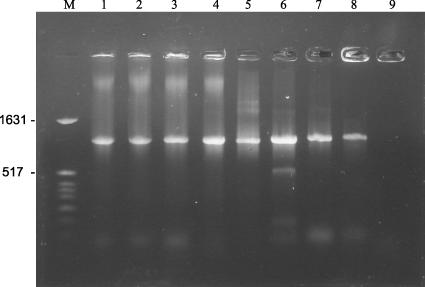
Stephen Marshall and coworkers (1) reported a simple PCR-restriction fragment length polymorphism (RFLP)-based method for the identification and differentiation of Campylobacter, Arcobacter, and Helicobacter spp. by use of crude lysates of cells or purified DNA from these bacteria. The method involved the construction of primers, CAH 16S 1a and CAH 16S 1b, that could amplify a 1,004-bp fragment within the coding region of the 16S rRNA gene in all these bacteria. For differentiation of these bacteria based on the restriction digestion pattern of the 1,004-bp amplicon, three restriction enzymes, namely, DdeI, TaqI, and BsrI, were used for Campylobacter, Arcobacter, and Helicobacter spp., respectively. Each enzyme could give a restriction pattern specific to its respective genus. Out of the three enzymes, TaqI could digest the 1,004-bp PCR amplicon of isolates from the genus Arcobacter only and could not digest that from Campylobacter or Helicobacter. Thus, this method was reported to be useful in the identification and differentiation of isolates belonging to the three said genera and relies on the specificity of the primers reported for the three genera mentioned. In the present study, PCR amplification of the 16S rRNA gene fragment by use of the primer set CAH 16S 1a and CAH 16S 1b was checked for the ability to differentiate various bacterial strains. Campylobacter jejuni, Arcobacter butzleri, and Comamonas aquatica (30 isolates) strains used in this study were isolated from poultry meat samples and identified using 16S rRNA gene sequencing. Salmonella enterica serovar Typhi, Bacillus subtilis, Escherichia coli, and Staphylococcus aureus were obtained from MTCC. When crude lysates of all the bacterial strains were subjected to PCR amplification as the DNA source by use of the primers CAH 16S 1a and CAH 16S 1b, an amplicon of 1,004 bp was obtained for Campylobacter jejuni and Arcobacter butzleri as expected, but similarly sized amplicons were obtained from Salmonella enterica serovar Typhi, Bacillus subtilis, Escherichia coli, and Comamonas aquatica but not from Staphylococcus aureus (Fig. 1). These results are at variance with those reported previously (1), since the primers should have amplified 16S rRNA segments within the three reported genera only. The 1,004-bp amplicon obtained was subjected to complete sequencing, and the results indicated that the amplified segment belonged to 16S rRNA gene segments of the respective genera tested (data not shown). This indicates that the primers reported for this method are not specific to the three genera Campylobacter, Arcobacter, and Helicobacter but also can amplify 16S rRNA gene fragments of bacteria belonging to other genera.
FIG. 1.
PCR product amplified using primers CAH 16S 1a and CAH 16S 1b. Lanes: M, molecular ladder; 1, C. jejuni; 2 to 4, Comamonas sp. isolates; 5, A. butzleri; 6, S. enterica serovar Typhi; 7, E. coli; 8, B. subtilis; 9, S. aureus. The numbers to the left are molecular sizes in kilobases.
It was also reported that this method could be used to identify new species if unique RFLP fingerprints were obtained. The authors did not fail to mention that the potential interspecies variability of this method could be confirmed only after a large number of isolates were tested. However, the nonspecific amplification caused by the primers reported could be a misleading feature, since isolates belonging to other genera may give an RFLP pattern different from that previously reported, indicating new species. In our opinion, the PCR-RFLP method reported cannot be used for identification and differentiation. Since the amplification was found to be nonspecific, we did not carry out RFLP analysis with all restriction enzymes as Marshall et al. did (1). Rather, only the TaqI enzyme was used to check the digestion of the 1,004-bp amplicon obtained from all the bacterial strains subjected to PCR. It was observed that although the amplicon of Campylobacter jejuni was not digested by the restriction enzyme, the amplicons belonging to all other strains were digested and showed different restriction patterns. This indicates that amplicons of strains belonging to genera other than Arcobacter would also show restriction digestion and could lead to their misidentification as Arcobacter spp. strains. An additional exercise was carried out to check the homology of the primer sequence to the genomes of different bacteria by use of NCBI BLAST. The results indicated a homology between primers and 16S rRNA genes of many gram-positive and gram-negative bacteria. This indicates that the primer can bind to 16S rRNA genes of many bacteria other than Campylobacter, Arcobacter, and Helicobacter spp. We therefore state that the primers reported by Stephen Marshall and coworkers (1) are not specific to these three genera. Thus, the use of the reported PCR-RFLP method for crude lysates or DNA of test bacteria should not be considered as the sole method for the identification of Campylobacter, Arcobacter, and Helicobacter spp. The primers designed are not specific to these three genera, and additional identification of test isolates by biochemical and molecular techniques should occur before they are subjected to PCR-RFLP analysis as reported.
Ed. Note: The authors of the published article did not respond.
REFERENCE
- 1.Marshall, S. M., P. L. Melito, D. L. Woodward, W. M. Johnson, F. G. Rodgers, and M. R. Mulvey. 1999. Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J. Clin. Microbiol. 37:4158-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]