Abstract
Cosmid and BAC contig maps have been constructed across two Fugu genomic regions containing the orthologs of human genes mapping to human chromosome 20q. Contig gene contents have been assessed by sample sequencing and comparative database analyses. Contigs are centered around two Fugu topoisomerase1 (top1) genes that were initially identified by sequence similarity to human TOP1 (20q12). Two other genes (SNAI1 and KRML) mapping to human chromosome 20 are also duplicated in Fugu. The two contigs have been mapped to separate Fugu chromosomes. Our data indicate that these linkage groups result from the duplication of an ancestral chromosome segment containing at least 40 genes that now map to the long arm of human chromosome 20. Although there is considerable conservation of synteny, gene orders are not well conserved between Fugu and human, with only very short sections of two to three adjacent genes being maintained in both organisms. Comparative analyses have allowed this duplication event to be dated before the separation of Fugu and zebrafish. Our data (which are best explained by regional duplication, followed by substantial gene loss) support the hypothesis that there have been a large number of gene and regional duplications (and corresponding gene loss) in the fish lineage, possibly resulting from a single whole genome duplication event.
[Reagents, samples, and unpublished information freely provided by D. Barnes and I.D. Hickson.]
INTRODUCTION
The pufferfish (Fugu rubripes) is now an established tool for comparative gene and genome analyses. Fugu is only a distant relative to human and has one of the smallest vertebrate genomes, but herein lies its success as a comparative analysis tool (for review, see Venkatesh et al. 2000). The genomic features that are well conserved between the two species are the ones that are fundamental to vertebrate existence—the genes and functional noncoding elements. Because of its compact size (400 Mb), and relative scarcity of repeat sequences, gene hunting and sequencing in Fugu is relatively simple (Brenner et al. 1993). Whereas the pufferfish has been used for comparative gene and short-range regional comparisons, the regions studied to date have generally been over distances covered by single BACs and cosmids.
Although the pufferfish genome is ∼7.5× smaller than the 3000-Mb human genome (Brenner et al. 1993; Elgar et al. 1999), estimates of gene numbers for Fugu are similar to those for human (Brenner et al. 1993). The extent of genomic data available for the pufferfishes and human is, however, vastly different. At the time of writing this letter, both Fugu, via a sequencing consortium (http://fugu.hgmp.mrc.ac.uk/PFW/Other/consortium.html), and the freshwater pufferfish Tetraodon nigroviridis (http://www.genoscope.cns.fr/externe/tetraodon) are the subject of genomic shotgun sequencing projects, with ∼2× genomic coverage currently available for each. Both, however, lack detailed physical and genetic maps. For human, both physical and genetic mapping data are extensive, and the complete sequence, although unfinished in places, is all but complete.
The only other fish species that has been so well studied is the zebrafish (Danio rerio). Work on zebrafish has focused on the examination of genetic mutants and pathways (for review, see Mullins et al. 1994; Driever et al. 1996) and on the creation of expressed sequence tag (EST) resources and radiation hybrid and genetic linkage maps (Gates et al. 1999; Geisler et al. 1999; Shimoda et al. 1999; Woods et al. 2000). Although the zebrafish is also being sequenced, this effort has yet to make real headway, and the types of data available for Fugu and zebrafish are therefore quite different. The short sections of assembled pufferfish sequence that are available have not been mapped to individual linkage groups. In zebrafish, long-range linkage data lacks complementary short-range analyses of gene content and orders. The types of genomic and evolutionary analyses that are possible between both fishes and human are therefore still relatively limited. The analysis presented here is an attempt to compare gene content and gene orders for regions of both fish genomes that share synteny with one region of the human genome, the long arm of chromosome 20.
Comparisons of gene content and orders in different species are helping us dissect the physical processes that have allowed the development of complex vertebrate genomes. Analyses of vertebrate gene content with species similar to those present at the base of the vertebrate lineage have, for example, indicated that there were two rounds of whole genome duplication in the early evolution of vertebrates. In particular, studies of hox gene complexes have shown that although there is one hox gene cluster in the cephalochordate amphioxus, there are four paralogous copies in human and mouse (for review, see Holland et al. 1994). Opinions do, however, differ, and recent analyses of the timing of the expansion of vertebrate gene families have also been used to argue against two rounds of simultaneous (whole genome) duplication (e.g., see Hughes et al. 2001).
Within the vertebrate lineage, analyses of different genomes can also highlight similarities and differences in genome structures. Recent analyses from zebrafish, for example, have indicated that an additional whole genome duplication may have occurred in zebrafish evolution. In zebrafish, seven hox complexes have been identified (Amores et al. 1998), and recent analyses of gene families have indicated that many other gene families also have further additional copies in the fish lineage (Wittbrot et al. 1998; Robinson-Rechavi et al. 2001).
Recent analyses in zebrafish have also identified conserved synteny groups between the zebrafish and human genomes (Barbazuk et al. 2000; Postlethwait et al. 2000). Instead of one orthologous chromosomal segment being present for each human chromosomal region, however, two paralogous chromosome segments have been commonly found in the zebrafish. In many cases, these paralogous segments each contain one copy of a single copy human gene. Knowledge of this kind is of particular importance to researchers using model organisms to dissect gene functions and structures. Functions may be shared or divided between duplicated genes, or duplicates may have adopted new roles within the organism. As a whole, the zebrafish data indicates that an additional genome duplication may have occurred in the fish lineage, but it is currently unclear whether this duplication occurred before the separation of Fugu and zebrafish. There is currently little Fugu data on gene duplication or paralogous linkage groups, although an analysis of the Fugu hox clusters has indicated that at least one has been duplicated in the Fugu lineage (Aparicio et al. 1997).
In this study, we have generated some of the longest sections of Fugu contig data available to date and used these to conduct comparative analyses between Fugu and human as well as between Fugu and zebrafish. In doing so, we have found both duplicated genes and paralogous genomic segments. The two Fugu regions studied were initially selected for their shared synteny with human 20q11.2–12, a region commonly deleted in human myeloid malignancies. When a human 20q gene probe for topoisomerase 1 (TOP1) was used, however, two Fugu genes were identified. We have focused on creating gene contigs and examining the extent of Fugu and human shared synteny around these Fugu genes. Both contigs form Fugu paralogous linkage groups, each sharing synteny with human 20q. The contigs, which span 65 identified Fugu genes in total, have allowed cross-species comparisons of shared synteny, gene content (including copy numbers), and gene orders. These two regions (containing duplicated Fugu genes) are also comparable to a pair of paralogous regions in the zebrafish, which indicates they were duplicated before the separation of the two fish lineages. They provide evidence that the two genomes have undergone similar evolutionary processes and suggest that the Fugu genome may also have been duplicated.
RESULTS
Two Fugu Contigs Sharing Synteny with Human 20q
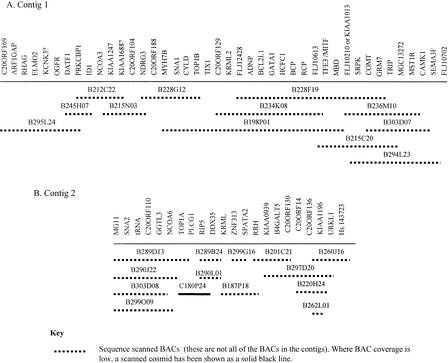
We have used BAC and cosmid clones to construct two Fugu contigs across regions sharing synteny with human 20q. Clones from both Fugu contigs have been sequence scanned to allow the examination of gene content (Fig. 1). Our experience has shown that this is generally an effective method of determining the vast majority of genes present (Elgar et al. 1999). In general, the cosmid and BAC coverage is good, although in some places only one clone was available for the sequence scanning process. Genes have been named according to the orthologous human gene unless they have been previously named in Fugu. Where official names for the human genes did not exist, these genes are referred to by the corresponding Unigene cluster. Gene orders have been determined by use of a combination of complete sequencing, sequence scanning, and sequence tagged site (STS) mapping techniques. In most cases, the sequence scanning and STS mapping techniques have allowed the definitive ordering of the identified genes, whereas the depth of the sequence scanning data has allowed the identification of the probable human orthologs of the Fugu genes present. In some cases, however, a Fugu gene has been found to have close sequence similarity to more than one human gene or a number of gene family members. In these cases, it has been difficult to determine the true human ortholog and its corresponding map location.
Figure 1.
Tiling paths for scanned BACs in the two Fugu contigs. An additional 14 cosmids have been scanned to generate data for Fugu contig 1 and a further 13 cosmids for Fugu contig 2. In total, 64 BACs and 77 BACs were used in the STS mapping of the genes on contig 1, whereas 43 BACs and 25 cosmids were used to create the contig 2 gene map.
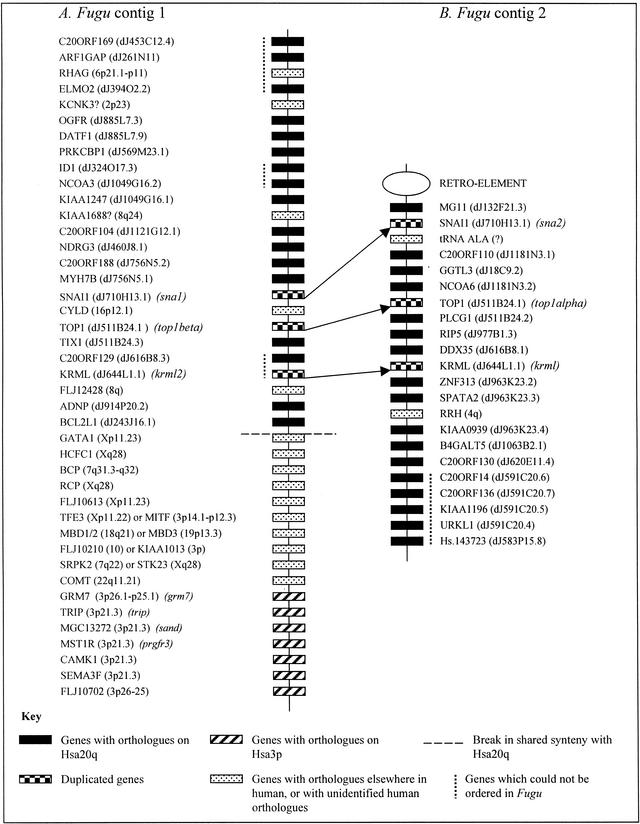
The contigs have been physically placed onto Fugu chromosomes by FISH. The larger contig, covering ∼400 kb of genomic sequence, is composed of 64 BACs and 77 cosmids of which 14 BACs (of which two were coligated) and 15 cosmids have been sequence scanned (Fig. 1A). In total, 42 genes have been identified, of which 20 have orthologs on Hsa20q (Fig. 2A). In addition to the section of this contig, which shares synteny with human 20q, this contig contains a region containing seven genes with orthologs on human 3p21 and a short section of genes with putative orthologs predominantly on human chromosomes X and 7. A number of the Fugu genes in this latter segment of DNA are members of gene families with high sequence similarity. One of the genes, for example, is a member of the methyl CpG binding domain (MBD) family. This has made it difficult to determine orthologous human genes, especially when there appears to be no unique human genome segment sharing synteny with this region of Fugu DNA. In Figure 2A, therefore, alternative human orthologs are shown for a number of genes. Elsewhere in both contigs, assigning probable orthologs has been less problematic, but when a human ortholog could not be assigned with a high degree of confidence, this is denoted with a question mark following the gene name in Figure 2.
Figure 2.
Gene content on the two Fugu contigs. Genes are named according to the nomenclature for the orthologous human gene. Human mapping locations are shown in italics after the gene name. For genes mapping to Hsa20, locations are shown as positions on bacterial clones in the Hsa20 contig map. In contig 1 there is a clear break in shared synteny with human 20q, whereas in contig 2 synteny with 20q is conserved along the entire contig length. If a gene is duplicated in Fugu, or the Fugu gene has already been named elsewhere, the corresponding Fugu gene name is shown in italics. The relative positions of the duplicated Fugu genes are indicated by arrows.
The smaller of the two contigs spans ∼250 kb and contains 43 BACs and 25 cosmids, of which 13 cosmids and 13 BACs (Fig. 1B) have been sequence scanned. Despite the presence of two genes with probable mapping locations elsewhere in the human genome, synteny with human 20q is well conserved across the entire contig (which contains 22 genes) (Fig. 2B). One end of this contig is flanked by an uncharacterized retroelement. Verifying the contig assembly across this element has proved problematic, but our additional data (not presented here) indicate that synteny with human 20q is not conserved after this point. At the other end of the contig, all of the genes present have orthologs on Hsa20q, and it is likely that shared synteny with human 20q would continue if the contig were extended further in this direction.
Given the size of the contigs, and the numbers of genes present, this equates to a gene every 10 kb. Gene distribution on the two contigs appears relatively uniform, with approximately five genes detected on each individual cosmid and no large gene voids. If this figure were extended to the whole Fugu genome, this would give a total of ∼40,000 genes in the 400-Mb sequence. This figure is comparable to current estimates of gene content in the human genome and is slightly lower than some of the previous (65,000–70,000) estimates for Fugu (Brenner et al. 1993; Elgar et al. 1999).
Duplicated Genes
Sequence analyses of two of the duplicated genes, the Fugu top1 (Fugu top1alpha and Fugu top1beta) (Smith et al. 2000) and snail genes (Fugu sna1 and Fugu sna2) (Smith et al. 2001), have already shown that these genes were duplicated in the fish lineage. In the case of the snail genes, this duplication can be placed before the separation of the Fugu and zebrafish lineages. The lack of full-length top1 gene data from other fish species has meant that we have been unable to date the top1 duplication, but degenerate PCR analysis has also shown the presence of two top1 gene fragments in zebrafish (Smith et al. 2001).
The third duplicated gene found on both Fugu contigs is a maf gene. These genes are members of the basic region/leucine zipper (b-Zip) superfamily and are transcription factors associated with the regulation of cell differentiation (Blank and Andrews 1997). Members of the maf superfamily can be further divided into the small and large maf proteins. In human, one of the large maf genes, KRML, a member of the mafb gene family, has been mapped to human 20q (Wang et al. 1999) and has been placed on PAC dJ644L1 ∼400 kb away from human TOP1 and PLCG1 in the draft human sequence. The ortholog of this gene in zebrafish (krml1/val) has been mapped to linkage group 23 on the Oregon gene map, again close to plcg1 (Moens et al. 1998; Woods et al. 2000). In zebrafish, a second homolog, krml2, has also been identified. This gene has been mapped to zebrafish linkage group 11 (Schvarzstein et al. 1999).
The coding regions of the two maf genes have been sequenced in Fugu, and phylogenetic analyses (not shown) show that the KRML gene on the Fugu top1alpha contig is the probable ortholog of zebrafish val and that these are both members of the mafb gene family. The second Fugu KRML is similar to zebrafish krml2, but neither of these genes have high sequence similarity to other members of the mafb gene family.
Chromosomal Locations of the Contigs
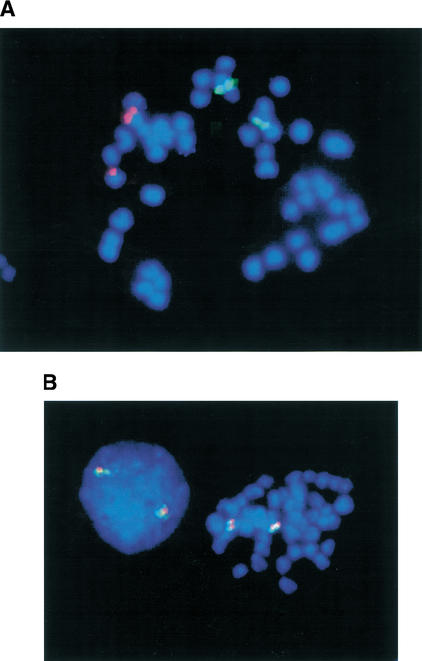
To determine whether the two paralogous Fugu regions are linked or separated in the genome, BACs from each contig were mapped onto Fugu metaphase chromosomes. BACs B299O09 (from the contig containing Fugu top1 alpha, Fig. 1A) and B294L23 (from the contig containing Fugu top1beta, Fig. 1B) were found to map to different Fugu chromosomes (Fig. 3A).
Figure 3.
(A) FISH mapping of nonsyntenic BACs B294L23 (contig 1) and B299O09 (contig 2) to Fugu metaphase chromosomes. The two Fugu contigs are therefore not linked in the Fugu genome. (B) FISH mapping of syntenic BACs B234K08 and B294L23 (both contig 1) to Fugu metaphase chromosomes. These BACs, which span the breakpoint in shared synteny between contig 1 and human 20q, show no evidence of chimerism.
The second Fugu contig has at least two apparent breakpoints in shared synteny with the human genome. To check the integrity of this contig, BAC clones were selected for FISH mapping to Fugu metaphase chromosomes. BAC B234K08 (Fig. 1A), which spans the genes KRML to TFE3, and B294L23 (Fig. 1A), which spans much of the region sharing synteny with 3p21, were both confirmed as being syntenic and nonchimeric (Fig. 3B).
Comparisons with Human 20q
On both contigs, within the segments sharing synteny with human 20q, there are a small number of genes that appear to map elsewhere in human. Given the amount of human sequence data now available for 20q, it is thought unlikely that more similar genes will be found on this chromosome. It is likely that these genes have been translocated onto this chromosome segment in the Fugu lineage or, conversely, have been moved from this region in human.
The three duplicated genes with orthologs on human 20q are colinear on the two Fugu genomic segments and in the same order (krml-top1-sna) as they are found in human. The order of these genes does not therefore seem to have been disrupted by inversions in either lineage. The orders of many of the other 20q genes are different in the Fugu and human lineages (Fig. 4), indicating that other gene orders have been disrupted by localized inversions. The dramatic differences in gene orders between the two species may be clearly seen by comparing gene orders between the two Fugu contigs and smaller sections of human 20q. A comparison of the region spanning 20q11.22–20q12, for example (Fig. 5), shows that genes present on this section of human 20q are split between the two contigs in Fugu. This indicates a pattern of gene duplication in the Fugu lineage followed (in the majority of cases) by loss of one of the two gene copies. There does not appear to be any pattern associated with the loss of the second gene copy, as the retained genes are distributed quite evenly between the two Fugu regions.
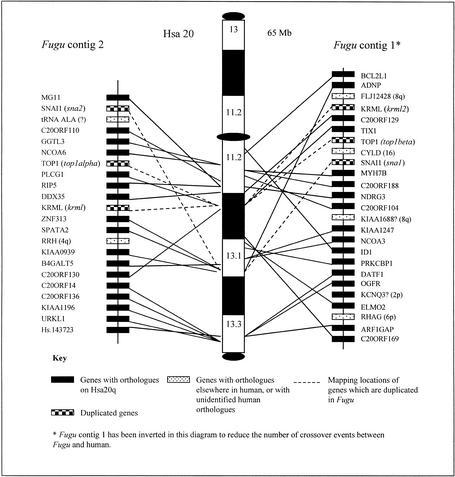
Figure 4.
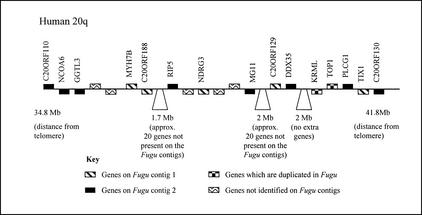
Fugu/Human 20q comparative gene maps. The positions of the human 20q orthologs of genes identified on the two Fugu contigs are shown on an ideogram of human chromosome 20. Mapping locations for the human genes span the whole of human 20q. Genes are named according to the nomenclature for the orthologous human gene. If a gene is duplicated in Fugu, the corresponding Fugu gene name is shown in italics. The positions of the Fugu and human orthologs are linked by black lines. Genes that have been identified as being present in two copies in Fugu are linked to the mapping position of the corresponding human gene by dashed lines. Although Fugu contig 1 has been inverted in this diagram to reduce the number of crossover events, the number of changes in gene orders between the Fugu contigs and human 20q remains high. This indicates that gene orders in the two lineages have been disrupted by a number of localized rearrangements.
Figure 5.
Fugu/human 20q11.2–12 comparative gene map. This ideogram provides a more detailed comparison between the two Fugu contigs and a 7-Mb section of human 20q (a section that contains the human orthologs of two of the genes, KRML and TOP1, known to be duplicated in Fugu). The orthologs of ∼25% of the genes mapping to this region are represented on one or another of the two Fugu contigs. The distribution of the genes between the two Fugu contigs, however, appears to be random, indicating a pattern of duplication in the Fugu lineage followed by the loss (in the majority of cases) of one of the additional gene copies.
When comparing the Fugu and human gene maps, it is clear that many genes mapping to human 20q are not represented on the two assembled Fugu contigs. This is characteristic of the two contigs, which contain only a subsection of the genes now known to map to human 20q. It is, however, apparent that the human orthologs of identified Fugu genes tend to be clustered in short blocks separated by longer tracts of genes for which no Fugu orthologs have been identified. This indicates that the ancestral chromosome segment that gave rise to these two Fugu contigs lacked the additional genes now seen on human 20q. It is not clear whether these genes were, however, present in the same ancestral linkage group. If this were the case, it is likely that additional genes with orthologs on human 20q could be found by further extension of these regions in Fugu.
Comparisons with Zebrafish
To determine whether these linkage groups are conserved in both Fugu and zebrafish, we have tried to identify the mapping locations of the zebrafish orthologs of the identified Fugu genes. In some cases this data was already published. In others, in silico searches of EST mapping and cluster databases could be used to find mapping locations for orthologous zebrafish genes. This information is summarized in Table 1.
Table 1.
Conserved Linkage Groups in Fugu and Zebrafish
| Human gene name | Human mapping location | Fugu contig location | Mapped zebrafish EST, or gene, name | Zebrafish linkage group |
|---|---|---|---|---|
| ARF1GAP | 20q | Contig 1 | fj34h02.y1 | 11 |
| RCP | Xq28 | Contig 1 | rdopsa | 11 |
| GATA1 | Xp11 | Contig 1 | gata1a | 11 |
| KIAA1688b | iq24 | Contig 1 | fa99h06.x1 | 11 |
| C20ORF188 | 20q | Contig 1 | taplaa | 11 |
| DATF1 | 20q | Contig 1 | Fi35b04.x1 | 11 |
| ADNP | 20q | Contig 1 | fi35b06.x1 | 11 |
| SNAI1 | 20q | Contig 2/1 | sna1/sna2a | 23/11 |
| KRML | 20q | Contig 2/1 | val/krml2a | 23/11 |
| TOP1 | 20q | Contig 2/1 | fd15f07.y1/fk31a05.y1 | 23/11 |
| PLCG1 | 20q | Contig 2 | plcg1a | 23 |
| RIP5 | 20q | Contig 2 | fi26d09.y1 | 23 |
| GGTL3 | 20q | Contig 2 | fi41a10.x1 | 23 |
| C20ORF14 | 20q | Contig 2 | toma | 23 |
Mapping locations from Woods et al. 2000 and http://zfin.org.
Most similar human gene.
The ten genes from the larger Fugu contig, which have also been mapped in zebrafish (including genes such as gata1 and rcp , which do not have orthologs mapping to human 20q), all map to zebrafish linkage group 11 (Table 1). We were unable to find zebrafish mapping locations for any of the genes from this contig that have orthologs on human 3p21. The published mapping data for zebrafish linkage group 11 do, however, indicate that this linkage group also shares synteny with Hsa3p21 (Woods et al. 2000).
The seven genes from the smaller Fugu contig that have also been mapped in zebrafish all map to zebrafish linkage group 23.
Our data therefore indicate that (at least for these regions of the genome) there have been relatively few interchromosomal translocations in the 150 million years since the divergence of Fugu and zebrafish. Detailed comparisons of the Fugu and zebrafish gene orders have not been possible because of the fact that the zebrafish genes have been separately localized onto a number of different mapping panels.
DISCUSSION
These mapping data have allowed the comparison of a large section human 20q and two chromosome segments sharing conserved synteny in Fugu. Large-scale mapping data are generally not yet available in Fugu, and previous studies have focused on comparatively mapping short regions of the human genome. Despite this, there are other Fugu data that indicate additional paralogous linkage groups containing duplicated genes do exist in Fugu.
Studies of genes mapping to human 9q34, for example, have shown that the Fugu orthologs of 9q34 genes are also divided between two different Fugu chromosomes, and that at least one of the 9q34 genes studied (DNM1) is duplicated in Fugu (Bouchireb et al. 2001).
In addition to the duplicated genes that we have identified as part of this project, two further genes (from the contig region sharing synteny with 3p21) are also duplicated in Fugu. In a study of the plasminogen-related growth factor (PRGF) genes in Fugu, Cottage and colleagues (1999) identified two paralogs, prgfr2 and prgfr3, which both mapped to regions sharing synteny with 3p21. Phylogenetic analysis indicated that the duplication was specific to the fish lineage. One of these two genes, prgfr3, is now known to be present on the larger of the two contigs discussed here and is positioned next to a calmodulin-binding protein, a probable homolog of human CAMK1 (also from Hsa3p21). Cottage et al. (1999) identified a second CAMK1 homolog neighboring, prgfr2. Thus the region sharing synteny with 3p21 is also known to be duplicated. Although the mapping location in Fugu for these genes is unknown, our data would indicate that prgfr2 and the neighboring calmodulin-binding protein will map to the same Fugu linkage group as the smaller of the two Fugu contigs presented here.
These data show that Fugu, like zebrafish, contains pairs of linkage groups that share synteny with single chromosome segments in human. These contain the semiorthologs (both equally related but not necessarily equally similar) to single copy human genes (for definitions of orthology and paralogy, see Sharman 1999). The presence of duplicated genes on these paralogous linkage groups, in addition to their presence on different chromosomes, indicates that they were initially generated by an ancestral duplication, rather than a translocation event, followed by subsequent gene loss. The patterns of gene distribution, as well as gene loss and retention, between the paralogous linkage groups that we examined in Fugu and zebrafish indicate that most gene loss following the duplication event occurred before the ancestral separation of these two fish species ∼150 million years ago (for a review of the evolution of bony fishes, see Young 1995). We have not tried to detect duplicates of any of the other 20q genes that we have identified; therefore, we are unable to say whether or not any of these are also present in duplicate in Fugu. Hence it has not been possible to estimate the retention rates for duplicated genes.
These patterns of duplication of genomic segments and the partial retention of additional gene copies are similar to those seen in the zebrafish genome. It therefore seems likely that Fugu and zebrafish have undergone similar duplication events. If this is the case, we would expect to find increasing numbers of Fugu linkage group pairs that share synteny with single human chromosomes. This is not at odds with the Fugu data generated to date. In general, Fugu gene searches have been focused around single Fugu genes of interest. Single gene probes have been used to select genomic libraries for the target gene, and the examination of synteny has been a secondary rather than a primary focus. Unless the gene probe used were for a duplicated Fugu gene, paralogous Fugu linkage groups would not be identified.
METHODS
Identification of Cosmid and BAC Clones
A Fugu genomic cosmid library available from the UK HGMP Resource Centre (http://www.hgmp.mrc.ac.uk) was probed with a human chromosome 20 TOP1 gene probe (A. Bench, Department of Haematology, University of Cambridge). This identified two sets of nonoverlapping cosmids each containing a Fugu top1 gene. Contigs were extended with cosmids from the same library; subsequently, BAC clones (Incyte Genomics) were also added.
Sample Sequencing
Gene content on the cosmids and BACs was assessed by sequence scanning. Approximately 50 random sequences were generated from each cosmid and ∼150 sequences from each BAC or PAC were generated with the methods of Elgar et al. (1999).
Complete Sequencing and Analyses of Duplicated Genes
The complete sequencing of the duplicated snail and top1 genes has been detailed in Smith et al. (2000) and Smith et al. (2001). Comparative analyses of the Fugu maf sequences were facilitated by assembling the two sets of genomic maf fragments in GAP4. Gaps in sequence coverage were closed by the design of primers at fragment ends and the use of PCR across gaps to produce templates for dye terminator sequencing.
STS Mapping of Individual BACs and Cosmids
Primers for STS mapping were primarily designed from Fugu sequences showing sequence similarity to known human genes. Additional STSs were designed from sequences showing no high similarity to known genes in regions of low gene density. Sequence templates were selected for high quality (as shown by a low percentage of ambiguous bases); where possible, primers were designed for regions where sequence data were confirmed by other sequenced subclones. Primers were designed with PRIME (Wisconsin Package, Genetics Computer Group Ltd.).
STS primers were used on individual BACs and cosmids in the two Fugu contigs. Data were ordered manually and displayed with ExCelsior (UK HGMP Resource Centre). This allowed the ordering of both genes and genomic clones.
Preparation of BAC Pools for STS Mapping
A Fugu genomic BAC library (InCyte Genomics), stored in 111 384-well microtiter plates, was used to prepare DNA pools for use as STS mapping templates. DNA was prepared from a single 384-well microtiter plate by inoculating four separate cultures with individual sets of 96 clones with a 96-pin replicating tool. Cultures were grown overnight and DNA from each 96-clone pool extracted with a standard alkaline lysis technique. All four DNA pools from a 384-well plate were then recombined to produce a single DNA pool representing the genomic content of a single 384-well microtiter plate. Ninety six of the 384-well plate DNA pools, representing plates 193 to 271 (79 plates) and plates 287 to 303 (17 plates), of the library were stored in 96-well microtiter plate format and used as templates for STS mapping.
BAC Fingerprinting
Cosmids and BACs were mini-prepped in 96-well plate format with an alkaline lysis technique. Clones were digested with HindIII enzyme and the fragments separated on agarose gels. Gels were stained with SyBr Gold (Cambridge Bioscience) and scanned on a Typhoon 8600 phosphoimager (Molecular Dynamics).
Gel images were tracked and bands called with Image (Sanger Centre). Clone fingerprints were then assembled into contigs with FPC (Soderland et al. 1997).
Comparative Sequence Analyses
Genomic sequences were used in BLASTv.2 (Altschul et al. 1997) sequence similarity searches of the SWISS-PROT (Bairoch and Apweiler 1997), TREMBL (Bairoch and Apweiler 1997), and EMBL (Rodriguez-Tomé et al. 1996) databases. Search results were filtered through MSP-Crunch (Sonnhammer and Durbin 1994) and viewed via the Fugu web pages (http://fugu.hgmp.mrc.ac.uk) (Elgar et al. 1999) at the UK HGMP Resource Centre. Sequence similarities to database entries were assessed on an individual basis, taking account of the sequence quality, organism, and sequence type matched, and the quality and length of the match (including ‘Score’ and ‘Expect’ values). The numbers of different matches to the same database entry were also considered. In some cases, in which BLAST matches were to ESTs, these were used to search the human UniGene database (www.ncbi.nlm.nih.gov/Schuler/UniGene/) to allow assignment of matches to specific human UniGene clusters. Cases in which ESTs in the same UniGene cluster had been mapped also allowed human chromosomal locations to be found, even for uncharacterized human genes. Localizations for well-characterized human genes were also obtained by use of gene names in searches of GeneMap (www.ncbi.nlm.nih.gov/genemap/) and LocusLink (www.ncbi.nlm.nih.gov/LocusLink/). BLAST similarity to human genomic PAC clones sequenced as part of the whole genome sequencing effort allowed more precise chromosomal localizations to be established for specific genes.
Where BLAST similarity was to a number of similar human genes, probable human orthologs were identified through examination of the extent of shared synteny around the Fugu and human genes.
These searches were repeated at frequent intervals to allow for comparisons with new database entries. These new searches became increasingly informative, allowing easier identification of probable human orthologs as human whole genome sequence data were submitted to the databases in increasing quantities. Final locations for human chromosome 20 genes (including locations on human bacterial clones) were found with Ensembl (http://ensembl.ebi.ac.uk/).
Human gene names used are those found in LocusLink. Genes that have no official name have been identified according to the corresponding Unigene EST cluster.
Fugu BLAST EST data were also used to identify zebrafish ESTs with high sequence similarity to Fugu subclones. The names of the ESTs identified were used to search the zebrafish EST mapping data of the WashU-Zebrafish Genome Resources Project (http://zfish.wustl.edu/). This allowed the identification of zebrafish EST clusters for the corresponding zebrafish genes. Cases in which radiation hybrid mapping data were available for one or more of the clustered ESTs also allowed the probable mapping location of the zebrafish gene to be found. If an identified zebrafish EST was not in a mapped cluster but data were available for its paired read from the other end of the cDNA, this mapping location was used.
Tissue Culture, Chromosome Preparation, and Banding
Metaphase chromosomes of Fugu rubripes were prepared from a fibroblast line (Bradford et al. 1997) maintained in Ham's F-12 medium supplemented with 15% fetal calf serum, 1 mM L-glutamine, and antibiotics. Cells were grown at 27°C in a 5% CO2 incubator.
For chromosome preparation, Colcemid (final concentration, 0.1 μg/mL medium) was added to the culture medium at least 4 h before harvest to arrest cells in metaphase. Cells were removed from the flask by gentle trypsinization. The cell pellet was resuspended in a hypotonic solution consisting of 50 mM KCl for 20 min. Cells were fixed overnight in a 3 : 1 mixture of methanol : acetic acid. Slides were prepared with the conventional ‘drop-splash’ technique.
DNA Probes and FISH
DNA probes were labeled with either biotin-16-dUTP or digoxigenin-11-dUTP by nick translation.
Standard FISH protocols were followed. Briefly, the slides were treated with 100 μg/mL RNase A in 2× SSC, pH 7.0 at 37°C for 30 min and with 0.01% pepsin in 10 mM HCl at 37°C for 10 min. After refixing the preparations for 10 min in 1× PBS, 50 mM MgCl2, 1% formaldehyde, they were dehydrated in an ethanol series (70%, 80%, 100%). Slides were denatured for 1 min at 90°C in 70% formamide, 2× SSC, pH 7.0 and again dehydrated in an alcohol series. For hybridization of one slide, 400 ng of biotinylated and/or digoxigenated probe DNA were coprecipitated with 50–100 μg sheared Fugu genomic DNA (as competitor for single-copy probes) and 10–20 μg sheared human placental DNA (as carrier) and redissolved in 50% formamide, 10% dextran sulphate, 2× SSC. The hybridization mixture was denatured for 10 min at 80°C. Preannealing of repetitive sequences DNA was performed for 30 min at 37°C. Then the hybridization mixture was applied to each slide and sealed under a coverslip. The slides were hybridized for at least 3 d in a moist chamber at 37°C. The slides were then washed three times for 5 min in 50% formamide, 2× SSC at 42°C and once for 5 min in 0.1× SSC, pH 7.0 at 60°C, before being blocked with 4× SSC, 3% BSA, 0.1% Tween 20 at 37°C for 30 min. Probes were detected with fluorescein isothiocyanate (FITC)-conjugated avidin (Vector Laboratories) and Cy3-conjugated antidigoxin antibody (Dianova). Chromosomes and cell nuclei were counterstained with 1 μg/mL 4‘,6-diamidino-2-phenylindole (DAPI) in 2× SSC for 1 min. The slides were mounted in 90% glycerol, 0.1 M Tris-HCl, pH 8.0, and 2.3% 1,4-diazobicyclo-2,2,2-octane.
Digital Imaging Microscopy
Images were taken with a Zeiss epifluorescence microscope equipped with a thermoelectrically cooled charge-coupled device camera (Photometrics CH250) controlled by an Apple Macintosh computer. Vysis imaging software was used to capture gray scale images and to superimpose the source images into a color image.
WEB SITE REFERENCES
http://ensembl.ebi.ac.uk; Ensembl home page.
http://fugu.hgmp.mrc.ac.uk/PFW/Other/consortium.html; Fugu Genomics Projects.
http://fugu.hgmp.mrc.ac.uk; UK HGMP Resource Centre Fugu Group home page.
http://zfish.wustl.edu; WashU-Zebrafish Genome Resources Project home page.
http://www.genoscope.cns.fr/externe/tetraodon; Genoscope Tetraodon nigroviridis home page.
http://www.ncbi.nlm.nih.gov; NCBI home page.
http://zfin.org; the Zebrafish Information Network home page.
Acknowledgments
The authors thank Dr. I. D. Hickson of the Institute of Molecular Medicine, University of Oxford, for providing the human TOP1 probe, and David Barnes from ATCC for providing the Fugu cell line.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sfsmith@hgmp.mrc.ac.uk, FAX (44)1223-494512.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.221802. Article published online before print in April 2002.
REFERENCES
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Aparicio S, Hawker K, Cottage A, Mikawa Y, Zuo L, Venkatesh B, Chen E, Krumlauf R, Brenner S. Organisation of the Fugu Hox clusters: Evidence for continuing evolution of vertebrate Hox complexes. Nat Genet. 1997;16:79–83. doi: 10.1038/ng0597-79. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence data bank and its supplement TrEMBL. Nucleic Acids Res. 1997;25:31–36. doi: 10.1093/nar/25.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk W, Korf I, Kadavi C, Heyen J, Tate S, Wun E, Bedell J, McPherson J, Johnson S. The syntenic relationship of the zebrafish and human genomes. Genome Res. 2000;10:1351–1358. doi: 10.1101/gr.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank V, Andrews N. The Maf transcription factors: Regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- Bouchireb N, Gruetzner F, Haaf T, Stephens RJ, Elgar G, Clark MS. Comparative mapping of the human 9q34 region in Fugu rubripes. Cyto Genet Cell Genet. 2001;94:173–179. doi: 10.1159/000048811. [DOI] [PubMed] [Google Scholar]
- Bradford C, Miller A, Toumadje A, Nishiyama K, Shirahata S, Barnes D. Characterization of cell cultures derived from Fugu, the Japanese pufferfish. Mol Mar Biol Biotechnol. 1997;6:279–288. [PubMed] [Google Scholar]
- Brenner S, Elgar G, Sandford R, Macrae A, Venkatesh B, Aparicio S. Characterization of the pufferfish (Fugu) genome as a compact model vertebrate genome. Nature. 1993;366:265–268. doi: 10.1038/366265a0. [DOI] [PubMed] [Google Scholar]
- Cottage A, Clark M, Hawker K, Umrania Y, Wheller D, Bishop M, Elgar G. Three receptor genes for plasminogen related growth factors in the genome of the puffer fish Fugu rubripes. FEBS Lett. 1999;443:370–374. doi: 10.1016/s0014-5793(99)00011-3. [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stanier D Y, Zwartkruis F, Abedelilah S, Rangini Z, et al. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 1996;123:37–46. doi: 10.1242/dev.123.1.37. [DOI] [PubMed] [Google Scholar]
- Elgar G, Clark MS, Meek S, Smith S, Warner S, Edwards YJ, Bouchireb N, Cottage A, Yeo GS, Umrania Y, et al. Generation and analysis of 25 Mb of genomic DNA from the pufferfish Fugu rubripes by sequence scanning. Genome Res. 1999;9:960–971. doi: 10.1101/gr.9.10.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates M, Kim L, Egan E, Cardozo T, Sirotkin H, Dougan S, Lashkari D, Abagyan R, Schier A, Talbot W. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 1999;9:334–347. [PubMed] [Google Scholar]
- Geisler R, Rauch GJ, Baier H, van Bebber F, Brobeta, Dekens MP, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Holland PW, Garcia-Fernandez J, Williams NA, Sidow A. Gene duplications and the origin of vertebrate development. Development Suppl. 1994;1994:125–133. [PubMed] [Google Scholar]
- Hughes A, da Silva J, Friedman R. Ancient genome duplications did not structure the human hox-bearing chromosomes. Genome Res. 2001;11:771–780. doi: 10.1101/gr.160001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens C, Cordes S, Giorgianni M, Barsh G, Kimmel C. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Haffter P, Nusslein-Volhard C. Large-scale mutagenesis in the zebrafish: A search for genes controlling development in a vertebrate. Curr Biol. 1994;4:189–202. doi: 10.1016/s0960-9822(00)00048-8. [DOI] [PubMed] [Google Scholar]
- Postlethwait J, Woods I, Ngo-Hazelett P, Yan Y, Kelly P, Chu F, Huang H, Hill-Force A, Talbot W. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- Robinson-Rechavi M, Marchand O, Escriva H, Bardet PL, Zelus D, Hughes S, Laudet V. Euteleost fish genomes are characterized by expansion of gene families. Genome Res. 2001;11:781–788. doi: 10.1101/gr.165601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Tomé P, Stoehr PJ, Cameron GN, Flores TP. The European Bioinformatics Institute (EBI) databases. Nucleic Acids Res. 1996;24:6–12. doi: 10.1093/nar/24.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvarzstein M, Kirn A, Haffter P, Cordes S. Expression of Zkrml2, a homologue of the Krml/val segmentation gene, during embryonic patterning of the zebrafish (Danio rerio) Mech Dev. 1999;80:223–226. doi: 10.1016/s0925-4773(98)00220-2. [DOI] [PubMed] [Google Scholar]
- Sharman AC. Some new terms for duplicated genes. Semin Cel Dev Biol. 1999;10:561–563. doi: 10.1006/scdb.1999.0338. [DOI] [PubMed] [Google Scholar]
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. Zebrafish linkage map with 2000 microsatellite markers. Genomics. 1999;58:219–232. doi: 10.1006/geno.1999.5824. [DOI] [PubMed] [Google Scholar]
- Smith S, Metcalfe JA, Elgar G. Identification and analysis of two snail genes in the pufferfish (Fugu rubripes) and mapping of human SNA to 20q. Gene. 2000;247:119–128. doi: 10.1016/s0378-1119(00)00110-4. [DOI] [PubMed] [Google Scholar]
- Smith S, Metcalfe J, Elgar G. Characterisation of two topoisomerase 1 genes in the pufferfish (Fugu rubripes) Gene. 2001;265:195–204. doi: 10.1016/s0378-1119(01)00366-3. [DOI] [PubMed] [Google Scholar]
- Soderland C, Longden I, Mott R. FPC: A system for building contigs from restriction fingerprinted clones. CABIOS. 1997;13:523–535. doi: 10.1093/bioinformatics/13.5.523. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E, Durbin R. A workbench for large-scale sequence homology analysis. Comput Appl Biosci. 1994;10:301–307. doi: 10.1093/bioinformatics/10.3.301. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Gilligan P, Brenner S. Fugu: A compact vertebrate reference genome. FEBS Lett. 2000;476:3–7. doi: 10.1016/s0014-5793(00)01659-8. [DOI] [PubMed] [Google Scholar]
- Wang P, Eisenbart J, Cordes S, Barsh G, Stoffel M, Le Beau M. Human KRML (MAFB): cDNA cloning, genomic structure, and evaluation as a candidate tumour suppressor gene in myeloid leukemias. Genomics. 1999;59:275–281. doi: 10.1006/geno.1999.5884. [DOI] [PubMed] [Google Scholar]
- Wittbrot J, Meyer A, Schartl M. More genes in fish? Bioessays. 1998;20:511–515. [Google Scholar]
- Woods I, Kelly P, Chu F, Ngo-Hazelett P, Yan Y, Huang H, Postlethwait J, Talbot W. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JZ. The life of vertebrates. 3rd ed. Oxford, UK: Oxford University Press; 1995. The evolution of bony fishes; pp. 189–199. [Google Scholar]