype 4 C/E1 is presented in Fig. 1d and shows the close relationship between subtype 4b isolate Z1 and QC264. QC264 was obtained from a patient originating from Congo. Thirteen variants could not be assigned to a previously recognized subtype. QC124, QC127, QC132, QC147, QC253, QC256, and QC282 grouped independently from each other and were distinct from previously described strains. QC108 and QC109, isolated from patients originating from Congo, clustered together and were most closely related in NS5B to G4MP166, obtained from a Cameroonian patient living in France (data not shown) (29). Finally, QC126, QC170, QC218, and QC254 formed an independent cluster and were most closely related to newly recognized subtype 4q (43). The average genetic distance observed in C/E1 between isolates of subtype 4q and of this cluster fell short compared to that observed between 4a and 4c sequences (Fig. 1d). Consequently, it was considered premature to assign this cluster to a new subtype. These variants and those of subtype 4q were obtained from patients originating from Rwanda or Burundi and did not display specific 5′UTR sequence motifs. The C/E1 tree topology was found to be analogous in NS5B.
Genotype 5 variants.
Genotype 5a was the least prevalent genotype, occurring in only 0.6% of patients. Patients with genotype 5a showed an overrepresentation of females (P < 0.001) and were significantly older (P < 0.001) than patients infected by other genotypes (Table 3). Clustering of strains by sex was not observed (data not shown).
Genotype 6 variants.
The most prevalent subtype of genotype 6 was 6e (40.4%), followed by 6a (20.5%) (Table 2). Although subtype 6e-infected patients had a lower male-to-female ratio and a higher mean age than subtype 6a-infected patients, this did not reach statistical significance. Variants belonging to subtypes 6f, 6h, 6l, and 6o to 6q were also identified (Fig. 1e). Genotype 6 had the highest proportion (12.3%) of variants that did not explicitly classify within existing subtypes. QC81, QC112, QC148, QC150, QC191, QC197, QC251, QC266, QC269, QC271, QC273, and QC323 classified independently from each other and were distinct from other strains. QC226 and QC327, obtained from patients of Vietnamese origin, clustered together and were distantly related to subtype 6k isolate VN405. QC255 in C/E1 grouped distantly from other subtype 6h strains and for this reason was not presently assigned to this subtype (data not shown). QC255 was isolated from a patient originating from Laos. Two independent clusters, the first composed of eight variants (QC117, QC120, QC174, QC245, QC247, QC291, QC295, and QC330) and the second composed of three variants (QC66, QC225, and QC292), were assigned subtypes 6r and 6s, respectively (Fig. 1e). Patients infected with subtypes 6s, 6r, and the newly recognized 6q (43) were of Cambodian origin. Finally, a cluster composed of QC131, QC145, and QC240, isolated from patients of Vietnamese origin, grouped independently from other genotype 6 variants. In C/E1, QC240 was found to be more distantly related to QC131 and QC145 than in NS5B (data not shown). Consequently, they were not assigned to a novel subtype. All but three of the genotype 6-infected patients were of Asian origin. The NS5B tree topology was found to be reproduced in C/E1.
Variants not classifying within genotypes 1 to 6.
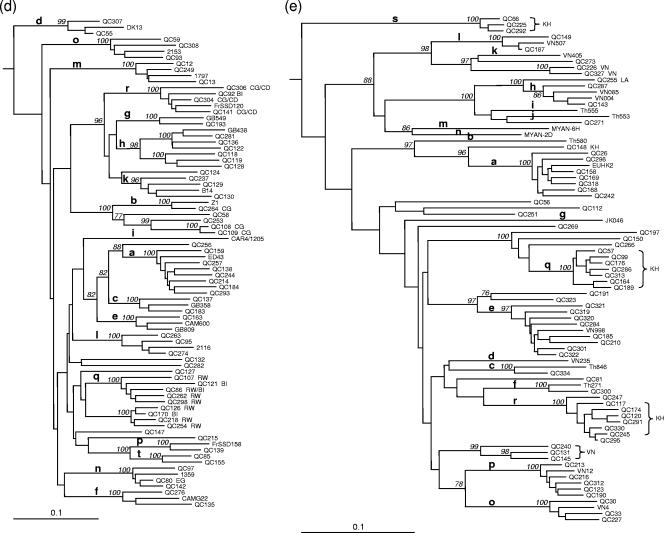
Two variants did not classify within the six genotypes in both NS5B (Fig. 2) and C/E1 (tree not shown). QC69 was obtained from a 48-year-old male, and QC272 was obtained from a 15-year-old female, both emigrants of the Democratic Republic of Congo. QC69 was found to be closely related to CS101285 and CS101300, while QC272 was not closely related to other isolates of HCV. QC272 contained a unique six-nucleotide insertion within E1. The 5′UTR sequences of both QC69 and QC272 were unique but displayed sequence polymorphisms present among genotype 2 sequences (Fig. 3a).
FIG. 2.
Unrooted neighbor-joining tree of NS5B sequences (340 bp) (positions 8276 to 8615) showing the relationship of QC69 and QC272 to representatives of genotypes 1 to 6, CS101285, and CS101300 sequences. The bar at the base of the tree shows the genetic divergence.
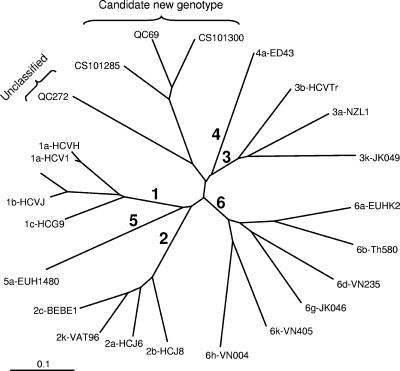
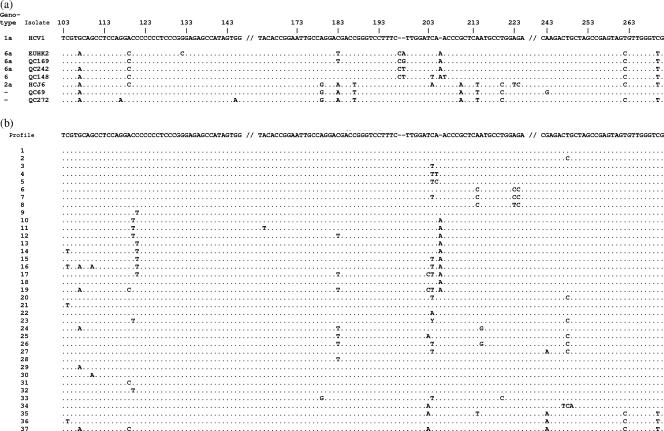
FIG. 3.
Comparison of sequences in the 5′UTR region (196 bp) (positions 96 to 291). (a) Comparison of QC69, QC272, genotype 6a QC169 and QC242, and genotype 6 QC148 sequences to HCJ6 (genotype 2a) and EUHK2 (genotype 6a) sequences. Nucleotides different from those of genotype 1a HCV-1 are indicated. (b) Thirty-seven genotype 6 sequence profiles obtained for 100 non-6a, non-6b variants. Nucleotides different from those of genotype 1b HCV-BK are indicated.
Infections with multiple genotypes.
The presence of numerous mixed bases interspaced on the electropherograms was suggestive of infection with two genotypes. Blood samples were repeatedly tested to confirm this observation. Sequences specific to each genotype were determined by direct sequencing of PCR products using genotype-specific primers (Table 1). Coinfections by two genotypes/subtypes were confirmed in 15 patients: 5 patients were coinfected with subtypes 1a and 1b, 5 were coinfected with subtypes 1a and 3a, 2 were coinfected with subtypes 1a and 2a, 2 were coinfected with subtypes 1a and 2b, and 1 was coinfected with subtypes 1b and 2a.
5′UTR sequences of genotype 6 variants.
Genotype 6a and 6b sequences show a dual nucleotide insertion between positions 197 and 198 that is not present in other genotypes. This insertion is generally attributed to residues C and A. However, we have identified one subtype 6a sequence displaying a CG (QC169) insertion and a second one displaying a CT (QC242) insertion (Fig. 3a). A third sequence, QC148, isolated from a patient of Cambodian origin, clustering closely but independently from subtypes 6a and 6b also displayed an atypical CT insertion.
Although subtypes 6a and 6b can be differentiated from genotype 1 on the basis of a dual nucleotide insertion in the 5′UTR, other genotype 6 variants show few nucleotide differences from genotype 1. The 5′UTR sequence profiles obtained for 100 variants other than those of subtypes 6a and 6b are presented in Fig. 3b and Table 4. Thirty-seven distinct nucleotide profiles were distinguishable, but no polymorphisms unique to genotype 6 could be identified. In 34 (34%) profiles, the 5′UTR sequences were similar to those of genotype 1b isolate HCV-BK (Table 4). These were comprised of 28 subtype 6e isolates and 6 variants (QC131, QC145, QC150, QC226, QC240, and QC271) not classifiable into the presently identified subtypes. Twelve sequences, comprised of 10 subtype 6e variants, QC149 (6l), and QC323 were identical to genotype 1b HCV-BK except for a C polymorphism at nucleotide position 248. Seven additional sequences, composed of four subtype 6e isolates, QC143 (6h), QC197, and QC255, showed a T polymorphism at nucleotide position 204. These sequences are not unique to genotype 6, as identical sequences are present among genotype 1b isolates. It should be pointed out that the 5′UTR sequences of newly assigned subtypes 6r and 6s shared unique polymorphisms distinct from other genotype 6 variants. Seven of the eight 6r variants displayed TT polymorphisms at positions 204 to 205, while the remaining 6r variant displayed a TC polymorphism at these positions (profiles 4 and 5) (Table 4). The 6s variants displayed a C polymorphism at position 214 and either CC or TC polymorphisms at positions 223 to 224 (profiles 6 to 8). Interestingly, three subtype 6o sequences (QC30, QC33, and QC227) and QC269 showed a single T polymorphism at position 121 (profile 9). Finally, the sequences that showed an A insertion following position 205 belonged to subtype 6c, 6p, or 6q (profiles 10 to 19).
TABLE 4.
Frequency of 5′UTR sequence profiles observed for genotype 6 subtypes other than subtypes 6a and 6b
| Profile(s) | No. with subtype:
|
Total no. of observations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6c | 6e | 6f | 6h | 6l | 6o | 6p | 6q | 6r | 6s | 6 | ||
| 1 | 28 | 6 | 34 | |||||||||
| 2 | 10 | 1 | 1 | 12 | ||||||||
| 3 | 4 | 1 | 2 | 7 | ||||||||
| 4 | 7 | 7 | ||||||||||
| 5 | 1 | 1 | ||||||||||
| 6-8 | 3 | 3 | ||||||||||
| 9 | 3 | 1 | 4 | |||||||||
| 10 | 3 | 3 | ||||||||||
| 11 and 12 | 2 | 2 | ||||||||||
| 13 | 2 | 2 | ||||||||||
| 14-18 | 5 | 5 | ||||||||||
| 19 | 1 | 1 | ||||||||||
| 20 | 1 | 1 | 2 | |||||||||
| 21-25 | 5 | 5 | ||||||||||
| 26 | 1 | 1 | ||||||||||
| 27 | 1 | 1 | ||||||||||
| 28 | 1 | 1 | ||||||||||
| 29-37 | 9 | 9 | ||||||||||
| Total no. of sequence profiles | 1 | 9 | 1 | 2 | 2 | 1 | 3 | 6 | 2 | 3 | 14 | |
DISCUSSION
We have successfully implemented nucleotide sequence analysis of the NS5B region for routine genotyping of HCV in a clinical reference laboratory. Genotypes were determined for >97% of samples, an efficiency similar to that reported previously by others (6, 7). For genotypes 1 to 5, a viral load below 4 log IU/ml was the key factor leading to the absence of amplicons, although this was not an absolute rule. Overall, approximately 3% of patient samples displayed viral loads below 4 log IU/ml. Amplification of genotype 6 sequences proved to be more challenging. We chose the NS5B region as the target since this coding region has received the most attention for the characterization of isolates worldwide. This allowed more precise genotype assignments and comparisons of isolates not presently assigned to a specific subtype. In this study, it became evident that the sequences targeted by the NS5B primers, although conserved for genotypes 1 to 5, were less conserved among genotype 6 isolates. This variability was greater for sequences targeted by the sense primer. Because of this variability, three different primer pairs were required to efficiently amplify all genotype 6 isolates. Since the NS5B region is not entirely conserved, novel HCV genotypes/subtypes can prove to be difficult to amplify due to inadequate primer sequences or a low viral load. For example, novel variants QC69 and QC272 yielded weak PCR products in NS5B. For the latter variant, this could be attributable to its low viral load (4.6 log IU/ml). Consequently, novel variants detectable only in the 5′UTR would likely go unrecognized.
Patients infected with genotypes 1a, 1b, 2a, 2b, 3a, and 5a were mostly of Canadian origin. On the other hand, subjects infected with other viral strains were generally found in emigrants. For genotype 2, subtypes 2c and 2i were found in 11% and 4% of patients, respectively. Patients infected with subtype 2c were mostly of Italian origin, while subtype 2i infections were identified in subjects originating from Morocco. Genotype 2c-infected patients displayed a mean age of 62 years. Interestingly, genotype 2c is most prevalent in Italy and in adults older than 60 years of age (1, 24). Infections by genotype 4 were accounted for by several subtypes. More than 80% of the infections were due to subtypes 4a, 4d, 4l, 4k, and 4r. A significant proportion of patients infected with subtypes 4a, 4d, and 4l were of Canadian origin, suggesting that genotype 4 has begun to spread within the local population. For genotype 6, all patients except three were of Southeast Asian origin. These three patients of Canadian origin were infected with subtypes 6a, 6e, and 6o (QC227). Associated risk factors were injection drug use in the former two and blood transfusion in the latter. In contrast to genotype 4, genotype 6 has not significantly spread within the local population.
Patients infected with genotypes 1a, 3a, 2b, and 4d were younger and showed an overrepresentation of males (except for subtype 4d) in comparison to other genotypes. In France and Italy, genotypes 1a and 3a have been associated with intravenous drug use predominantly infecting patients <45 years old (3, 41, 46). Genotype 4d has also been found among drug users in France (26). In our study, 58% of subtype 4d-infected patients were of Canadian origin. Therefore, the association of intravenous drug use in young patients suggests that this activity is likely to be a predominant risk factor for these four genotypes in our population.
Only 0.6% of patients were found to be infected with genotype 5a. This is in contrast to our previously reported frequency of 4.5% in blood donors and patients from 1992 to 1995 in Montreal (2, 27), where genotype 5a-infected patients were younger, with blood transfusion as the most frequent risk factor. A decrease in the latter means of acquired infections may explain the lower infection rate observed in the present study. This is corroborated by the difference in the mean age of genotype 5a-infected subjects between studies.
Genotyping assays based on sequencing analysis of coding region sequences have the advantage of being able to identify novel variants. The genotype consensus proposal described by Simmonds et al. (43) stipulates that NS5B and C/E1 sequences of at least three examples are required for a provisional designation of a new subtype. The provisional subtypes reported in this study comply with these guidelines. For genotype 1, two new subtypes (subtypes 1j and 1k) were identified. Subtype 1j- and 1k-infected patients were of Haitian origin. Interestingly, sequences grouping closely but independently of subtypes 1j and 1k were also obtained from patients of Haitian origin. This suggests the past introduction of a genotype 1 ancestor sequence in the Caribbean that gradually diversified over time. For genotype 2, two new subtypes (subtypes 2m and 2r) were recognized. Subtype 2m-infected patients originated from Southeast Asia, while subtype 2r-infected patients originated from the Caribbean. The finding of novel genotype 2 variants in diverse geographic areas further illustrates the ancient spread of this virus.
For genotype 3, seven variants belonging to a novel subtype were recognized upon sequencing of NS5B. Lole et al. (23) previously proposed the assignation of genotype 3i for isolates obtained from patients in India following an analysis of partial core sequences. To determine if these seven variants belonged to genotype 3i as proposed, the complete core gene sequence was determined for three of them. Comparisons of the sequences confirmed that these isolates and those described previously by Lole et al. (23) belonged to the same subtype (data not shown). Of particular interest was the finding of a 12-nucleotide insertion within E1 solely among subtype 3i-infected patients of Canadian origin. The epidemiological origin of this insertion is not known at the moment. However, in four patients, drug abuse was identified as the probable risk factor.
For genotype 4, one new subtype (subtype 4q) was recognized. In addition, an independent cluster composed of four isolates grouped closely to subtype 4q. All these patients originated from Rwanda or Burundi. The finding of additional novel sequences of genotype 4 in patients originating from Central Africa is indicative of the long-term presence of this genotype in this region. To date, one variant (Z1) of subtype 4b has been identified for which only 5′UTR, C, and E1 sequences are available (4). We have identified one variant, QC264, which showed 93% sequence homology to Z1 in C/E1. QC264 in NS5B did not group with previously identified genotype 4 sequences. This would be the first report of an NS5B-derived sequence for a genotype 4b strain.
For genotype 5, all isolates belonged to genotype 5a. The geographic origin of genotype 5a remains to be resolved (49). Since genotype 5 is comprised of a sole subtype, its dissemination is likely to be a more recent event. For genotype 6, three new subtypes (subtypes 6q, 6r, and 6s) were recognized in patients originating from Cambodia. The finding of additional subtypes indicates that genotype 6 has spread widely throughout Southeast Asia. To date, only one variant (Th846) corresponding to subtype 6c has been reported (48). In our genotype 6 phylogenetic reconstruction, we report a second subtype 6c variant identified in a patient of Southeast Asian origin following our study period.
In this study, two samples did not classify within the six genotypes. QC69 grouped with two additional isolates known to date. These three isolates would constitute a new genotype, genotype 7, of HCV. In contrast, QC272 did not group closely with any other HCV sequence presently available in the international databases. In NS5B, QC272 was found to be too distantly related to be classified within this putative genotype 7. However, in C/E1, it was found to be more closely related to QC69 than in NS5B. Additional sequence data from these novel isolates are required to accurately establish their genotype status. QC69 and QC272 were identified in emigrants from the Democratic Republic of Congo, suggesting the presence of hitherto-unrecognized HCV genotypes in Central Africa.
The number of patients identified as being infected by more than one genotype/subtype was relatively low (15/8,252). This may be related to the genotyping method employed, as sequencing analysis is a relatively insensitive method for detecting minor viral populations (21, 33). Sequencing also requires experienced staff for interpreting electropherograms indicative of an infection by more than one genotype/subtype. Hence, the level of mixed-HCV genotype infection in our population is likely to be higher than that observed.
Recently, intergenotypic (subtypes 2k/1b, 2b/1b, and 2i/6p) and intragenotypic (subtype 1b/1a) recombinants of HCV have been identified (9, 17, 18, 34). In this study, over 250 variants corresponding to common and less common genotypes have been sequenced at both the 5′ (C/E1) and the 3′ (NS5B) ends of their genomes. Phylogenetic analysis in both regions did not provide evidence for recombinant forms. However, until complete genome sequences become widely available, a recombinant nature for the less common genotypes cannot be entirely ruled out. Finally, as recombinations remain an infrequent event, routine genotyping in more than one subgenomic region for clinical use is not warranted.
The 5′UTR sequences of genotype 6 isolates belonging to subtypes other than subtypes 6a and 6b were similar to those of genotype 1. Since subtype 6a accounted for only 20% of genotype 6 isolates, 5′UTR-based genotyping assays would have significantly underestimated the true prevalence of genotype 6 in our population. The 5′UTR sequences of subtypes 6a and 6b display a two-nucleotide insertion following position 196 (25). This CA insertion is believed to be highly conserved. The finding of CG and CT insertions among subtype 6a isolates indicates that the CA insertion is not as highly conserved as previously anticipated. Further studies are required to determine the prevalence of these novel nucleotide polymorphisms.
This study showed that 5′UTR sequences within a given genotype are often shared among the different subtypes. This indicates that only the genotype (genotypes 1 to 6), and not the subtype, should be reported when determined by 5′UTR-based in-house or commercial assays. The possible exception is genotype 5, for which no subtype other than subtype 5a has been reported to date. Finally, the 5′UTR is not reliable to assess the genotype in patients originating from Southeast Asia. Genotypes from these patients should be determined in one of the coding regions, such as NS5B or C/E1.
In conclusion, sequence analysis of NS5B for genotyping of HCV provides precise genotype identification and an accurate epidemiological picture of circulating viral strains. This study showed that the high genetic diversity observed among HCV genotypes in Africa and Asia can also be observed in Western countries albeit not as frequently. Finally, the belief that a universally protective vaccine for HCV may require the inclusion of genotype-specific epitopes and that the activity of HCV antivirals may be compromised by the genotype of the infecting virus strain further indicates the importance of understanding the genetic diversity of HCV (37, 38).
Acknowledgments
We acknowledge Régis Cantin, Jasmine Chamberland, Lyne Désautels, Josée Dubuque, Andrée Falardeau, Carole Gagnon, Micheline Lortie, and Marie-France Sirois for their excellent technical assistance. We thank Patrick Gonin, Régis Cantin, and Michel Lamontagne for their assistance in running Phred, Phrap, and Gap4. We are grateful to Eric Frost, Claude Lachance, Dominique Manibal, Daniel Ménard, and Paul Plaisir for providing epidemiological data on patients and to Michel Couillard and Hugues Charest for their review of the manuscript.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Ansaldi, F., B. Bruzzone, S. Salmaso, M. C. Rota, P. Durando, R. Gasparini, and G. Icardi. 2005. Different seroprevalence and molecular epidemiology patterns of hepatitis C virus infection in Italy. J. Med. Virol. 76:327-332. [DOI] [PubMed] [Google Scholar]
- 2.Bernier, L., B. Willems, G. Delage, and D. G. Murphy. 1996. Identification of numerous hepatitis C virus genotypes in Montreal, Canada. J. Clin. Microbiol. 34:2815-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bourliere, M., J. M. Barberin, M. Rotily, V. Guagliardo, I. Portal, L. Lecomte, S. Benali, C. Boustière, H. Perrier, M. Jullien, G. Lambot, R. Loyer, O. LeBars, R. Daniel, H. Khiri, and P. Halfon. 2002. Epidemiological changes in hepatitis C virus genotypes in France: evidence in intravenous drug users. J. Viral Hepat. 9:62-70. [DOI] [PubMed] [Google Scholar]
- 4.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 5.Cantaloube, J. F., P. Biagini, H. Attoui, P. Gallian, P. de Micco, and X. de Lamballerie. 2003. Evolution of hepatitis C virus in blood donors and their respective recipients. J. Gen. Virol. 84:441-446. [DOI] [PubMed] [Google Scholar]
- 6.Cantaloube, J. F., S. Laperche, P. Gallian, F. Bouchardeau, X. de Lamballerie, and P. de Micco. 2006. Analysis of the 5′ noncoding region versus the NS5b region in genotyping hepatitis C virus isolates from blood donors in France. J. Clin. Microbiol. 44:2051-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., and K. E. Weck. 2002. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 40:3127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chinchai, T., J. Labout, S. Noppornpanth, A. Theamboonlers, B. L. Haagmans, A. D. M. E. Osterhaus, and Y. Poovorawan. 2003. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J. Virol. Methods 109:195-201. [DOI] [PubMed] [Google Scholar]
- 9.Colina, R., D. Casane, S. Vasquez, L. Garcia-Aguirre, A. Chunga, H. Romero, B. Khan, and J. Cristina. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J. Gen. Virol. 85:31-37. [DOI] [PubMed] [Google Scholar]
- 10.Corbet, S., J. Bukh, A. Heinsen, and A. Fomsgaard. 2003. Hepatitis C virus subtyping by a core-envelope 1-based reverse transcriptase PCR assay with sequencing and its use in determining subtype distribution among Danish patients. J. Clin. Microbiol. 41:1091-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 12.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 14.Germer, J. J., P. N. Rys, J. N. Thorvilson, and D. H. Persing. 1999. Determination of hepatitis C virus genotype by direct sequence analysis of products generated with the Amplicor HCV test. J. Clin. Microbiol. 37:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 16.Huang, G. M. 1999. High-throughput DNA sequencing: a genomic data manufacturing process. DNA Seq. 10:149-153. [DOI] [PubMed] [Google Scholar]
- 17.Kageyama, S., D. M. Agdamag, E. T. Alesna, P. S. Leano, A. M. L. Heredia, I. P. Abellanosa-Tac-An, L. D. Jereza, T. Tanimoto, J. I. Yamamura, and H. Ichimura. 2006. A natural inter-genotypic (2b/1b) recombinant of hepatitis C virus in the Philippines. J. Med. Virol. 78:1423-1428. [DOI] [PubMed] [Google Scholar]
- 18.Kalinina, O., H. Norder, S. Mukomolov, and L. O. Magnius. 2002. A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg. J. Virol. 76:4034-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos HCV Sequence Database. Bioinformatics 21:379-384. [DOI] [PubMed] [Google Scholar]
- 20.Kuiken, C., M. Mizokami, G. Deleage, K. Yusin, F. Penin, T. Shin-I, C. Charavay, N. Tao, D. Crisan, D. Grando, A. Dalwani, C. Geourjon, A. Agrawal, and C. Combet. 2006. Hepatitis C databases, principles and utility to researchers. Hepatology 43:1157-1165. [DOI] [PubMed] [Google Scholar]
- 21.Laperche, S., F. Lunel, J. Izopet, S. Alain, P. Deny, G. Duverlie, C. Gaudy, J. M. Pawlotsky, J. C. Plantier, B. Pozzetto, V. Thibault, F. Tosetti, and J. J. Lefrère. 2005. Comparison of hepatitis C virus NS5b and 5′ noncoding gene sequencing methods in a multicenter study. J. Clin. Microbiol. 43:733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., and C. M. Rice. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436:933-938. [DOI] [PubMed] [Google Scholar]
- 23.Lole, K. S., J. A. Jha, S. P. Shrotri, B. N. Tandon, V. G. M. Prasad, and V. A. Arankalle. 2003. Comparison of hepatitis C virus by 5′ noncoding region- and core-based reverse transcriptase PCR assay with sequencing and use of the assay for determining subtype distribution in India. J. Clin. Microbiol. 41:5240-5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maggi, F., M. L. Vatteroni, C. Fornai, A. Morrica, M. Giorgi, M. Bendinelli, and M. Pistello. 1997. Subtype 2c of hepatitis C virus is highly prevalent in Italy and is heterogeneous in NS5A region. J. Clin. Microbiol. 35:161-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mellor, J., E. A. Walsh, L. E. Prescott, L. M. Jarvis, F. Davidson, P. L. Yap, and P. Simmonds. 1996. Survey of type 6 group variants of hepatitis C virus in southeast Asia by using a core-based genotyping assay. J. Clin. Microbiol. 34:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morice, Y., D. Roulot, V. Grando, J. Stirnemann, E. Gault, V. Jeantils, M. Bentata, B. Jarousse, O. Lotholary, C. Pallier, and P. Deny. 2001. Phylogenetic analyses confirm the high prevalence of hepatitis C virus (HCV) type 4 in the Seine-Saint-Denis district (France) and indicate seven different HCV-4 subtypes linked to two different epidemiological patterns. J. Gen. Virol. 82:1001-1012. [DOI] [PubMed] [Google Scholar]
- 27.Murphy, D. G., B. Willems, J. Vincelette, L. Bernier, J. Côté, and G. Delage. 1996. Biological and clinicopathological features associated with hepatitis C virus type 5 infections. J. Hepatol. 24:109-113. [DOI] [PubMed] [Google Scholar]
- 28.Ndjomou, J., O. G. Pybus, and B. Matz. 2003. Phylogenetic analysis of hepatitis C virus isolates indicates a unique pattern of endemic infection in Cameroon. J. Gen. Virol. 84:2333-2341. [DOI] [PubMed] [Google Scholar]
- 29.Nicot, F., F. Legrand-Abravanel, K. Sandres-Saune, A. Boulestin, M. Dubois, L. Alric, J. P. Vinel, C. Pasquier, and J. Izopet. 2005. Heterogeneity of hepatitis C virus genotype 4 strains circulating in south-western France. J. Gen. Virol. 86:107-114. [DOI] [PubMed] [Google Scholar]
- 30.Njouom, R., C. Pasquier, A. Ayouba, A. Gessain, A. Froment, J. Mfoupouendoun, R. Pouillot, M. Dubois, K. Sandres-Saune, J. Thonnon, J. Izopet, and E. Nerrienet. 2003. High rate of hepatitis C virus infection and predominance of genotype 4 among elderly inhabitants of a remote village of the rain forest of South Cameroon. J. Med. Virol. 71:219-225. [DOI] [PubMed] [Google Scholar]
- 31.Njouom, R., C. Pasquier, A. Ayouba, M. C. Tejiokem, A. Vessiere, J. Mfoupouendoun, G. Tene, N. Eteki, M. M. Lobe, J. Izopet, and E. Nerrienet. 2005. Low risk of mother-to-child transmission of hepatitis C virus in Yaounde, Cameroon: the ANRS 1262 study. Am. J. Trop. Med. Hyg. 73:460-466. [PubMed] [Google Scholar]
- 32.Nolte, F. S. 2001. Hepatitis C virus genotyping: clinical implications and methods. Mol. Diagn. 6:265-277. [DOI] [PubMed] [Google Scholar]
- 33.Nolte, F. S., A. M. Green, K. R. Fiebelkorn, A. M. Caliendo, C. Sturchio, A. Grunwald, and M. Healy. 2003. Clinical evaluation of two methods for genotyping hepatitis C virus based on analysis of the 5′ noncoding region. J. Clin. Microbiol. 41:1558-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noppornpanth, S., T. X. Lien, Y. Poovorawan, S. L. Smits, A. D. M. E. Osterhaus, and B. L. Haagmans. 2006. Identification of a naturally occurring recombinant genotype 2/6 hepatitis C virus. J. Virol. 80:7569-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 36.Pawlotsky, J. M. 2002. Use and interpretation of virological tests for hepatitis C. Hepatology 36(Suppl. 1):S65-S73. [DOI] [PubMed] [Google Scholar]
- 37.Prince, A. M., B. Brotman, D. H. Lee, W. Pfahler, N. Tricoche, L. Andrus, and M. T. Shata. 2005. Protection against chronic hepatitis C virus infection after rechallenge with homologous, but not heterologous, genotypes in a chimpanzee model. J. Infect. Dis. 192:1701-1709. [DOI] [PubMed] [Google Scholar]
- 38.Reiser, M., H. Hinrichsen, Y. Benhamou, H. W. Reesink, H. Wedemeyer, C. Avendano, N. Riba, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2005. Antiviral efficacy of NS3-serine protease inhibitor BILN-2061 in patients with chronic genotype 2 and 3 hepatitis C. Hepatology 41:832-835. [DOI] [PubMed] [Google Scholar]
- 39.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 40.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 41.Silini, E., F. Bono, A. Cividini, A. Cerino, A. Maccabruni, C. Tinelli, S. Bruno, A. Bellobuono, and M. U. Mondelli. 1995. Molecular epidemiology of hepatitis C virus infection among intravenous drug users. J. Hepatol. 22:691-695. [DOI] [PubMed] [Google Scholar]
- 42.Simmonds, P., D. B. Smith, F. McOmish, P. L. Yap. J. Kolberg, M. S. Urdea, and E. C. Holmes. 1994. Identification of genotypes of hepatitis C virus by sequence comparisons in the core, E1 and NS-5 regions. J. Gen. Virol. 75:1053-1061. [DOI] [PubMed] [Google Scholar]
- 43.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, T. Shin-I, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 44.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132:115-130. [DOI] [PubMed] [Google Scholar]
- 45.Stuyver, L., A. Wyseur, W. Van Arnhem, F. Hernandez, and G. Maertens. 1996. Second-generation line probe assay for hepatitis C virus genotyping. J. Clin. Microbiol. 34:2259-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tamalet, C., P. Colson, H. Tissot-Dupont, M. Henry, C. Tourres, N. Tivoli, D. Botta, I. Ravaux, I. Poizot-Martin, and N. Yahi. 2003. Genomic and phylogenetic analysis of hepatitis C virus isolates: a survey of 535 strains circulating in southern France. J. Med. Virol. 71:391-398. [DOI] [PubMed] [Google Scholar]
- 47.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tokita, H., H. Okamoto, P. Luengrojanakul, K. Vareesangthip, T. Chainuvati, H. Iizuka, F. Tsuda, Y. Miyakawa, and M. Mayumi. 1995. Hepatitis C virus variants from Thailand classifiable into five novel genotypes in the sixth (6b), seventh (7c, 7d) and ninth (9b, 9c) major genetic groups. J. Gen. Virol. 76:2329-2335. [DOI] [PubMed] [Google Scholar]
- 49.Verbeeck, J., P. Maes, P. Lemey, O. G. Pybus, E. Wollants, E. Song, F. Nevens, J. Fevery, W. Delport, S. Van der Merwe, and M. Van Ranst. 2006. Investigating the origin and spread of hepatitis C virus genotype 5a. J. Virol. 80:4220-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeuzem, S. 2004. Heterogeneous virologic response rates to interferon-based therapy in patients with chronic hepatitis C: who responds less well? Ann. Intern. Med. 140:370-381. [DOI] [PubMed] [Google Scholar]