Abstract
Meningococcal disease is characterized by cyclic fluctuations in incidence, serogroup distribution, and antigenic profiles. In greater São Paulo, Brazil, there has been a constant increase in the incidence of serogroup C meningococcal disease since the late 1980s. To gain an understanding of changes in serogroup C meningococcal disease over three decades in greater São Paulo, Brazil, 1,059 invasive Neisseria meningitidis serogroup C isolates from 1976 and 2005 were analyzed. Three major clone complexes, sequence type (ST)-11, ST-8, and ST-103, were identified by multilocus sequence typing, and the isolates were characterized by serotyping and 16S rRNA typing. During the 30-year period, there were two major antigenic replacements: from 2a:P1.(5,2) to 2b:P1.3 and subsequently to 23:P1.14-6. All strains of clone ST-103 were characterized as serotype 23 and serosubtype P1.14-6. The origin of 23:P1.14-6 ST-103 complex strains is unknown, but efforts are needed to monitor its spread and define its virulence. The antigenic replacements we observed likely represent a mechanism to sustain meningococcal disease in the population as immunity to circulating strains accumulated.
Neisseria meningitidis is a leading cause of bacterial meningitis and septicemia in children and young adults worldwide (36; http://www.saude.gov.br/sinanweb). In greater São Paulo (GSP), São Paulo State, Brazil, four epidemic periods of meningococcal disease (MD) have been reported and documented in the last 80 years. MD was caused by N. meningitidis serogroup A in the early 1920s and late 1940s: the incidence of the disease was 12 and 24 cases per 100,000 inhabitants in 1923 and 1947, respectively (21). The third epidemic period occurred in the early 1970s, when there was overlap of two epidemics, one caused by N. meningitidis serogroup C that started in 1971 and a larger serogroup A epidemic that started in 1974 (22). After a vaccination campaign with a bivalent AC polysaccharide vaccine that achieved 95% vaccine coverage in GSP in 1975, the incidence of MD fell to 3.9 cases per 100,000 inhabitants in 1977 (8, 21, 22). This reduction in incidence was followed by an increase in incidence of disease caused by N. meningitidis serogroup B in 1977 (26).
During the period from 1978 to 1987, the incidence of MD remained low, at approximately two cases per 100,000, with a remarkable shift from serogroup C to serogroup B by the end of this period; in 1987, 71% of MD cases were caused by serogroup B and only 6.3% were caused by serogroup C (26). In 1988, the incidence of MD in GSP increased again, indicating the beginning of the fourth epidemic period, which extended through 2005. During this 18-year period, MD was mainly caused by (i) serogroup B between 1988 and 1989 (mean incidence of 5.0 cases per 100,000 inhabitants with 80% caused by serogroup B), (ii) serogroup B and serogroup C from 1990 to 2002 (mean incidence of 6.0 cases per 100,000 inhabitants with 59% and 36% caused by serogroup B and serogroup C, respectively), and (iii) serogroup C from 2003 to 2005 (mean incidence of 3.9 cases per 100,000 inhabitants with 65% caused by serogroup C) (16, 25, 26; unpublished official data from the São Paulo State Center for Epidemiologic Surveillance, conveyed by M. I. C. Gonçalves).
A sample of meningococcal isolates from GSP has been previously characterized phenotypically and genotypically (25, 26, 28). These studies have shown that until 1982, nonserotypeable (NT) and nonserosubtypeable (NST) serogroup B strains accounted for most of the serogroup B isolates. These serogroup B NT:NST strains were replaced by serogroup B 4:P1.19,15 strains, which accounted for about 50% to 75% of all serogroup B isolates from 1988 to 2005. Most of these serogroup B 4:P1.19,15 strains are of a single clone which belongs to the multilocus enzyme electrophoresis type (ET)-5 complex (26, 28; unpublished data). In 1990, the number of cases of MD caused by serogroup C strains started to increase, and serogroup C became the most prevalent serogroup from 2003 to 2005. Previous studies have shown that 65% of the serogroup C strains isolated from 1990 to 1996 were 2b:P1.3 (24; unpublished data). By the year 2002 there was a drastic change in serotype (SerT) distribution, and no serogroup C 2b:P1.3 strains were reported. During 2003 to 2005, only one serogroup C 2b:P1.3 strain was isolated, and NT:NST strains were responsible for 89% of all serogroup C cases (25).
Considering the high incidence of MD caused by serogroup C in GSP in the last 3 years (2003 to 2005) and planned widespread use of C conjugate vaccine, characterization of serogroup C isolates is critical to understand the dynamics of the disease in our region and to help in making decisions about the appropriate use and evaluation of this vaccine. The National Reference Center for Meningitis, at the Instituto Adolfo Lutz (IAL), designed a comprehensive study to better understand the characteristics of these serogroup C isolates in GSP, using phenotypic and genotypic approaches to characterize a collection of 1,059 serogroup C isolates recovered during the last 30 years, from 1976 to 2005. To accomplish this goal we (i) expanded the standard serotyping method by adding two novel monoclonal antibodies (MAbs) produced to characterize the serogroup C NT:NST strains and (ii) used molecular methods such as PorB and PorA variable-region (VR) typing, multilocus sequence typing (MLST), and 16S rRNA gene typing on a subset of these 1,059 isolates to determine their genetic relatedness.
MATERIALS AND METHODS
Epidemiologic data and bacterial isolates.
The epidemiological data on MD in GSP were colleted and analyzed at the Alexandre Vranjac Center for Epidemiological Surveillance, São Paulo, Brazil. The population data were obtained from the Brazilian Institute for Geography and Statistics (http://www.ibge.gov.br). The GSP area includes the capital city of São Paulo and 39 counties, which had a total population of 19.1 million in 2005; this constitutes 49% of the state's population, making it the most densely populated area in Brazil. During the period from 1976 to 2005, 20,818 cases of MD were reported in GSP. IAL received 1,059 serogroup C isolates for biochemical identification and serogroup confirmation, and these were used in the present study. Unfortunately, only limited clinical or epidemiological data were available for the 129 cases occurring prior to 1990. Of the 14,686 cases occurring between 1990 and 2005 63.9% (9,387 cases) were confirmed in the laboratory by either (i) culture (44.4%; 4,170 cases), (ii) detection of meningococcal antigens in cerebrospinal fluid or serum by counterimmunoelectrophoresis and/or latex agglutination (27.5%; 2,584 cases), or (iii) identification of the presence of gram-negative diplococci by Gram staining of cerebrospinal fluid (28%; 2,633 cases). Out of the 6,754 cases confirmed in the laboratory by culture or antigen detection, the serogroup was determined for 68.7% (4,640 cases). Of those, serogroup B and serogroup C isolates were responsible for 58.6% (2,721 cases) and 37.4% (1,738 cases), respectively. The remaining 4% of cases were due to serogroups W, Y, and 29E.
Serotyping.
Standard serotyping for all 1,059 isolates was performed by dot blotting using whole-cell suspensions as previously described (42). The term standard serotyping is used in this study to refer to both SerT and serosubtype (SerST) characterization by use of currently available MAbs. The resulting phenotype may describe a fully serotypeable isolate that includes the SerT and SerST (e.g., 2b:P1.3), a partially serotyped isolate for which SerT or SerST is not determined (e.g., 2b:NST or NT:P1.3), and a NT isolate for which both SerT and SerST are not determined (e.g., NT:NST). Standard serotyping was performed with a set of 17 PorB and 14 PorA murine MAbs specific for the VRs. MAbs for SerTs 2a (F12-7B7/1E10), 2b (F1-9H10/1B3), 4 (F10-2H7/1F7), 7 (F22-8B5/1D10), 9 (F24-11F5/3B4), 17 (F4-3C1/1A6), and 10 (F11-6D12/1C5) and for SerSTs P1.1 (F10-5G6/1B11), P1.4 (F11-2A9/1A4), P1.9 (F24-5E11/2H9), P1.15 (F8-8F12/1D6), and P1.22-1 (F4-1F1/1F3) were produced at IAL by us. MAbs for SerTs 8 (2725H6) and 15 (1951C8) and for SerST P1.2 (1649C7) were provided by C. E. Frasch, FDA, Bethesda, MD. MAbs for SerTs 2c (5-1-P2c), 5 (7BG5-H2), 11 (9-1-P11), and 19 (17-1-P19) and for SerSTs P1.3 (12-1), P1.16 (3-1-P1.16), and P1.19 (7A2-11) were provided by W. D. Zollinger, WRAIR, Washington, DC. The MAb for SerT 22 (ATIA5A7/5) was provided by P. Kriz, NIPH, Prague, Czech Republic. MAbs for SerTs 1 (MN3C6B-95/680), 14 (MN5C8C-95/688), and 21 (6B11-F2-B5-95/692); and SerSTs P1.5 (MN22A9.19-95/702), P1.7 (MN14C11.6-95/706), P1.10 (MN20F4.17-95/710), P1.12 (MN20A7.10-95/712), and P1.14 (MN21G3.17-95/716) were provided by the NIBSC, Potters Barr, England. The term expanded serotyping is used in this study to refer to SerT and SerST characterization, with the addition of two novel MAbs produced in this study, 23 and P1.14-6, respectively. These two new MAbs were tested against all 1,059 serogroup C isolates.
OMVs, SDS-PAGE, and immunoblotting.
Outer membrane vesicles (OMVs) were prepared and purified from a serogroup C NT:NST isolate (strain N.753/00), the isolate chosen for MAb production, as previously described (10, 27, 28, 40). This serogroup C spinal fluid isolate was recovered in 2000 from a 17-year-old female in GSP with a fatal case of MD. OMVs were dissolved in suspension buffer (0.1 M NaCl, 0.05 M EDTA, 0.05 M Tris, pH 8.0), and the total amount of protein was determined by the method of Lowry et al. (17). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 15% polyacrylamide minigels (14, 40). Purified OMVs were separated by SDS-PAGE to verify the presence of PorA (class 1) protein and PorB (class 2 or 3) protein and to prepare immunoblots used for screening new MAbs. Immunoblots were performed as previously described (42).
PorA expression on SDS-PAGE gels.
Based on serotyping results, 59 isolates were evaluated for the expression of PorA protein as bands on SDS-PAGE gels after staining by Coomassie brilliant blue R-250. The presence or absence of expression was determined by visual comparison to protein bands of serotyping reference strains H355 and M981, used as controls in each gel to test for the presence or absence of PorA bands, respectively. These same 59 isolates were tested on immunoblots against MAb P1.14-6.
CNBr cleavage of PorA protein.
To determine whether the binding site epitope of the newly produced MAb was located in PorA VR1 or VR2 regions, PorA protein band on SDS-PAGE from the isolate N.753/00 was removed and cleaved by use of cyanogen bromide (CNBr) as previously described (10, 35, 43). CNBr splits PorA at the single methionine located between VR1 and VR2, resulting in VR1 and VR2 being within two fragments of different sizes (about 16 and 25 kDa, respectively). These fragments were separated on SDS-PAGE and transferred to a nitrocellulose membrane (40). MAb reactivity with these two fragments was detected by immunoblotting.
MAb production.
The method used for MAb production was described previously (11). BALB/c mice were immunized intraperitoneally three times a week for 4 weeks with whole-cell suspensions of the isolate N.753/00. Fusion with P3UI murine myeloma cells was performed 3 days after the final immunization. MAbs obtained were screened by immunoblotting 10 to 15 days after fusion.
PorB and PorA VR typing.
For PorB VR typing the full length of the porB gene, which includes VR1 to VR4, was sequenced. For PorA VR typing only two regions of the porA gene, which include VR1 and VR2, were sequenced (29, 30). A total of 23 and 122 serogroup C isolates were selected based on serotyping results from PorB and PorA VR typing, respectively, obtained in the present study. In the present study, we have used PorA and PorB VR classification and designation as previously described (28; http://neisseria.org/nm/typing/).
MLST and 16S rRNA gene typing.
To determine the genetic relatedness among serogroup C isolates, we randomly selected 30% (321 of 1,059) of the isolates for MLST and 16S rRNA gene typing. MLST was performed according to the methods of Maiden et al. (18). The designation of sequence types (STs) is described on the MLST web site (http://pubmlst.org). STs were considered to belong to the same clonal complex if they share alleles at four or more of the seven MLST loci with the founder ST of that complex. Designation of N. meningitidis clones may include the MLST result plus its corresponding multilocus enzyme electrophoresis result, for example, ST-11 complex/ET-37 complex. 16S rRNA gene typing was carried out as previously described (32). In the present study, the term 16S rRNA complex was defined as a group of 16S rRNA sequences types with ≤5 nucleotide differences (∼99.7% similarity) among them.
Nucleotide sequence accession numbers.
For the isolate N.753/00 (isolate chosen for MAb production) the full lengths of the porA and porB genes were sequenced and submitted to the GenBank database with assigned accession numbers AF737715 and AY972814, respectively.
RESULTS
Standard serotyping.
The prevalence of SerTs, SerSTs, and the various combinations used with the standard serotyping MAb panel are shown in Table 1. Out of the 1,059 isolates 617 (58.3%) were serotypeable, and 559 (52.8%) were serosubtypeable. The most prevalent SerTs were 2b (n = 398; 37.6%), and 2a (n = 151; 14.2%); all other SerTs occurred in fewer than 3% of isolates. The most prevalent SerSTs were P1.3 (n = 286; 27.0%), P1.2 (n = 80; 7.5%, P1.5 (n = 58; 5.5%), and P1.5,2 (n = 55; 5.2%); all other SerSTs occurred in fewer than 3% of isolates. The most prevalent combination was 2b:P1.3 (n = 283; 26.7%); all other phenotypes presented less than 7% prevalence (Table 1).
TABLE 1.
Distribution of serotypes and serosubtypes of N. meningitidis serogroup C isolated from 1976 to 2005 in GSP, São Paulo, Brazil, using the standard set of serotyping MAbsa
| SerT | No. of strains with indicated SerST
|
Total no. of strains | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1.1 | P1.2 | P1.3 | P1.5 | P1.5,2 | P1.5,10 | P1.7 | P1.7,1 | P1.9 | P1.10 | P1.12 | P1.15 | P1.16 | P1.19,15 | P1.22-1,14 | NST | ||
| 2a | 69 | 1 | 20 | 47 | 1 | 1 | 12 | 151 | |||||||||
| 2b | 8 | 283 | 19 | 3 | 1 | 23 | 2 | 59 | 398 | ||||||||
| 4 | 1 | 1 | 9 | 3 | 5 | 19 | |||||||||||
| 4,1 | 1 | 1 | |||||||||||||||
| 4,7 | 1 | 1 | 2 | 1 | 15 | 3 | 23 | ||||||||||
| 4,10 | 1 | 1 | 2 | ||||||||||||||
| 4,21 | 3 | 1 | 4 | ||||||||||||||
| 10 | 1 | 1 | 2 | ||||||||||||||
| 15 | 1 | 1 | |||||||||||||||
| 17,10 | 1 | 1 | |||||||||||||||
| 19,1 | 1 | 1 | |||||||||||||||
| 19,7 | 1 | 1 | |||||||||||||||
| 19,10 | 1 | 1 | 1 | 3 | |||||||||||||
| 19,14 | 1 | 2 | 2 | 1 | 2 | 8 | |||||||||||
| 21 | 1 | 1 | 2 | ||||||||||||||
| NT | 1 | 2 | 1 | 16 | 3 | 3 | 1 | 1 | 1 | 1 | 412 | 442 | |||||
| Total | 1 | 80 | 286 | 58 | 55 | 1 | 4 | 4 | 7 | 24 | 1 | 12 | 1 | 20 | 5 | 500 | 1,059 |
The standard set of serotyping MAbs is described in Materials and Methods. It does not include MAbs 23 and P1.14-6, described in this study.
New MAb development, production, and characterization.
From the cell fusion two hybridomas, F291G1B4 and F298H11E11, were selected. Each one of the hybridomas produces MAbs that recognize PorB of class 2 or PorA protein of the isolate N.753/90 on immunoblots containing 0.25% of Empigen BB to restore the antibody binding activity (41). The porB gene sequence of isolate N.753/00 (the isolate chosen for MAb production) translates a PorB class 2 protein with VR1 type D, VR2 type Ed, VR3 type D, and VR4 type Db according to the proposed classification of PorB VR typing (29). In our tests, the F291G1B4 MAb did not react with any of the 17 different SerT reference strains. When this lack of reactivity was compared to previously described PorB VR types and the PorB VR types for the N.753/00 isolate, PorB VR3 type D was the only possible epitope location for the new MAb. Based on the sequential numerical order of the serotyping MAbs available, the new MAb was named 23. The porA gene sequence of isolate N.753/00 translates to a PorA protein with VR1 type 22 and VR2 type 14-6 (P1.22,14-6) (http://neisseria.org/nm/typing/). In our tests, the new F298H11E11 MAb did not react with any of the 14 different SerST reference strains. When this lack of reactivity was compared to that for previously described PorA VR types and the PorA VR types of the N.753/00 isolate, PorA VR2 type 14-6 was shown to be the only possible epitope location for the new MAb. In addition, PorA VR2 as the epitope location for this MAb is suggested by the binding of the F298H11E11 MAb to CNBr-cleaved PorA. The new MAb reacted on immunoblots with the 25-kDa fragment containing VR2 14-6 of the N.753/00 isolate. Based on these results, the new MAb was named P1.14-6.
Expanded serotyping.
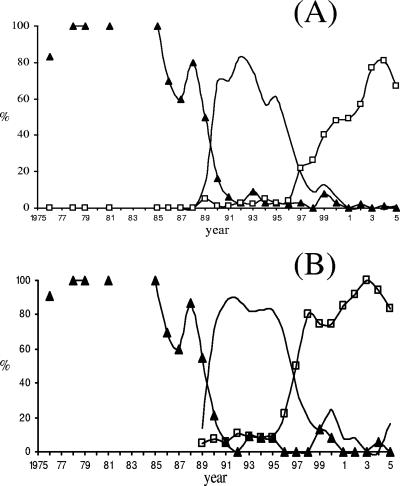
All 1,059 serogroup C isolates were screened with MAbs 23 and P1.14-6 by dot blotting. These MAbs did not react with any of the 617 or 559 isolates with previously characterized SerT or SerST, respectively. MAb 23 reacted with 419 (94.8%) of the 442 previously NT isolates, and MAb P1.14-6 reacted with 329 (65.8%) of the 500 previously NST isolates, increasing the percentages of serotypeable and serosubtypeables isolates to 97.8% and 83.9%, respectively (Table 2). Twenty-three (2.2%) and 171 (16.1%) isolates remained NT and NST, respectively. The most prevalent combinations were 23:P1.14-6 (n = 320; 30.2%), and 2b:P1.3 (n = 283; 26.7%); all other phenotypes presented less than 7% prevalence. Three hundred twenty of the 419 (76.4%) isolates of SerT 23 were also of SerST P1.14-6, and 320 of the 329 (97.3%) isolates of SerST P1.14-6 were also of SerT 23. The annual distribution of the most prevalent phenotypes is presented on Fig. 1A.
TABLE 2.
Comparison of phenotypes obtained using the standard and expanded sets of serotyping MAbsa
| Phenotype | Standard set
|
Expanded set
|
||
|---|---|---|---|---|
| No. of isolates | % | No. of isolates | % | |
| SerT | 617 | 58.3 | 1,036 | 97.8 |
| NT | 442 | 41.7 | 23 | 2.2 |
| SerST | 559 | 52.8 | 888 | 83.9 |
| NST | 500 | 47.2 | 171 | 16.1 |
| SerT:SerST | 529 | 45.6 | 869 | 82.1 |
| SerT:NST | 88 | 8.7 | 167 | 15.8 |
| NT:SerST | 30 | 2.6 | 18 | 1.7 |
| NT:NST | 412 | 43.1 | 5 | 0.5 |
The term expanded serotyping is used in this study to refer to standard serotyping including use of the two new MAbs produced in this study, MAbs 23 and P1.14-6.
FIG. 1.
Thirty-year historical distribution of the three major serogroup C clones circulating in GSP from 1976 to 2005. (A) Phenotypic characterization. Line with triangles, phenotype 2a:P1.(5,2); line, phenotype 2b:P1.3; line with squares, phenotype 23:P1.14-6. (B) Genotypic characterization. Line with triangles, ST-11/16S-13 complex; line, ST-8/16S-91 complex; line with squares, ST-103/16S-17 complex. Disconnected solid triangles indicate that there were no cases in that particular year.
NT and NST isolates.
To investigate the lack of reactivity of the new MAbs with the remaining 23 NT and 171 NST isolates, we selected all 23 NT isolates for PorB VR typing and 122 (71%) of the 171 NST isolates for PorA VR typing. As determined by DNA sequencing, 12 of the 23 NT isolates did not have PorB VR3 type 23, and for the remaining 11 isolates we were not able to amplify the porB gene with the external primers used (29). Out of the 122 NST isolates, 59 (48.3%) had PorA VR2 type P1.14-6, 49 (40.2%) had PorA types other than P1.14-6 for which there are no currently available MAbs, and for the remaining 14 (11.5%) isolates we were not able to amplify the porA gene. Out of the 59 NST isolates with PorA VR type P1.14-6, only 10 expressed PorA on SDS-PAGE gels and reacted with MAb P1.14-6 on immunoblots; this is a typical characteristic of some already-described PorA masked epitopes (44).
MLST and 16S rRNA gene typing.
To investigate the clonal characteristics of serogroup C in GSP, we randomly selected 321 (30%) of the 1,059 isolates for MLST and 16S rRNA gene typing (Table 3). Seventeen different STs were found; they were grouped into seven different clonal complexes (ST-11 complex [n = 115], ST-8 complex [n = 84], ST-103 complex [n = 102], ST-32 complex [n = 8], ST-5 complex [n = 1], ST-41/44 complex [n = 7], and ST-231 complex [n = 4]) (Table 3). Twelve different 16S rRNA gene types were found; they were grouped into seven different 16S complexes (4, 5, 13, 17, 25, 30, and 91) (Table 3). The annual distribution of the most prevalent genotypes is presented on Fig. 1B.
TABLE 3.
Correlation between 16S rRNA typing, MLST, and serotyping results on 321 N. meningitidis isolates
| 16S rRNA typing | MLST
|
SerT | SerST | No. of isolates | ||
|---|---|---|---|---|---|---|
| Complex | Type | Complex | Type | |||
| 13 | 13 | ST-11 complex/ET-37 complex | 11 | 2a | P1.2 | 37 |
| P1.5 | 18 | |||||
| P1.5,2 | 9 | |||||
| P1.9 | 1 | |||||
| NST (P1.5,2)a | 3 | |||||
| 574 | 2a | P1.2 | 1 | |||
| 3323 | NT (2a)b | P1.5,2 | 1 | |||
| 245 | 5121 | 2a | P1.2 | 9 | ||
| P1.5,2 | 34 | |||||
| NST (P1.5,2)a | 1 | |||||
| NT (2a)b | P1.5,2 | 1 | ||||
| 91 | 91 | ST-8 complex/cluster A4 | 8 | 2b | P1.2 | 1 |
| P1.3 | 69 | |||||
| P1.5 | 3 | |||||
| P1.5,2 | 1 | |||||
| NST (P1.3)a | 5 | |||||
| 153 | 2b | P1.10 | 2 | |||
| 131 | 153 | 2b | P1.10 | 2 | ||
| 242 | 8 | 2b | P1.3 | 1 | ||
| 17 | 17 | ST-103 complex | 3779 | 23 | P1.5 | 2 |
| P1.14-6 | 37 | |||||
| NST (P1.14-6)a | 4 | |||||
| 3780 | 23 | P1.14-6 | 29 | |||
| 5122 | 23 | P1.14-6 | 27 | |||
| NST (P1.14-6)a | 2 | |||||
| 5123 | 23 | P1.14-6 | 1 | |||
| 4 | 4 | ST-32 complex/ET-5 complex | 34 | 4,7 | P1.16 | 1 |
| 33, 34, 639 | P1.19,15 | 5 | ||||
| 34 | NST | 1 | ||||
| 9 | 34 | 4,21 | P1.12 | 1 | ||
| 5 | 5 | ST-5 complex/subgroup III | 203 | 4 | P1.9 | 1 |
| 25 | 41 | ST-41/44 complex/lineage 3 | 1362 | 19.1 | NST | 1 |
| 2a | P1.22-1,14 | 1 | ||||
| 19,14 | P1.22-1,14 | 1 | ||||
| 246 | 2042 | 10 | P1.9 | 1 | ||
| NST | 1 | |||||
| 17,1 | P1.9 | 1 | ||||
| 19,1 | NST | 1 | ||||
| 30 | 30 | ST-231 complex | 1011 | NT | P1.7,1 | 3 |
| NST | 1 | |||||
| Total | 321 | |||||
The PorA VR type is indicated by parentheses.
The PorB VR type is indicated by parentheses.
DISCUSSION
Phenotypic and molecular studies on N. meningitidis are essential for understanding the events associated with the epidemiological spread of this organism in the population and the temporal variation in disease incidence and severity. Some of the genetic characteristics of serogroup C isolated in GSP between 1976 and 1990 have been previously documented. Those results show that the majority of serogroup C isolates from 1976 to 1988 were SerT 2a, which was replaced by SerT 2b in 1989 and 1990 (25). Most of the serogroup C 2a and serogroup C 2b isolates in that study belong to two distinct complexes of clones defined by multilocus enzyme electrophoresis as ET-11 and ET-8, respectively (25). However, that study did not correlate those complexes with previously described ETs, making it (i) difficult to compare those findings with any other published data on clonal distribution of serogroup C isolates and (ii) impossible to understand their clonal relatedness. The present study was designed to fill that gap by providing enough phenotypic and genotypic data to generate a comprehensive historical clonal structure of serogroup C strains isolated in GSP, Brazil, since the end of the third epidemic period in 1976.
Since 41.7% and 47.2% of the serogroup C isolates between 1976 and 2005 were NT and NST, respectively, our first step was to expand the standard set of serotyping MAbs, which we accomplished by producing two additional MAbs to characterize the PorB and PorA of those NT and NST isolates. With the expanded set of MAbs the percentages of NT and NST isolates were reduced to 2.2% and 16.1%, respectively. The percentage of fully serotypeable strains increased from 45.6% to 82.1%, reducing the number of NT:NST strains to only 0.5%.
The percentage of fully serotypeable strains was not higher because of the presence of (i) strains with masked PorA P1.14-6 epitope, (ii) strains with no detectable expression of PorA type P1.14-6 epitope, and (iii) strains expressing PorA epitopes for which there are no currently available MAbs. Large amounts of capsular polysaccharide or lipopolysaccharide in a particular strain can mask PorA protein and could explain the inability of some MAbs to bind to its epitope on dot blots (31). The lack of expression of PorA in some isolates may be explained by phase variation of PorA due to variations in the length of homopolymeric tracts of G or T residues in the putative −35 and −10 domains of the porA promoter region, or by length variation of a homopolimeric A tract in the porA coding region (37, 39). PorA expression may also be affected by other genetic events, such as mutations, the presence of a premature stop codon, or the presence of an insertion element, such as insertion sequence 1301, in the porA coding region (3, 23, 38, 39). In this study we did not attempt to further investigate the reasons for the lack of PorA expression.
Genotypic characterization by MLST and 16S rRNA gene typing were 100% in agreement in assigning these isolates into complexes. Genotypic and phenotypic results were also in agreement, and when combined, they revealed that 94% of the 1,059 serogroup C strains circulating in GSP over the last 30 years belong to three major clones: 2a:P1.(5,2) (including the three combinations P1.5, P1.2, and P1.5,2) ST-11 and 16S-13 complexes, 2b:P1.3 ST-8 and 16S-91 complexes, and 23:P1.14-6 ST-103 and 16S-17 complexes (Fig. 1B).
The serogroup C isolates responsible for numerous epidemics and outbreaks in the United States, Canada, and Europe since the early 1990s have generally belonged to the ST-11 complex. These hyperinvasive stains have predominantly affected adolescents and young adults, requiring frequent, massive public health investigations and interventions (7, 45). In the ST-11 clonal complex, PorA and PorB proteins have been fairly homogeneous, with most of the ST-11 complex strains having the 2a:P1.(5,2) phenotype (7). These phenotypes are usually associated with serogroup C and W135 isolates but are quite uncommon on serogroup Y and B strains isolated during the last 30 years in GSP (unpublished data).
In our studies, 2a:P1.(5,2) ST-11 complex strains have been isolated in GSP since 1976, and although there are no serogroup C strains isolated before 1976 for comparison, 2a ET-11 complex strains have been accepted as being responsible for the third epidemic, which reached an incidence of 17.7 cases in 1972 (7, 22, 25). After the mass vaccination campaign in GSP in 1975 the number of serogroup C cases fell dramatically, and serogroup B 4,7:P1.19,15 ET-5 complex strains were responsible for over 70% of the cases during the period from 1977 to 1989.
Evidence of horizontal transfer of siaD genes encoding polysialyltransferases has been shown to result in capsular serogroup switching in vitro and in vivo (2, 9, 13, 34). Capsular switching may occur naturally, and immunization with vaccines that target serogroup capsules could potentially favor the emergence of serogroup B ST-11 complex strains as reported in Canada and Spain, where serogroup B ST-11 complex strains have emerged since the introduction of serogroup C conjugate vaccines (1, 24, 33). In GSP, only two serogroup B 2a:P1.5,2 ET-5 complex strains were found, 16 and 20 years after the AC vaccination campaign, when similar prevalence of MD caused by serogroups B and C was reported (26). Based on these results, it seems like the immunization campaign targeting serogroup A and C capsules in GSP had little or no selective effect on capsule switching from serotype C to serotype B. However, we found five serogroup C 4,7:P1.19,15 ST-32 complex isolates in this study that could have emerged by a capsular switch from serogroup B to serogroup C. These five strains were isolated during a period of increasing prevalence of serogroup C in the years 1987, 1989, and 1997, and two strains were isolated in 2001, when serogroup C prevalence was higher than that of serogroup B. It is important to note that in GSP vaccination for serogroup C was performed using a plain polysaccharide vaccine that should have minor influence compared to conjugate vaccines in selection of capsule switching events, since the first type of vaccine does not affect carriers as the second one does.
The second most prevalent serogroup C clone found in GSP belongs to the ST-8 complex and has the phenotype/genotype 2b:P1.3. ST-8 complex strains have also been associated with epidemics and hyperendemic waves of disease. This clone may be related to serogroup B and C organisms generally characterized phenotypically as 2b:P1.10 (7). ST-8 complex strains have been a common cause of disease in the 1970s in the United States, Canada, the United Kingdom, Iceland, and many other European countries, and was responsible for a severe serogroup B epidemic in Cape Town, South Africa, that started in 1979 (4, 5, 6). But in contrast to these reports, serogroup C ST-8 complex strains in GSP have the peculiar phenotype 2b:P1.3, which has never been reported elsewhere as being epidemic associated (25).
Serogroup C strains isolated from areas where the disease is endemic or from outbreaks usually belongs to the ST-11 or ST-8 complex (19). Our data show that strains from another MLST complex, the ST-103 complex, have been present in GSP since 1989 and became associated with the serogroup C epidemic in the 2000s. According to the MLST website (http://neisseria.org/nm/typing/mlst) 70 different STs are included in the ST-103 complex, with a total of 84 strains isolated in 17 different countries in Europe, North and South America, and Asia. We found only four different STs among serogroup C ST-103 complex strains isolated in GSP. Two of them (ST-3779 and ST-3780) have been previously described for four serogroup C strains isolated in other Brazilian states in 2003 and 2004 (http://neisseria.org/nm/typing/mlst). The other two STs (ST-5122 and ST-5123) are newly described in this study. The four serogroup C strains isolated in other Brazilian states were not available for this study, but they are described as having the PorA VR type P1.22,14-6, the same PorA VR type found in ST-103 complex strains isolated in GSP. These data suggest that serogroup C epidemic-related ST-103 complex strains circulating in GSP are also present in other Brazilian states, and additional studies are needed to evaluate the extent of this dissemination.
Characterization of N. meningitidis strains by serogrouping and serotyping alone may not reveal the pathogenic potential and clonal structure of the circulating strains in a defined area. Lately, MLST has been used to recognized virulent clonal groups such as ST-11, ST-8, ST-103, ST-32, and others. This study shows that methods that are more affordable and easier to perform, like serotyping, may provide information similar to that obtained using more laborious and expensive genotyping methods based on DNA sequencing (Fig. 1). Recently, the value of serotyping has been documented in the emergence of serogroup B 17:P1.19 of ST-269 in Québec, Canada, during 2004 and 2005 (15). When used together, phenotypic and genotypic methods may also complement each other, as shown with the serogroup W135 Hajj outbreak investigation in 2000 in Saudi Arabia (20). Consequently, the choice of subtyping methods to be used in a particular investigation may be based on the type and extent of individual outbreaks and on each laboratory's requirements and capacities.
This study shows that in the last 30-year period, there was a sequential appearance of three major serogroup C clones circulating in GSP, and they were related to distinct epidemiological periods. These three clones could be equally characterized by serotyping, 16S rRNA typing, or MLST. This is the first report of serogroup C of ST-103 complex strains associated with an epidemic situation. All strains belonging to this complex that have been reported to date have particular PorB and PorA variants that can be characterized using the MAbs produced in this study as 23 and P1.14-6. The strain replacements demonstrated in this study, from 2a:P1.(5,2) to 2b:P1.3 and subsequently to 23:P1.14-6, may render populations susceptible to infection, and combined with the high virulence attributed to the circulating strains may explain the constant increase in incidence of MD caused by serogroup C strains in GSP since the late 1980s.
A similar phenomenon was recently described in Spain when, during 1996 and 1997, an epidemic wave of MD caused by serogroup C 2b:P1.5,2 showed displacement of the ET-37 cluster by strains belonging to the A4 cluster (2). The observed antigenic changes occurring with strains of different genetic lineages in both situations suggested clonal replacement rather than true antigenic shift.
A study in which an increasing incidence of disease caused by serogroups C and Y was associated with major changes in the outer membrane protein antigenic profile was recently carried out in Maryland (12). In that setting, antigenic profile changes occurred within strains of the same genetic backbone, namely ST-11 for serogroup C and ST-23 for serogroup Y, suggesting that the antigenic changes were mediated by horizontal gene transfer. In contrast, in the present study, we observed antigenic changes occurring with strains of different genetic lineages, suggesting strain replacement rather than true antigenic shift. Taken together, it appears that both strain replacement and antigenic shift are mechanisms used by N. meningitidis to maintain the ability to cause invasive disease in populations.
The origin of the serogroup C 23:P1.14-6 ST-103 complex is unknown, but efforts are needed to monitor it spread and define its virulence, as well as to understand the possible effects of the upcoming use of serogroup C conjugate vaccine on the dissemination and evolution of this clone in GSP. This will require ongoing public health surveillance for MD and phenotypic and genotypic characterization of invasive meningococcal isolates.
Acknowledgments
We thank Jonas Umeoka Yamauchite for technical assistance on identification and serogrouping of the isolates, the hospitals of GSP for providing the meningococcal isolates, Telma Carvalhana Marques and Helena Aparecida Barbosa for providing the epidemiological data, Martha Tanizaki for helping with the CNBr cleavage of the PorA protein, and Leonard Mayer and Lee Harrison for excellent suggestions and critical review of the manuscript.
This study was supported in part by a Fogarty International Center Global Infectious Diseases Research Training Program grant from the National Institutes of Health (5D43TW006592).
Footnotes
Published ahead of print on 21 February 2007.
REFERENCES
- 1.Alcalá, B., L. Arreaza, C. Salcedo, M. J. Uría, L. De La Fuente, and J. A. Vázquez. 2002. Capsule switching among C:2b:P1.2,5 meningococcal epidemic strains after mass immunization campaign, Spain. Emerg. Infect. Dis. 8:1512-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcalá, B., C. Salcedo, L. Arreaza, S. Berrón, L. De La Fuente, and J. A. Vázquez. 2002. The epidemic wave of meningococcal disease in Spain in 1996-1997: probably a consequence of strain displacement. J. Med. Microbiol. 51:1102-1106. [DOI] [PubMed] [Google Scholar]
- 3.Arhin, F. F., F. Moreau, J. W. Coulton, and E. Mills. 1998. Sequencing of porA from clinical isolates of Neisseria meningitidis defines a subtyping scheme and its genetic regulation. Can. J. Microbiol. 44:56-63. [PubMed] [Google Scholar]
- 4.Caugant, D. A., L. F. Mocca, C. E. Frasch, L. O. Froholm, W. D. Zollinger, and R. K. Selander. 1987. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J. Bacteriol. 169:2781-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caugant, D. A., W. D. Zollinger, L. F. Mocca, C. E. Frasch, T. S. Whittam, L. O. Froholm, and R. K. Selander. 1987. Genetic relationships and clonal population structure of serotype 2 strains of Neisseria meningitidis. Infect. Immun. 55:1503-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caugant, D. A., P. Bol, E. A. Hoiby, H. C. Zanen, and L. O. Froholm. 1990. Clones of serogroup B Neisseria meningitidis causing systemic disease in the Netherlands, 1958-1986. J. Infect. Dis. 162:867-874. [DOI] [PubMed] [Google Scholar]
- 7.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106:505-525. [PubMed] [Google Scholar]
- 8.Fonseca, C., J. C. Moraes, and R. B. Barata. 2004. O livro da meningite: uma doença sob a luz da cidade. Segmento Farma, São Paulo, SP, Brazil.
- 9.Frosch, M., and T. E. Meyer. 1992. Transformation-mediated exchange of virulence determinants by co-cultivation of pathogenic Neisseriae. FEMS Microbiol. Lett. 79:345-349. [DOI] [PubMed] [Google Scholar]
- 10.Hames, B. D. 1990. One-dimensional polyacrilamide gel electrophoresis, p. 1-147. In B. D. Hames and D. Rickwood (ed.), Gel electrophoresis of proteins: a practical approach, 2nd ed. IRL Press, Oxford, United Kingdom.
- 11.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual, p. 139-318. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 12.Harrison, L. H., K. A. Jolley, K. A. Shutt, J. W. Marsh, M. O'Leary, L. T. Sanza, and M. C. J. Maiden. 2006. Antigenic shift and increased incidence of meningococcal disease. J. Infect. Dis. 193:1266-1274. [DOI] [PubMed] [Google Scholar]
- 13.Kertesz, D. A., M. B. Coulthart, J. A. Ryan, W. M. Johnson, and F. E. Ashton. 1998. Serogroup B, eletrophoretic type 15 Neisseria meningitidis in Canada. J. Infect. Dis. 177:1754-1757. [DOI] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Law, D. K. S., M. Lorange, L. Ringuette, R. Dion, M. Giguère, A. M. Henderson, J. Stoltz, W. D. Zollinger, P. De Wals, and R. S. W. Tsang. 2006. Invasive meningococcal disease in Québec, Canada, due to an emerging clone of ST-269 serogroup B meningococci with serotype antigen 17 and serosubtype antigen P1.19 (B:17:P1.19). J. Clin. Microbiol. 44:2743-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemos, A. P. S., A. P. Brandão, M. C. O. Gorla, M. V. Paiva, V. Simonsen, and C. E. A. Melles. 2006. Phenotypic characterization of Neisseria meningitidis strains isolated from invasive disease in Brazil from 1990 to 2001. J. Med. Microbiol. 55:751-757. [DOI] [PubMed] [Google Scholar]
- 17.Lowry, O. H., J. N. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 18.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russel, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiden, M. C. J. 2002. Population structure of Neisseria meningitidis, p. 150-169. In C. Ferreirós, M. T. Criado, and J. A. Vasquez (ed.), Emerging strategies in the fight against meningitis: molecular and cellular aspects. Horizon Scientific Press, Wymondham, United Kingdom.
- 20.Mayer, L. W., M. W. Reeves, N. Al-Hamdan, C. T. Sacchi, M. K. Taha, G. W. Ajello, S. E. Schimink, C. A. Noble, M. L. C. Tondella, A. W. Whitney, Y. Al-Mazrou, M. Al-Jefri, A. Mishkhis, S. Sabban, D. A. Caugant, J. Lingappa, N. Rosenstein, and T. Popovic. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but expansion within the electrophoretic type-37 complex. J. Infect. Dis. 185:1596-1605. [DOI] [PubMed] [Google Scholar]
- 21.Moraes, J. C., and R. B. Barata. 2005. Meningococcal disease in São Paulo, Brazil, in the 20th century: epidemiological characteristics. Cad. Saúde Pública 21:1458-1471. [DOI] [PubMed] [Google Scholar]
- 22.Morais, J. S., R. S. Munford, J. B. Risi, E. Antezana, and A. Feldman. 1974. Epidemic disease due to serogroup C Neisseria meningitidis in São Paulo, Brazil. J. Infect. Dis. 129:568-571. [DOI] [PubMed] [Google Scholar]
- 23.Newcombe, J., K. Cartwright, S. Dyer, and J. McFadden. 1998. Naturally occurring insertional inactivation of the porA gene of Neisseria meningitidis by integration of IS1301. Mol. Microbiol. 30:453-454. [DOI] [PubMed] [Google Scholar]
- 24.Pérez-Trallero, E., D. Vicente, M. Montes, and R. Cisterna. 2002. Positive effect of meningococcal C vaccination on serogroup replacemet in Neisseria meningitidis. Lancet 360:953. [DOI] [PubMed] [Google Scholar]
- 25.Sacchi, C. T., R. C. Zanella, D. A. Caugant, C. E. Frasch, N. T. Hidalgo, L. G. Milagres, L. L. Pessoa, S. R. Ramos, M. C. C. Camargo, and C. E. A. Melles. 1992. Emergence of a new clone of serogroup C Neisseria meningitidis in São Paulo, Brazil. J. Clin. Microbiol. 30:1282-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacchi, C. T., L. L. Pessoa, S. R. Ramos, L. G. Milagres, M. C. C. Camargo, N. T. Hidalgo, C. E. A. Melles, D. A. Caugant, and C. E. A. Frasch. 1992. Ongoing group B Neisseria meningitidis epidemic in São Paulo, Brazil, due to increased prevalence of a single clone of the ET-5 complex. J. Clin. Microbiol. 30:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sacchi, C. T., A. P. S. Lemos, M. C. O. Gorla, and C. E. Frasch. 1995. Monoclonal antibody to serotype 17 of Neisseria meningitidis and their prevalence in Brazilian states. Rev. Inst. Med. Trop. São Paulo 37:1-5. [DOI] [PubMed] [Google Scholar]
- 28.Sacchi, C. T., A. P. S. Lemos, M. C. C. Camargo, A. M. Whitney, C. E. A. Melles, C. A. Solari, C. E. Frasch, and L. W. Mayer. 1998. Meningococcal disease caused by Neisseria meningitidis serogroup B serotype 4 in São Paulo, Brazil, 1990 to 1996. Rev. Inst. Med. Trop. São Paulo 40:65-70. [DOI] [PubMed] [Google Scholar]
- 29.Sacchi, C. T., A. P. S. Lemos, A. M. Whitney, C. A. Solari, M. E. Brandt, C. E. A. Melles, C. E. Frasch, and L. W. Mayer. 1998. Correlation between serological and sequencing analysis of the PorB outer membrane protein in the Neisseria meningitidis serotyping system. Clin. Diagn. Lab. Immunol. 5:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sacchi, C. T., A. P. S. Lemos, M. E. Brandt, A. M. Whitney, C. E. A. Melles, C. A. Solari, C. E. Frasch, and L. W. Mayer. 1998. Proposed standardization of Neisseria meningitidis PorA variable region typing nomenclature. Clin. Diagn. Lab. Immunol. 5:845-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sacchi, C. T., A. M. Whitney, T. Popovic, D. S. Beall, M. W. Reeves, B. D. Plikaytis, N. Rosenstein, B. A. Perkins, M. L. C. Tondella, and L. W. Mayer. 2000. Diversity and prevalence of PorA types in Neisseria meningitidis serogroup B in the United Sates, 1992-1998. J. Infect. Dis. 182:1169-1176. [DOI] [PubMed] [Google Scholar]
- 32.Sacchi, C. T., A. M. Whitney, M. W. Reeves, L. W. Mayer, and T. Popovic. 2002. Sequence diversity of Neisseria meningitidis 16S rRNA genes and use of 16S rRNA gene sequencing as a molecular subtyping tool. J. Clin. Microbiol. 40:4520-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stefanelli, P., C. Fazio, A. Néri, T. Sofia, and P. Mastrantonio. 2003. First report of capsule replacement among electrophoretic type 37 Neisseria meningitidis strains in Italy. J. Clin. Microbiol. 41:5783-5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Swartley, J. S., A. A. Marfin, S. Edupuganti, L. J. Liu, P. Cieslak, B. Perkins, J. D. Wenger, and D. S. Stephens. 1997. Capsule switching of Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 94:271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai, C. M., and C. E. Frasch. 1980. Chemical analysis of major outer membrane proteins of Neisseria meningitidis: comparison of serotypes 2 and 11. J. Clin. Microbiol. 141:169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tzeng, Y. L., and D. S. Stephens. 2000. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2:687-700. [DOI] [PubMed] [Google Scholar]
- 37.van der Ende, A., C. T. P. Hopman, S. Zaat, B. D. O. Essink, B. Berkhout, and J. Dankert. 1995. Variable expression of class 1 outer membrane protein in Neisseria meningitidis is caused by variation in the spacing between the −10 and −35 regions of the promoter. J. Bacteriol. 177:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van der Ende, A., C. T. P. Hopman, and J. Dankert. 1999. Deletion of PorA recombination between clusters of repetitive extragenic palindromic sequences in Neisseria meningitidis. Infect. Immun. 67:2928-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Ende, A., C. T. P. Hopman, and J. Dankert. 2000. Multiple mechanisms of phase variation of PorA in Neisseria meningitidis. Infect. Immun. 68:6685-6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wedege, E., and L. O. Froholm. 1986. Human antibody response to a group B serotype 2a meningococcal vaccine determined by immunoblotting. Infect. Immun. 51:571-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wedege, E., K. Bryan, and L. O. Froholm. 1988. Restoration of antibody binding to blotted meningococcal outer membrane proteins using various detergents. J. Immunol. Methods 113:51-59. [DOI] [PubMed] [Google Scholar]
- 42.Wedege, E., E. A. Hoiby, E. Rosenqvist, and L. O. Froholm. 1990. Serotyping and subtyping of Neisseria meningitidis isolates by co-agglutination, dot-blotting and ELISA. J. Med. Microbiol. 31:195-201. [DOI] [PubMed] [Google Scholar]
- 43.Wedege, E., D. A. Caugant, A. Musacchio, N. B. Saunders, and W. D. Zollinger. 1990. Redesignation of a purported P1.15 subtype-specific meningococcal monoclonal antibody as a P1.19-specific reagent. Clin. Diagn. Lab. Immunol. 6:639-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wedege, E., R. Dalseg, D. A. Caugant, J. T. Poolman, and L. O. Froholm. 1993. Expression of an inaccessible P1.7 subtype epitope on meningococcal class 1 protein. J. Med. Microbiol. 38:23-28. [DOI] [PubMed] [Google Scholar]
- 45.Whalen, C. M., J. C. Hockin, A. Ryan, and F. Ashton. 1995. The changing epidemiology of invasive meningococcal disease in Canada, 1985 through 1992. Emergence of a virulent clone of Neisseria meningitidis. JAMA 273:390-394. [PubMed] [Google Scholar]