Abstract
Newcastle disease viruses isolated from Hong Kong live bird markets (LBMs) were not detected by a USDA-validated matrix gene real-time reverse transcription-PCR (RT-PCR) assay. Based upon phylogenetic analysis of the fusion gene, these viruses were related to lentogenic class I viruses found in U.S. LBMs and wild waterfowl. An alternative real-time RT-PCR assay which complements the matrix gene assay was developed to efficiently detect class I viruses.
Newcastle disease is a significant disease of poultry worldwide and is caused by virulent forms of avian paramyxovirus type 1, also known as Newcastle disease virus (NDV). Causing a spectrum of clinical symptoms, NDVs are categorized by virulence. Lentogens cause mild respiratory disease or subclinical infections in young or immunocompromised birds; mesogens produce moderate respiratory disease; and velogens (virulent viruses) replicate systemically, exhibiting tropism for the gastrointestinal tract and/or the nervous system (3). Currently, virulent NDVs (mesogens and velogens) are notifiable pathogens and must be reported to the World Organization for Animal Health (OIE).
NDV belongs to the genus Avulavirus (15) in the family Paramyxoviridae (12); it is a single-stranded, negative-sense, nonsegmented RNA virus of approximately 15.2 kb. From the NDV genome (3′ to the 5′ terminus), six proteins are produced: nucleocapsid (N gene), phosphoprotein (P gene), matrix (M gene), fusion (F gene), hemagglutinin-neuraminidase (HN gene), and the RNA-dependent RNA polymerase (L gene). NDVs have previously been grouped into either genotypes (5) or genetic lineages (1) under one serotype. Recent analysis of the genome size as well as sequence analysis of the F and L genes has revealed two distinct classes, class I and class II, within the NDV serotype 1 (6). Not commonly reported, class I NDVs include viruses from wild waterfowl and U.S. live bird markets (LBMs) (18). Class I NDVs are also found in domestic poultry, where at least one virulent isolate has been documented (1, 2). The class II viruses, which comprise the vast majority of the sequenced viruses and include poultry (gallinaceous birds), pet bird, and waterfowl viruses, are further categorized into genotypes I to IX. For the purpose of the discussion here, the class/genotype nomenclature will be used.
Sensitive rapid diagnostic assays for the detection of NDV exist, such as the M-gene-targeted real-time reverse transcription (RT)-PCR (M-gene assay) (20), and have been validated for use in the United States. However, recent data revealing significant heterogeneity in the genomes of these viruses suggests that some highly divergent NDVs may escape detection (6). This group, previously designated “lineage 6” by Aldous et al. in 2003 (1), has recently been renamed class I by Czeglédi et al. (6). To the authors' knowledge, no current assay consistently detects class I viruses.
Characterization of Hong Kong isolates.
The location, source, and date of collection for the Hong Kong class I isolates are listed in Table S1 in the supplemental material. The majority of the isolates (20/21) were obtained from swabs collected from healthy bird fecal samples and the environment during the 2003 to 2005 LBM surveillance performed by the Agriculture, Fisheries and Conservation Department of Hong Kong SAR (class I, n = 21 isolates; class II, n = 2 isolates). The swabs were inoculated into the allantoic cavities of 9- to 11-day old specific-pathogen-free chicken embryos and incubated at 35°C for two passages of 4 days each. Allantoic fluids from the incubated eggs were harvested either when the embryos were killed or after the completion of two passages and were then assayed for hemagglutination activity by following the OIE protocols for NDV (4); the presence of NDV was confirmed by hemagglutination inhibition assay with NDV-specific polyclonal antiserum.
Fusion gene sequencing.
The Hong Kong isolates and a class I isolate from waterfowl surveillance in Alaska [Northern Pintail/US(AK)/196/1998 (NOPI/98); GenBank accession number EF027165] were sequenced at the USDA ARS Southeast Poultry Research Laboratory and submitted to GenBank (see Table S1 in the supplemental material for the sequences of the class I and Table S2 for those of the class II viruses). Double-stranded nucleotide sequencing reactions were performed with fluorescent dideoxynucleotide terminators in an automated sequencer (ABI 3700 automated sequencer; Applied Biosystems Inc., Foster City, CA). The 5′ end of the fusion gene was sequenced by using primers targeting a conserved region 374 bp in length which included the fusion cleavage site (forward primer 4331F, 5′-GAG GTT ACC TCY ACY AAG CTR GAG A-3′; reverse primer 5090R. 5′-TCA TTA ACA AAY TGC TGC ATC TTC CCW AC-3′). Nucleotide sequence editing and analysis were conducted with the LaserGene sequence analysis software package (LaserGene, version 5.07; DNAStar, Inc., Madison, WI).
Evaluation of the M-gene assay for detection of class I NDVs and phylogenetic analysis.
Evaluation of the validated USDA M-gene assay included testing of Hong Kong class I (n = 21) and class II (n = 2) viruses, the class I Alaska virus, and 14 previously characterized class II viruses (NCBI accessions numbers AF419408, NDVFHN, NC_002617, NDVFPE, NDVNDVF, AF241537, NDU22286, AY246050, NDU22265, AY008325, NDU22274, AY562989, AY630414, and AY288994) (16, 18) by using the USDA-validated M-gene assay (20). The RNA used for testing was extracted from the samples by using the TRIzol LS reagent (Invitrogen, Carlsbad, CA), according to the manufacturer's protocol.
To investigate why the M-gene assay often failed to recognize class I NDV isolates, alignment and comparative nucleotide analysis with the ClustalW program (19), followed by manual editing with the BioEdit sequence alignment editor (8) of the 24-bp M-gene assay probe site, were performed with available class I (n = 4) and class II (n = 70) GenBank sequences (query date, 31 June 2006, n = 74; accession numbers with duplicate virus identifications were excluded). This analysis revealed substantial genomic variability between the two classes (see Table S3 in the supplemental material). All class I isolates diverged from the M-gene assay probe-site sequence by at least 25%, revealing a minimum of six mismatches along the probe site compared to the class II isolate sequences. The majority of the mismatches occurred at the 3′ end of the probe. In contrast, 93% of the class II isolates (65/70) had only one to two mismatches, with 66% (46/70) having a sequence identical to that of the M-gene assay probe.
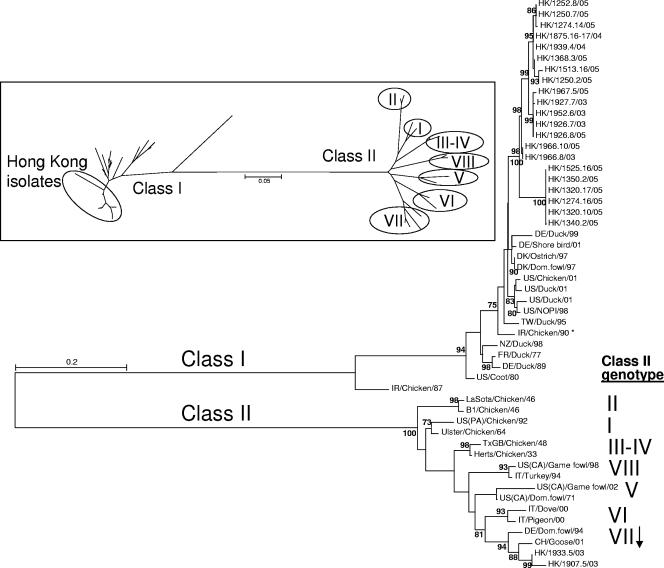
To further characterize these isolates, phylogenetic analysis of a 374-bp region of the F gene for class I (n = 36) and class II (n = 16) isolates was performed (Fig. 1). The phylogenetic tree illustrates the considerable distance between the two classes at this fusion gene region. Phylogenetic analysis included tree construction by the maximum-likelihood method as well as an unrooted neighbor-joining method, with comparable topologies obtained for each. The Hong Kong isolates clustered in four groups that were clearly separate from the other class I viruses. Among the class I viruses, Chicken/Ireland/AV2038-85 Espey/1987 (IR/CKN/87; GenBank accession number AY135759) appeared to be the most divergent isolate overall. The amino acid identities at the 125-amino-acid fusion protein fragment among the Hong Kong class I isolates ranged from 90.3 to 100% (data not shown). There were 3.3 to 13% amino acid differences between the Hong Kong isolates and other class I viruses, whereas there were 28.3 to 35.5% amino acid differences between the Hong Kong isolates and class II viruses. These data demonstrate that the class I viruses diverged markedly from class II viruses and that the Hong Kong isolates clustered separately from the other class I viruses.
FIG. 1.
Phylogenetic tree construction of 52 NDV isolates (class I, n = 36; class II n = 16) for the 374 bp corresponding to the 5′ end of the fusion gene-coding region. The maximum-likelihood tree (bootstrap values ≥70 are represented) and an unrooted neighbor-joining tree (inset; with class II genotypes denoted in Roman numerals) produced comparable topologies. The asterisk denotes the virulent virus in class I. Accession numbers are listed in Table S2 in the supplemental material.
Each of the class I Hong Kong isolates contained an amino acid fusion cleavage site consistent with a lentogenic virus (none exhibited the multiple basic amino acids at the fusion cleavage site found among virulent NDVs, and each had a leucine in place of a phenylalanine at position 117) and therefore would not be expected to cause clinical disease (see Table S2 in the supplemental material). There were three different cleavage site motifs among the class I Hong Kong isolates, with the majority (10/21) represented by GGERQERL (amino acids 110 to 117). The motifs for the class I Hong Kong isolates diverge from the classic vaccine motif of GGGRQGRL of the B1 and LaSota strains at positions 112 and 115, where glutamate has replaced glycine.
L-gene assay development for class I viruses.
Due to the sequence variability of the class I viruses at the M-gene assay probe site, other potential genomic regions for real-time RT-PCR assay development were evaluated by calculating the proportion of invariant sites to determine the degree of conservation within each of the six genes of NDVs (data not shown). The L gene exhibited the highest degree of conservation among the six genes (P [invariate] = 0.55). By using an alignment of 38 NDV L-gene sequences at the 5′ end with the Duck/US/119535/2001 sequence (Duck/01; GenBank accession number AY626266) as the reference strain, a conserved region was selected for use as the assay probe (positions 8817 to 8838 on Duck/01; see Table S4 in the supplemental material). This alignment showed a high degree of conservation among the sequenced viruses, with only one to four mismatches present between class I and II viruses. The primers and the probe were designed by using a combination of software and manual selection (PrimerExpress, ABI Prism, version 2.0) at the 5′ region of the L gene.
The selection of the L-gene assay primers and probe started with screening primers by using standard RT-PCR with a panel of nine NDV isolates from class I (n = 3) and class II (n = 6) and then proceeded to real-time PCR to test known concentrations of plasmid DNA containing a 689-bp L-gene insert from NOPI/98 (data not shown). Nuclease-free water was used as the negative control for all PCRs. For the real-time PCR and the real-time RT-PCRs in this study, cycle threshold (CT) values ≥35 were considered suspect, and a CT value of 0 indicated a negative result.
The final primer/probe set for the L-gene assay (primer L+8784, 5′CGTTCTGAGGAATTTGACAGYMT 3′; primer L−8868, 5′GRAGCCATGCGAAYTTGG 3′; and probe L+8817, 5′-6-carboxyfluorescein-CCGGCATTCTGGTTTCACTCAA-Black Hole Quencher 1-3′) was optimized based on real-time RT-PCR sensitivity and specificity by using 10-fold serial dilutions of NOPI/98 in vitro-transcribed RNA from a starting concentration of 125 pg/μl (1.86 × 108 copies/μl). The optimization included annealing temperature, primer/probe concentrations and ratios, magnesium chloride concentration, and cycle times. For the optimized L-gene assay, 25-μl reaction mixtures (17 μl master mixture added to 8 μl template) were used with the QIAGEN One-Step RT-PCR kit. The master mix contained 1 μl of kit-supplied enzyme mix (including Hot Start Taq polymerase and reverse transcriptase), 5 μl of kit-supplied buffer (5×), 13.4 pmol of the reverse primer, 26.8 pmol of the forward primer, 6 pmol of probe, 0.8 μl of kit-supplied deoxynucleoside triphosphates (final concentration, 320 μM each), 1.25 μl of 25 mM MgCl2 (combined with MgCl2 in the kit-supplied buffer; final concentration, 3.75 mM), and 13 U of RNase inhibitor (Promega). The RT step was 30 min at 50°C, followed by 15 min at 95°C. The cycling conditions were 40 cycles of 10 s of denaturation at 94°C, 30 s of annealing at 52°C, and extension at 72°C for 10 s.
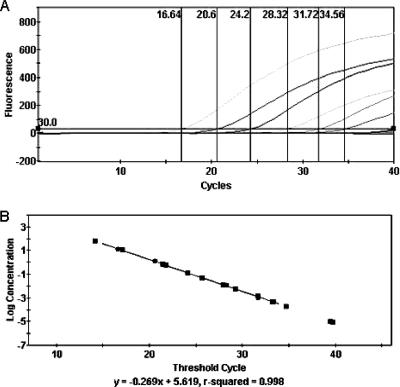
To verify the validity of the initial testing by the real-time PCR, the amplification curves obtained from the NOPI/98 L-gene plasmid DNA were compared to those obtained with the corresponding transcribed RNA by real-time RT-PCR and were found to be comparable in slope and amplitude, as determined by the Cepheid SmartCycler software, version 2.0d (equation for real-time PCR, y = −0.285x + 4.093 [R2 = 0.99]; equation for real-time RT-PCR, y = −0.286x + 6.073 [R2 = 0.99]) (11). This assay is capable of detecting 102.3 copies (<1 fg/μl) of the target gene from the in vitro-transcribed RNA of NOPI/98 (Fig. 2).
FIG. 2.
(A) Polymerase gene-targeted real-time RT-PCR with 10-fold serial dilutions of transcribed RNA from Northern Pintail/US(AK)/196/1998 (GenBank accession number EF027165). The first dilution detected was 12.5 pg/μl (107.3 copies); the last dilution detected was 0.125 fg/μl (102.3 copies). (B) Standard curve of dilutions; the line equation was calculated with Cepheid SmartCycler, version 2.0d.
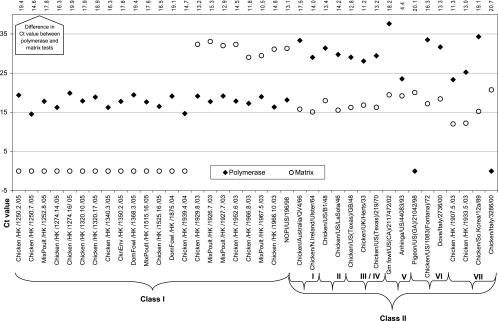
A panel of 38 isolates (n = 22 class I isolates; n = 16 class II isolates) were tested by both the L- and M-gene assays (Fig. 3). For the class I isolates, the L-gene assay detected 100% of isolates (22/22), and the M-gene assay detected 36.4% (8/22) but revealed high CT values (CTs ≥ 29). The L-gene assay proved to be more sensitive and specific for class I isolates, detecting them an average of 16.0 CT values earlier than the M-gene assay. For class II isolates, the M-gene assay detected 100% of isolates, while the L-gene assay detected 81.3% (13/16); on average, the M-gene assay detected class II isolates an average of 13.4 CT values earlier than the L-gene assay. RNA samples from Duck/01 and two other class I isolates (GenBank accession number AY626267 for the isolate from a duck and GenBank accession number AY626268 for the isolate from a chicken) were not available from the USDA National Veterinary Services Laboratory and therefore were not tested.
FIG. 3.
Comparison of the L-gene and M-gene real-time RT-PCR assays with selected isolates from both NDV class I (n = 22) and NDV class II (n = 16). CT value interpretation: lower values indicate earlier detection; values above 35 are considered suspect; and a CT value of 0 is a negative result by the assay. The difference in CT values between the L- and M-gene assays is listed at the top of the chart.
The Hong Kong viruses reported here do not appear to be phylogenetically related to the viruses in the primary vaccines used locally, such as the B1, LaSota, Clone 30, or other vaccine viruses of Australian origin, suggesting that the source of these class I viruses is not related to the shedding of vaccine viruses. However, because these viruses appear to be related to isolates from both domestic and waterfowl populations, increased surveillance for both types of populations in markets and poultry facilities followed by epidemiological analysis will be necessary to determine the origin of the class I viruses isolated in Hong Kong.
The potential for endemic waterfowl NDVs to become adapted to domestic poultry upon replication and the possibility of mutation toward virulence both highlight the need for the accurate identification of these isolates. Although there is no evidence that class I viruses have become established in domestic poultry, the previous identification of these viruses in U.S. LBMs (18) suggests that domestic poultry can be infected and can shed these viruses. In Australia, endemic lentogenic viruses circulated among domestic poultry for over 30 years, but then unknown conditions caused these viruses to undergo genomic changes that resulted in selection toward a virulent genotype (7). In Ireland, a virulent class I NDV isolate was identified in an outbreak affecting laying hens during 1990 (2). Due to our incomplete understanding of the evolutionary forces and selection pressures upon NDV, active surveillance for viruses of waterfowl origin and the development of more sensitive detection methods are the most realistic strategies for the prevention and control of outbreaks caused by endemic NDVs.
Development of a single rapid assay for the detection of all NDVs has proven to be challenging due to the genetic diversity present in the genome, with viruses of class I being among the most genetically divergent (7, 9, 10, 13, 14, 17). Here, an L-gene assay that complements the USDA-validated M-gene assay was developed to allow the rapid identification of these viruses. Phylogenetic analysis suggests that the L-gene assay would also detect class I isolates previously identified in U.S. LBMs. In addition, primers that permit the phylogenetic characterization and effective determination of the virulence potential by sequencing the genomic region encoding the fusion protein cleavage site of the Hong Kong class I NDVs were developed. These tools may prove to be useful for the future identification and characterization of other class I viruses isolated in the United States and worldwide.
Nucleotide sequence accession numbers.
Table S1 in the supplemental material contains the sequences of the class I viruses, which can be found in the GenBank database under accession numbers EF027142 to EF027155, EF027157 to EF027161, and EF027163 to EF027164, and Table S2 contains those of the class II viruses, which can be found in the GenBank database under accession numbers EF027156 and EF027162.
Supplementary Material
Acknowledgments
We gratefully acknowledge Dawn Williams-Coplin, Tim Olivier, Pamela Li, and Clara Li for technical assistance; the South Atlantic Area Sequencing Facility for nucleotide sequencing; Erica Spackman and Bruce Seal for scientific insights; and Michael Wege for providing the Northern Pintail/US(AK)/196/1998 isolate.
These studies were supported by USDA-ARS CRIS project number 6612-32000-038 and Homeland Security Funds number 6612-32000-041.
Footnotes
Published ahead of print on 7 February 2007.
Supplemental material for this article may be found at http:/jcm.asm.org.
REFERENCES
- 1.Aldous, E. W., J. K. Mynn, J. Banks, and D. J. Alexander. 2003. A molecular epidemiological study of avian paramyxovirus type 1 (Newcastle disease virus) isolates by phylogenetic analysis of a partial nucleotide sequence of the fusion protein gene. Avian Pathol. 32:239-256. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, D. J., G. Campbell, R. J. Manvell, M. S. Collins, G. Parsons, and M. S. McNulty. 1992. Characterisation of an antigenically unusual virus responsible for two outbreaks of Newcastle disease in the Republic of Ireland in 1990. Vet. Rec. 130:65-68. [DOI] [PubMed] [Google Scholar]
- 3.Alexander, D. J., and R. E. Gough. 2003. Newcastle disease, other avian paramyxoviruses, and pneumovirus infections, p. 63-92. In Y. M. Saif, H. J. Barnes, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Disease of poultry. Iowa State University Press, Ames.
- 4.Anonymous. 2006. Chapter 2.1.15. In OIE manual of diagnostic tests and vaccines for terrestrial animals. OIE World Organisation for Animal Health, Paris, France.
- 5.Ballagi-Pordany, A., E. Wehmann, J. Herczeg, S. Belak, and B. Lomniczi. 1996. Identification and grouping of Newcastle disease virus strains by restriction site analysis of a region from the F gene. Arch. Virol. 141:243-261. [DOI] [PubMed] [Google Scholar]
- 6.Czeglédi, A., D. Ujvari, E. Somogyi, E. Wehmann, O. Werner, and B. Lomniczi. 2006. Third genome size category of avian paramyxovirus serotype 1 (Newcastle disease virus) and evolutionary implications. Virus Res. 120:36-48. [DOI] [PubMed] [Google Scholar]
- 7.Gould, A. R., J. A. Kattenbelt, P. Selleck, E. Hansson, A. la-Porta, and H. A. Westbury. 2001. Virulent Newcastle disease in Australia: molecular epidemiological analysis of viruses isolated prior to and during the outbreaks of 1998-2000. Virus Res. 77:51-60. [DOI] [PubMed] [Google Scholar]
- 8.Hall, T. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 9.Herczeg, J., E. Wehmann, R. R. Bragg, P. M. Travassos Dias, G. Hadjiev, O. Werner, and B. Lomniczi. 1999. Two novel genetic groups (VIIb and VIII) responsible for recent Newcastle disease outbreaks in southern Africa, one (VIIb) of which reached southern Europe. Arch. Virol. 144:2087-2099. [DOI] [PubMed] [Google Scholar]
- 10.Ke, G. M., H. J. Liu, M. Y. Lin, J. H. Chen, S. S. Tsai, and P. C. Chang. 2001. Molecular characterization of Newcastle disease viruses isolated from recent outbreaks in Taiwan. J. Virol. Methods 97:1-11. [DOI] [PubMed] [Google Scholar]
- 11.Kim, L. M., C. L. Afonso, and D. L. Suarez. 2006. Effect of probe-site mismatches on detection of virulent Newcastle disease viruses using a fusion-gene real-time reverse transcription polymerase chain reaction test. J. Vet. Diagn. Investig. 18:519-528. [DOI] [PubMed] [Google Scholar]
- 12.Lamb, R., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. Knipe and P. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 13.Lee, Y. J., H. W. Sung, J. G. Choi, J. H. Kim, and C. S. Song. 2004. Molecular epidemiology of Newcastle disease viruses isolated in South Korea using sequencing of the fusion protein cleavage site region and phylogenetic relationships. Avian Pathol. 33:482-491. [DOI] [PubMed] [Google Scholar]
- 14.Mase, M., K. Imai, Y. Sanada, N. Sanada, N. Yuasa, T. Imada, K. Tsukamoto, and S. Yamaguchi. 2002. Phylogenetic analysis of Newcastle disease virus genotypes isolated in Japan. J. Clin. Microbiol. 40:3826-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayo, M. A. 2002. Virus taxonomy—Houston 2002. Arch. Virol. 147:1071-1076. [DOI] [PubMed] [Google Scholar]
- 16.Pedersen, J. C., D. A. Senne, P. R. Woolcock, H. Kinde, D. J. King, M. G. Wise, B. Panigrahy, and B. S. Seal. 2004. Phylogenetic relationships among virulent Newcastle disease virus isolates from the 2002-2003 outbreak in California and other recent outbreaks in North America. J. Clin. Microbiol. 42:2329-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seal, B. S., D. J. King, D. P. Locke, D. A. Senne, and M. W. Jackwood. 1998. Phylogenetic relationships among highly virulent Newcastle disease virus isolates obtained from exotic birds and poultry from 1989 to 1996. J. Clin. Microbiol. 36:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seal, B. S., M. G. Wise, J. C. Pedersen, D. A. Senne, R. Alvarez, M. S. Scott, D. J. King, Q. Yu, and D. R. Kapczynski. 2005. Genomic sequences of low-virulence avian paramyxovirus-1 (Newcastle disease virus) isolates obtained from live-bird markets in North America not related to commonly utilized commercial vaccine strains. Vet. Microbiol. 106:7-16. [DOI] [PubMed] [Google Scholar]
- 19.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wise, M. G., D. L. Suarez, B. S. Seal, J. C. Pedersen, D. A. Senne, D. J. King, D. R. Kapczynski, and E. Spackman. 2004. Development of a real-time reverse-transcription PCR for detection of Newcastle disease virus RNA in clinical samples. J. Clin. Microbiol. 42:329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.