Abstract
Twelve strains of gram-negative, nonfermenting rods recovered mainly from septicemic patients were studied using conventional and molecular methods. The phenotypic profiles of these strains most closely resembled Psychrobacter phenylpyruvicus. They produced catalase, oxidase, urease, and H2S (lead acetate paper) but did not produce indole, reduce nitrate or nitrite, or hydrolyze gelatin or esculin. No acid production was observed in a King's oxidation-fermentation base containing d-glucose, d-xylose, d-mannitol, sucrose, lactose, or maltose. All strains were nonmotile and nonpigmented. Most strains produced green discoloration on blood agar. All strains grew at 25°C and 35°C and most grew on MacConkey agar. They shared a common cellular fatty acid (CFA) profile characterized by large amounts (56% to 90%) of 18:1ω7c and the presence of 3-OH-10:0, 16:1ω7c, 16:0, and 19:0cycω8c that overall was most similar to that of Rhodobacter species but was quite distinct from that of P. phenylpyruvicus. The MICs for most β-lactams, fluoroquinolones, aminoglycosides, and carbapenems were low. MICs for aztreonam and piperacillin were higher, with MICs for some strains of > 64 mg/liter and > 128 mg/liter, respectively. Polyphasic analysis of these strains, including morphological, biochemical, CFA composition, DNA-DNA hybridization, 16S rRNA gene sequencing, and percent guanine-plus-cytosine (G+C) content analysis, demonstrated that these strains and Rhodobacter massiliensis represent a new genus, “Haematobacter” (proposed name), with the species H. missouriensis (type strain H1892T = CCUG 52307T = CIP 109176T) and H. massiliensis comb. nov. (type strain FramboiseT = CCUG 47968T = CIP 107725T) and an unnamed genomospecies.
The Special Bacteriology Reference Laboratory (SBRL) of the Centers for Disease Control and Prevention receives for identification and classification bacterial isolates from reference laboratories throughout the United States. Between 1997 and 2003, SBRL received 12 phenotypically similar unidentified clinical isolates from eight U.S. state health departments. The phenotypic profiles of these strains resemble that of Psychrobacter phenylpyruvicus. 16S rRNA gene sequence analysis showed that the SBRL strains cluster with the species Rhodobacter massiliensis (5), which is represented by a single isolate from the nose of a patient with aspiration pneumonia.
Although a relatively small number of isolates were studied, the most common source of isolation for these strains was blood, suggesting the potential to cause invasive disease. In this report we present a polyphasic analysis of the SBRL strains, including morphological, biochemical, cellular fatty acid (CFA) composition, DNA-DNA hybridization, 16S rRNA gene sequencing, percent guanine-plus-cytosine (G+C) content, and in vitro antimicrobial susceptibility determinations. These results demonstrate that these strains represent a new genus, “Haematobacter” (proposed name), with the species H. missouriensis and H. massiliensis comb. nov. and an unnamed genomospecies.
MATERIALS AND METHODS
Bacterial strains.
The strains studied, along with their sources and geographic origins, are presented in Table 1. Type strains of Rhodobacter capsulatus (ATCC 11166T), R. blasticus (ATCC 33485T), and R. sphaeroides (ATCC 17023T) were kindly provided by Jane Tang, American Type Culture Collection, Manassas, VA. In addition, the type strain of R. massiliensis (CCUG 47968T) was kindly provided by Enevold Falsen, Culture Collection of the University of Goteborg (CCUG), Goteborg, Sweden. Unless otherwise indicated, strains were cultured on heart infusion agar supplemented with 5% rabbit blood agar (RBA) (BBL Microbiology Systems, Cockeysville, MD) and incubated aerobically at 35°C. All strains were stored as suspensions in defibrinated rabbit blood in liquid nitrogen.
TABLE 1.
Sources and demographic and clinical information for strains of Haematobacter and Rhodobacter in this study
| Strain | Yr received | Location of isolation | Patient's sex, agea | Source | GenBank accession no. |
|---|---|---|---|---|---|
| H0445 | 1997 | Texas | F, 2 mo | Blood | DQ342316 |
| H1892 | 2001 | Missouri | M, 88 yr | Blood | DQ342315 |
| H1925 | 2001 | Indiana | M, 81 yr | Blood | DQ342318 |
| H2098 | 2001 | Alabama | M, 72 yr | Blood | DQ342311 |
| H2136 | 2001 | Alabama | F, 70 yr | Blood | DQ342308 |
| H2178 | 2001 | New Hampshire | F, 4 yr | Blood | DQ342314 |
| H2195 | 2002 | Michigan | M, 83 yr | Blood | DQ342317 |
| H2240 | 2002 | Colorado | F, NG | Blood | DQ342319 |
| H2381 | 2002 | Michigan | M, 67 yr | Blood | DQ342313 |
| H2443 | 2002 | Michigan | F, 65 yr | Traumatic wound | DQ342310 |
| H2639 | 2003 | Illinois | F, 41 yr | Blood | DQ342307 |
| H2646 | 2003 | Alabama | M, 63 yr | Blood | DQ342312 |
| ATCC 11166T, R. capsulatus | 2003 | DQ342320 | |||
| ATCC 33485T, R. blasticus | 2003 | Freshwater pond, England | DQ342322 | ||
| ATCC 17023T, R. sphaeroides | 2003 | DQ342321 | |||
| CCUG 47968T, R. massiliensis | 2003 | Amoebal coculture of nose swab, France | DQ342309 |
M, male; F, female; NG, not given.
Phenotypic tests.
Biochemical testing was done using conventional methods (10) and commercial systems using API 20NE and API ZYM (bioMérieux Canada, Inc., St. Laurent, Québec, Canada) and Microscan (Dade Behring, Mississauga, Ontario, Canada). All biochemical tests were performed at 35°C in an aerobic incubator except for the optimum growth temperature tests and biochemical tests for R. blasticus (ATCC 33485T) and R. capsulatus (ATCC 11166T), which were performed at 25°C. The oxidase, catalase, and growth temperature tests were read after 1 day of incubation. All other tests were read after 1, 2, and 7 days of incubation.
CFA analysis.
Cells from 2-day-old cultures were saponified, and the liberated fatty acids were methylated and analyzed by capillary gas-liquid chromatography (10). Identification of fatty acids was performed using a commercially available system (MIDI, Newark, DE).
DNA relatedness and percent G+C determination.
All strains were cultured on 20 to 30 RBA plates and incubated for 24 h at 35°C. Cells were harvested and lysed, and the DNA was isolated and purified according to the method of Brenner et al. (1). DNA from strains H0445 and H1892 was labeled using [32P]dCTP and a commercial nick translation kit (Bethesda Research Laboratories, Inc., Gaithersburg, MD) and tested for reassociation to unlabeled DNA from the same strain (homologous reaction) as well as to other strains and to the type and reference strains studied (heterologous reactions). Relative binding ratios and percent divergence were calculated as described previously (1). All reactions were done in duplicate at the optimal temperature of 65°C. The percent G+C was determined for strains H0445 and H1892 by thermal denaturation (7).
16S rRNA gene sequencing.
DNA was extracted and purified with a QIAamp DNA Mini kit (QIAGEN Inc., Valencia, CA) and amplified using primers fD1 and rP2 (9). Cycle sequencing was performed with BigDye version 1.1 dye terminator chemistry (Applied Biosystems, Foster City, CA), and reaction products were sequenced on an ABI 3100 sequencer (Applied Biosystems).
Sequence data were edited and compiled using the Wisconsin Sequence Analysis Package (Genetics Computer Group, Madison, Wis.). The 16S rRNA gene sequences determined in this study were aligned with sequences from GenBank using Clustal in MEGA version 3.1 (6). The multiple sequence alignment was edited by hand and used to derive a neighbor-joining tree with 1,000 bootstraps and the Kimura 2-parameter model.
In vitro antimicrobial susceptibility tests.
Antimicrobial susceptibility testing of 13 Haematobacter isolates, including the 12 clinical isolates and the type strain of R. massiliensis, was performed using the broth microdilution method and MIC trays prepared in house. All procedures were performed according to standard methods (2). Inocula were prepared by the direct inoculum method from 48-h growth on Trypticase soy agar plates with 5% defibrinated sheep blood (BBL, Cockeysville, MD) and adjusted to a 0.5 McFarland standard. Suspensions were inoculated into trays containing cation-supplemented Mueller-Hinton broth (Difco brand; BD), and trays were incubated in ambient air at 35°C. MIC endpoints were determined at 24 h and 48 h. The following antimicrobial agents were tested alone or in combination: amikacin (Sigma, St. Louis, MO), ampicillin (Sigma), amoxicillin (Sigma), aztreonam (Sigma), cefazolin (Sigma), cefepime (Bristol-Myers Squibb, Wallingford, CT), cefotaxime (Sigma), cefoxitin (Sigma), ceftazidime (Sigma), ceftriaxone (Sigma), chloramphenicol (Sigma), ciprofloxacin (Bayer, West Haven, CT), clavulanic acid (GlaxoSmithKline, Research Triangle Park, NC), gatifloxacin (Bristol-Myers Squibb), gentamicin (Sigma), imipenem (USP, Rockville, MD), levofloxacin (Pfizer, Groton, CT), meropenem (AstraZeneca, Wilmington, DE), piperacillin (Wyeth-Ayerst, Pearl River, NY), tazobactam (Wyeth-Ayerst), tetracycline (Sigma), ticarcillin (Sigma), and tobramycin (Sigma). Compounds were purchased from Sigma and USP; the other antimicrobial agents were supplied gratis from their respective manufacturer as listed above. The quality control strains for antimicrobial susceptibility testing were Enterococcus faecalis ATCC 29212, Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 29213, and Escherichia coli ATCC 35218.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences for the 12 Haematobacter strains and R. massiliensis (CCUG 47968T) were submitted to GenBank and assigned accession numbers DQ342307 to DQ342319 (Table 1).
RESULTS
The sources of all strains studied are listed in Table 1. The 12 clinical isolates were received from eight different state public health laboratories in the United States between 1997 and 2003. No single geographic area predominated. All but one of the isolates was recovered from blood cultures. The ages of the patients ranged from 2 months to 88 years (median, 57 years); six of the patients were female and six were male.
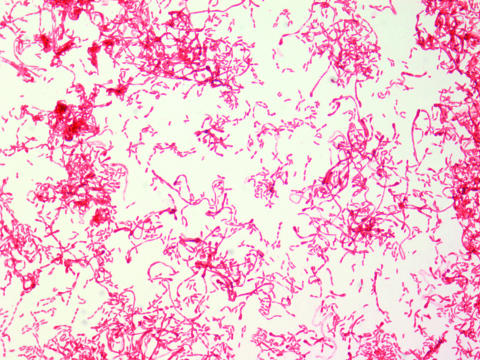
All 12 strains and R. massiliensis CCUG 47968T shared common phenotypic characteristics (Table 2). They were obligately aerobic, nonmotile rods and nonhemolytic on RBA, and some (5/12) strains failed to grow on MacConkey agar. There was no growth on Salmonella Shigella agar or on cetrimide agar. They produced catalase, oxidase, urease, phenylalanine deaminase, o-nitrophenyl-β-d-galactopyranoside (weak), and H2S (lead acetate paper) but did not produce indole, reduce nitrate or nitrite, or hydrolyze gelatin. No acid production in the slant or butt of triple-sugar iron agar was noted. All strains utilized sodium acetate as a carbon source. Isolated colonies on RBA were translucent, convex, entire, and 0.5 to 1 mm in diameter. Most strains produced green discoloration under areas of moderate to heavy growth on blood agar plates. Cells grown on heart infusion agar at 35°C for 18 to 24 h were gram-negative, serpentine pleomorphic rods with filaments (Fig. 1). Growth in API 20NE and Microscan systems was insufficient to allow identification. In API ZYM strips, all Haematobacter strains and R. massiliensis produced moderate to strong activity of the enzymes alkaline phosphatase, acid phosphatase, C4 esterase, C8 esterase lipase, and leucine arylamidase and weak to moderate activity of naphthol-AS-BI-phosphatase. P. phenylpyruvicus produced C4 esterase, C8 esterase lipase, leucine arylamidase, and weak activity of naphthol-AS-BI-phosphatase but did not produce alkaline phosphatase or acid phosphatase. Table 2 lists phenotypic characteristics that are useful in differentiating between Rhodobacter species, Haematobacter species, and P. phenylpyruvicus.
TABLE 2.
Differentiation of Haematobacter species, Psychrobacter phenylpyruvicus, and Rhodobacter speciesa
| Test | Result for species
|
||||||
|---|---|---|---|---|---|---|---|
| H. massiliensisc (n = 7) | H. missouriensis (n = 5) | Haematobacter genomospecies 1 (n = 1) | Psychrobacter phenylpyruvicus (n = 50) | R. blasticus (n = 1) | R. capsulatus (n = 1) | R. sphaeroides (n = 1) | |
| Acid from (OFb base): | |||||||
| d-Glucose | − | − | − | − | + | + | + |
| d-Xylose | − | − | − | − | + | + | + |
| d-Mannitol | − | − | − | − | + | − | + |
| Lactose | − | − | − | − | − | − | − |
| Sucrose | − | − | − | − | + | + | − |
| Maltose | − | − | − | − | + | +w | − |
| Growth on MacConkey agar | 5/7d | 2/5 | − | 40(3)/50 | − | − | + |
| Nitrate | − | − | − | 34/50 | − | − | − |
| Indole | − | − | − | − | − | − | − |
| Alkalinization of litmus milk | 2/7 | 1/5 | − | 10, 2W(9)/46 | − | − | − |
| Urea | + | + | + | + | − | − | − |
| Production of growth pigment | − | − | − | − | Pink-red | Pink | Pink-red |
Symbols and abbreviations: −, negative; +, positive; NG, no growth; w, weak; (), late reaction at 3 to 7 days.
OF, oxidation-fermentation.
Includes R. massiliensis CCUG 47968T.
Fractions indicate number of strains positive per total number of strains tested.
FIG. 1.
Gram stain (original magnification, ×1,000) of Haematobacter missouriensis strain H2646.
The CFA compositions of saponified whole cells of Haematobacter species, R. massiliensis, R. capsulatus, R. blasticus, R. sphaeroides, and P. phenylpyruvicus are shown in Table 3. All strains of Haematobacter species shared a common CFA profile characterized by large amounts (56% to 90%) of 18:1ω7c and the presence of lesser amounts (0% to 20%) of 3-OH-10:0, 16:1ω7c, 16:0, 18:0, 18:1ω9c, and 19:0cycω8c. Strain H2443 can be distinguished from other Haematobacter species by the presence of an additional 11% 17:1ω6c and 12% 17:0.
TABLE 3.
Cellular fatty acid compositions of Haematobacter species, P. phenylpyruvicus, R. capsulatus, R. blasticus, R. sphaeroides, and R. massiliensis
| Organism | % Fatty acida
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3-OH-10:0 | 12:1ω7c | 16:1ω7c | 16:0 | 17:1ω6c | 17:0 | 18:1ω9c | 18:1ω7c | 18:0 | 19:0cycω8c | 10-CH3-19:0 | 3-OH-18:0 | |
| Hematobacter spp. | ||||||||||||
| H0445 | 2 | − | 1 | 15 | − | T | 70 | 2 | 10 | − | − | |
| H1892 | 2 | − | T | 2 | − | − | T | 90 | T | 3 | − | − |
| H1925 | 11 | − | 1 | 5 | − | − | − | 60 | − | 20 | − | − |
| H2098 | 1 | − | 1 | 3 | − | − | 1 | 73 | 1 | 13 | − | − |
| H2136 | 2 | − | 1 | 3 | − | − | − | 79 | − | 14 | − | − |
| H2178 | 2 | − | 1 | 4 | − | − | − | 81 | 1 | 11 | − | − |
| H2195 | 2 | − | 1 | 2 | − | − | − | 90 | T | 3 | − | − |
| H2240 | 4 | − | 2 | 5 | − | − | − | 65 | 1 | 20 | − | − |
| H2381 | 4 | − | T | 3 | − | − | − | 80 | T | 10 | − | − |
| H2443 | 2 | − | − | 3 | 11 | 12 | − | 56 | 1 | 13 | − | − |
| H2639 | 3 | − | 1 | 7 | − | − | − | 71 | 1 | 14 | − | − |
| H2646 | 2 | − | 1 | 4 | − | − | − | 77 | 1 | 13 | − | − |
| P. phenylpyruvicusb | − | − | 11 | 11 | − | − | 22 | − | 4 | − | − | − |
| R. capsulatus ATCC 11166T | 6 | 4 | 9 | 2 | − | − | − | 74 | − | − | 2 | 3 |
| R. blasticus ATCC 33485T | 1 | − | 1 | 2 | − | − | 2 | 81 | 2 | − | 2 | 7 |
| R. sphaeroides ATCC 17023T | 1 | − | 1 | 3 | − | − | 1 | 85 | 6 | − | − | − |
| R. massiliensis CCUG 47968T | 3 | − | 1 | 3 | − | − | − | 84 | 1 | 4 | − | − |
The number before the colon indicates the number of carbons; the number after the colon is the number of double bonds; OH, a hydroxy group at the 3(ß)- position from the carboxyl end; ω, the position of the double bond counting from the hydrocarbon end of the carbon chain; c, cis isomer; cyc, a cyclopropane ring structure; CH3, a methyl group; T, 0.4% to 0.6%; −, not detected,. Values are percentages of total fatty acids and are arithmetic means of two consecutive growths of each strain.
Data for P. phenylpyruvicus were from Weyant and coworkers (10). P. phenylpyruvicus also contains 5% 10:0, 2% i-11:0, 4% 11:0, 3% 12:0, 1% 3-OH-11:0, 1% 12:1ω9c, 8% 3-OH-12:0, 1% 14:0, 23% 18:2, and 2% 20.4, which are not found in Haematobacter spp.
Results of DNA relatedness studies are shown in Table 4. Using the established molecular criteria for species-level relatedness (strains whose DNAs are 70% or more related at optimal conditions and whose related sequences show 5% or less divergence) (8), three species-level hybridization groups were identified among the 12 strains.
TABLE 4.
DNA relatedness of Haematobacter strains and Rhodobacter species
| Source of unlabeled DNA | Results of reaction with labeled DNA from straina:
|
|||
|---|---|---|---|---|
| H0445
|
H1892
|
|||
| RBR | % D | RBR | % D | |
| H. massiliensis H0445 | 100 | 0.0 | ||
| H. massiliensis H1925 | 76 | 1.0 | ||
| H. massiliensis H2098 | 82 | 1.5 | ||
| H. massiliensis H2136 | 86 | 1.5 | ||
| H. massiliensis H2443 | 86 | 1.5 | ||
| H. massiliensis H2639 | 99 | 0.5 | ||
| H. missouriensis H1892 | 40 | 6.5 | 100 | 0.0 |
| H. missouriensis H2178 | 42 | 6.0 | 75 | 0.0 |
| H. missouriensis H2195 | 54 | 7.0 | 100 | 0.0 |
| H. missouriensis H2381 | 52 | 7.0 | 88 | 0.0 |
| H. missouriensis H2646 | 55 | 6.5 | 92 | 0.5 |
| Haematobacter genomospecies 1 H2240 | 63 | 5.0 | 64 | 5.5 |
| Rhodobacter capsulatus ATCC 11166T | 12 | 10.0 | 7 | |
| Rhodobacter blasticus ATCC 33485T | 12 | 9.5 | 7 | |
| Rhodobacter sphaeroides ATCC 17023T | 21 | 7.0 | 9 | |
| Rhodobacter massiliensis CCUG 47968T | 100 | 0.0 | 49 | 7.5 |
RBR, relative binding ratio; % D, percent divergence. Reactions were performed at 65oC.
The first group consisted of H1892, H2178, H2195, H2381, and H2646. The relatedness within this group was greater than 74%, and the divergence was less than 1.0%. The second group contained strains H0445, H1925, H2098, H2136, H2443, H2639, and R. massiliensis CCUG 47968T. This group was more diverse, with relatedness and divergence greater than 75% and less than 2.0%, respectively. Strain H2240 was not related at the species level with any of the other 11 strains or reference strains included in this study. The percent G+C was determined for a representative strain of each DNA relatedness group, and values ranged from 65% for strain H0445 to 65.5% for strain H1892.
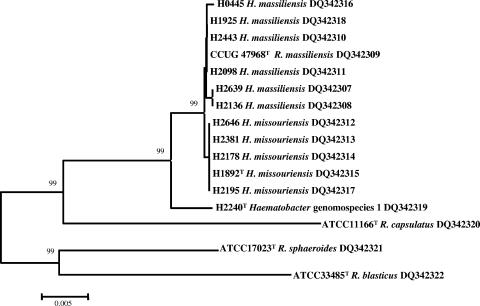
The 16S rRNA gene sequences of 12 clinical isolates and the reference strains were determined and aligned. The resulting phylogenetic tree (Fig. 2) demonstrated the close relationships between the 12 strains and R. massiliensis. The 16S rRNA gene sequence findings supported the DNA-DNA hybridization results. The closest 16S rRNA gene sequence relationship was between H. massiliensis and R. capsulatus, which differed by 52 bp and four gaps. In contrast, P. phenylpyruvicus was only distantly related, with 77% to 78% homology to 16S rRNA gene sequences of the Haematobacter strains (data not shown).
FIG. 2.
Phylogenetic tree based on 1,389-bp 16S rRNA gene sequences, showing the positions of Haematobacter and Rhodobacter strains. Bootstrap analysis was done with 1,000 resamplings; bootstrap values are indicated at some branch points. The scale bar represents 0.5% difference (7 bp) in DNA sequences. GenBank accession numbers for the sequences are shown.
A number of antimicrobial agents appear to be active against Haematobacter species. There was sufficient growth after 24 h of incubation to determine MIC endpoints, and there were no significant differences in MICs determined at 24 h and 48 h (data not shown). Aminoglycosides, fluoroquinolones, carbapenems, tetracycline, and chloramphenicol appeared to have good activity against these isolates. Ampicillin, amoxicillin-clavulanic acid, and cephalosporin MICs were also low. MICs to aztreonam and piperacillin were higher, with MICs > 64 mg/liter and > 128 mg/liter, respectively, for some strains. Currently there are no interpretive MIC breakpoints for isolates of this genus.
DISCUSSION
We have characterized isolates from patients that phenotypically resembled P. phenylpyruvicus but represent a new genus, Haematobacter, that may be differentiated simply and rapidly from P. phenylpyruvicus by microscopic appearance in Gram-stained smears. Comparisons of the sequences of 16S rRNA genes of Haematobacter strains and P. phenylpyruvicus demonstrated that these species are distinct. In addition, the CFA composition was useful in distinguishing Haematobacter species from P. phenylpyruvicus. Haematobacter strains differ from P. phenylpyruvicus by the presence of major amounts of 18:1ω7c and 19:0cycω8c, which are absent in P. phenylpyruvicus, and by the absence of 18:2 (0% versus 23%) and presence of smaller amounts of 18:1ω9c (0% to 3% versus 22%) (10).
The overall CFA profile of Haematobacter species was most similar to those of R. massiliensis, R. capsulatus, R. blasticus, and R. sphaeroides. However, Haematobacter species differed from R. capsulatus, R. blasticus, and R. sphaeroides by the presence of 19:0cyc ω8c in Haematobacter species. In addition, 10-CH3-19:0, 3-OH-18:0, and 12:1ω7c were absent in Haematobacter species. However, 10-CH3-19:0 and 3-OH-18:0 were found in R. capsulatus and R. blasticus, while 12:1ω7c was present only in R. capsulatus. Additional strains of each species must be tested to determine whether these differences are sufficient for differentiation by CFA alone.
The species of Haematobacter are defined by DNA-DNA hybridization and can be differentiated by 16S rRNA gene sequencing but not by phenotypic tests commonly used in diagnostic laboratories. For smaller laboratories without access to sequence analysis, the identification of isolates as Haematobacter spp. may be sufficient to merit referral to a larger laboratory for specific identification.
Most of the isolates we studied were recovered from blood cultures, but this is not necessarily evidence of a pathogenic role. An additional isolate of R. massiliensis was recovered from the blood of a patient in France (3). It is possible that Haematobacter species may be opportunistic pathogens.
The 16S rRNA gene sequences of Haematobacter species are distinct from those of Rhodobacter spp. other than R. massiliensis, and Haematobacter species are readily distinguished from Rhodobacter species by biochemical characteristics. Phylogenetic analysis using 16S rRNA gene sequences places the genus Haematobacter, including R. massiliensis, in close association with the genus Rhodobacter within the Rhodobacteraceae (5). The G+C ratio of Haematobacter is consistent with this taxonomic position (4).
The first isolation of Haematobacter predates the description of R. massiliensis. Accordingly, we have proposed the transfer of this strain to Haematobacter, as H. massiliensis. We propose the genus Haematobacter, with species H. missouriensis, H. massiliensis, and Haematobacter genomospecies 1; the latter contains only a single strain at this time and we have not proposed a specific epithet for this taxon.
Description of Haematobacter gen. nov.
Haematobacter (hae.ma'to.bac'ter. N.L. masc. n. haemat, blood, N.L. masc. n. bacter rod, N.L. masc. n. Haematobacter rod from blood). Members of the genus are gram-negative, nonmotile, nonsporing, nonfermentative, pleomorphic rods. Colonies on solid media are nonpigmented. They grow aerobically on 5% RBA, discoloring the medium, and some strains grow on MacConkey agar. No growth on Salmonella Shigella agar or on cetrimide agar is seen. Strains produce catalase, oxidase, and urease and grow at 25°C and 35°C but not at 42°C. Esculin and gelatin are not hydrolyzed. Strains utilize acetate, but not citrate, as a sole carbon source. Strains deaminate phenylalanine. Strains produce alkaline phosphatase, acid phosphatase, C4 esterase, C8 esterase lipase, and leucine arylamidase. All strains of Haematobacter species share a common cellular fatty acid profile characterized by large amounts (56% to 90%) of 18:1ω7c and the presence (0% to 20%) of 3-OH-10:0, 16:1ω7c, 16:0, 18:0, 18:1ω9c, and 19:0cycω8c. The G+C content of the DNA is 65 mol%. Haematobacter spp. are potentially pathogenic for humans, having been isolated from blood and from wound infections. Three genomospecies are recognized which can be differentiated by 16S rRNA gene sequence analysis. The type species is Haematobacter missouriensis.
Description of Haematobacter missouriensis sp. nov.
Haematobacter missouriensis (mis.souri.en'sis N.L. adj., referring to the state of Missouri, where the type strain was isolated). H. missouriensis exhibits all of the characteristics of the genus. Colonies on solid media are nonpigmented. They grow aerobically on 5% RBA, and some strains grow on MacConkey agar. Strains produce catalase, oxidase, and urease and grow at 25°C and 35°C but not at 42°C. Esculin and gelatin are not hydrolyzed. The type strain is H1892T = CCUG 52307T = CIP 109176T from human blood.
Description of Haematobacter massiliensis comb. nov.
Haematobacter massiliensis (from Massilia, old Roman name for Marseilles, where the type strain was isolated). H. massiliensis exhibits all of the characteristics of the genus. Colonies on solid media are nonpigmented. They grow aerobically on 5% RBA, and some strains grow on MacConkey agar. Strains produce catalase, oxidase, and urease and grow at 25°C and 35°C but not at 42°C. Esculin and gelatin are not hydrolyzed. The type strain is strain FramboiseT = CCUG 47968T = CIP 107725T from an amoebic coculture.
Description of Haematobacter genomospecies 1 sp. nov.
Haematobacter genomospecies 1 exhibits all of the characteristics of the genus. Colonies on solid media are nonpigmented. They grow aerobically on 5% RBA, and some strains grow on MacConkey agar. Strains produce catalase, oxidase, and urease and grow at 25°C and 35°C but not at 42°C. Esculin and gelatin are not hydrolyzed. The only strain is H2240 = CCUG 52313 = CIP 109177 from human blood.
Acknowledgments
We thank Enevold Falsen for his generous assistance with depositing cultures in collections and Hans Trüper for his kind advice regarding correct Latin etymology.
Footnotes
Published ahead of print on 7 February 2007.
REFERENCES
- 1.Brenner, D. J., A. C. McWhorter, J. K. L. Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard M7-A7, 7th ed. Clinical and Laboratory Standards Institute, Wayne, PA.
- 3.Drancourt, M., P. Berger, and D. Raoult. 2004. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J. Clin. Microbiol. 42:2197-2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garrity, G. M., J. A. Bell, and T. Lilburn. 2005. Family I. Rhodobacteraceae, p. 161-167. In D. J. Brenner, N. R. Krieg, and J. T. Staley (ed.), Bergey's manual of systematic bacteriology, vol. 2. Springer, New York, NY. [Google Scholar]
- 5.Greub, G., and D. Raoult. 2003. Rhodobacter massiliensis sp. nov., a new amoebae-resistant species isolated from the nose of a patient. Res. Microbiol. 154:631-635. [DOI] [PubMed] [Google Scholar]
- 6.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 7.Mandel, M., L. Igambi, J. Bergendahl, M. L. Dodson, and E. Scheltgen. 1970. Correlation of melting temperature and cesium chloride buoyant density of bacterial deoxyribonucleic acid. J. Bacteriol. 101:333-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the ad hoc committee on reconciliation of the approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 9.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weyant, R. S., C. W. Moss, R. E. Weaver, D. G. Hollis, J. G. Jordan, E. C. Cook, and M. I. Daneshvar. 1996. Identification of unusual pathogenic gram-negative aerobic and facultative anaerobic bacteria, 2nd ed. Williams & Wilkins, Baltimore, MD.