Abstract
We surveyed methicillin-resistant coagulase-positive staphylococcus (MRCPS) strains from 57 (26 inpatient and 31 outpatient) dogs and 20 veterinary staff in a veterinary teaching hospital. From the staff, three MRCPS strains were isolated, and two were methicillin-resistant Staphylococcus aureus (MRSA). In contrast, 18 MRCPS strains were detected in both inpatient (12 of 26 [46.2%]) and outpatient (6 of 31 [19.4%]) dogs. Among them, only one strain was MRSA. Using direct sequencing of sodA and hsp60 genes, the 18 MRCPS strains other than MRSA from a staff and 17 dogs, were finally identified as Staphylococcus pseudintermedius, a novel species of Staphylococcus from a cat. All of the methicillin-resistant S. pseudintermedius (MRSP) strains were multidrug resistant to erythromycin, clindamycin, trimethoprim-sulfamethoxazole, and levofloxacin. Most of the MRSP strains showed high-level resistance to oxacillin (≥128 μg/ml, 15 of 18 [83.3%]), and 10 of 15 (66.7%) high-level oxacillin-resistant MRSP strains carried type III SCCmec. DNA fingerprinting of MRSP strains by pulsed-field gel electrophoresis yielded eight clusters: clone A with four subtypes, clone B with four subtypes, clone C with three subtypes, and five other different single clones. MRSP strains from the staff and some inpatient and outpatient dogs shared three major clones (clones A, B, and C), but the strains of the other five different clusters were distributed independently among inpatient or outpatient dogs. This genetic diversity suggested that the MRSP strains were not only acquired in this veterinary teaching hospital but also acquired in primary veterinary clinics in the community. To our knowledge, this is the first report of MRSP in dogs and humans in a veterinary institution.
Methicillin-resistant Staphylococcus aureus (MRSA) is one of the most important antimicrobial-resistant nosocomial pathogens of humans worldwide and has also emerged recently in patients without established risk factors in the community (8). MRSA has also been detected in veterinary medicine, especially in cows, poultry, horses, and pet dogs and cats (39). In human hospitals, healthcare-associated outbreaks of MRSA are well documented (8). Recently, suspected cases of human-to-animal or animal-to-human transmission of MRSA have been reported sporadically (29, 36, 41, 42, 48, 50). In methicillin resistance of coagulase-positive staphylococci (CPS) other than S. aureus, which are predominant in animals, there have been also a few reports of methicillin-resistant S. intermedius (MRSI) strains obtained from dogs (15, 20, 23, 49). However, MRSI has been uncommon as a nosocomial pathogen in a veterinary hospital.
Six species of CPS other than S. aureus—S. intermedius, S. schleiferi, S. hyicus, S. lutrae, S. delphini, and S. pseudintermedius—have previously been described (11). Since biochemical characteristics of S. schleiferi and S. hyicus are different from those of S. intermedius, S. delphini, and S. pseudintermedius (11), it is easy to identify S. schleiferi and S. hyicus. Commercial kits such as Rapid ID32 Staph (bioMérieux), VITEK2 ID-GPC, and ID-GP identification card (bioMérieux), or Phoenix ID panel PMIC/ID-13 (BD) are also available for identification of the latter two species (26). However, the phenotypic characteristics of S. intermedius, S. delphini, and S. pseudintermedius resemble one another, and commercial kits are not available for differentiation of these species (11). Furthermore, 16S rRNA genes of these species are >99% identical and could not be used to differentiate the species (1, 11, 43). The discriminating power of sodA or hsp60 sequencing is superior to phenotypic characterization or 16S rRNA sequencing for species identification of staphylococci (24, 25, 38). Although S. intermedius has been reported to be prevalent among dogs, cats, horses, goats, and pigeons (7, 13), these S. intermedius strains with different biotypes might have included S. delphini and S. pseudintermedius.
In the present study, we screened CPS strains from dogs and staffs in a veterinary teaching hospital to determine the prevalence of methicillin-resistant CPS (MRCPS) and study their mode of nosocomial spread among dogs and humans. Furthermore, for definite species identification of CPS strains, we performed sodA and hsp60 sequencing.
MATERIALS AND METHODS
Sampling from dogs and veterinary staffs, isolation, and phenotypic characterization of MRCPS.
Samples from both the nostrils of 57 dogs (26 inpatients and 31 outpatients) and median septum of both nostrils of 20 veterinary staff members were collected using Seed Swab (Eiken Chemical Co., Ltd., Tokyo, Japan) from January to March 2006 at Nippon Veterinary and Life Science University Hospital. Informed consent of the owners and veterinary staffs were obtained prior to sampling. The Ethical Association of Nippon Veterinary and Life Science University approved this study. All patient dogs in the Nippon Veterinary and Life Science University Hospital were transferred from primary care animal hospitals in the community. Swabs were immediately inoculated on mannitol-salt agar (Nissui Co., Ltd., Tokyo, Japan) directly and incubated at 37°C for 48 h for selective isolation of staphylococci. Catalase-positive, gram-positive cocci that were presumptively identified as staphylococci by colonial morphology were subcultured on Trypticase soy agar II with 5% sheep blood (TSAB; BD Japan, Co., Ltd., Tokyo, Japan). Tube coagulase tests with rabbit plasma (Denka Seiken Co., Ltd., Tokyo, Japan) were performed, and only CPS strains were selected for further investigation (12). Species identification of CPS was carried out by using the latex agglutination test for clumping factor and protein A (PS Test; Eiken Chemistry Co., Ltd, Tokyo, Japan), DNase test, and rapid ID32 Staph (bioMérieux, Marcy l'Etoile, France) (12, 26, 37). A strain of S. intermedius identified phenotypically was designated S. intermedius group (SIG) and was composed of S. intermedius, S. delphini, and S. pseudintermedius presumptively because it was very difficult to differentiate these three species by biochemical reactions (11). These strains were stored in 10% skim milk at −85°C until use and maintained on TSAB.
Antimicrobial susceptibility testing.
Methicillin resistance was determined by measurement of MIC of oxacillin using the agar dilution method for all CPS strains (9). MIC tests for other antimicrobial agents were performed by broth microdilution method using the MIC-2000 system (Dry plate Eiken DP22; Eiken). The antibiotics tested included penicillin, ampicillin, oxacillin, cefazolin, cefotiam, cefaclor, flomoxef, imipenem, gentamicin, arbekacin, erythromycin, clindamycin, minocycline, levofloxacin, fosfomycin, vancomycin, teicoplanin, and trimethoprim-sulfamethoxazole. The MICs were interpreted as susceptible or resistant according to the Clinical and Laboratory Standards Institute (formerly National Committee for Clinical Laboratory Standards) recommendations (9). Breakpoints for human coagulase-negative staphylococci were applied to SIG strains according to recommendations of Bemis et al. (4). Oxacillin resistance was classified as low level (MIC of 0.5 to 64 μg/ml) or high level (MIC of ≥128 μg/ml).
DNA extraction for amplification.
A single colony was suspended to a McFarland 1.0 standard in 100 μl of TE buffer (20 mM Tris, 2 mM EDTA [pH 7.5]) with 10 U of achromopeptidase (Wako Chemical, Co., Ltd., Osaka, Japan), and the suspension was incubated at 55°C for 10 min. After centrifugation at 18,500 × g for 5 min, the supernatants were used as crude DNA extracts for PCR (22).
Genetic identification of CPS.
To confirm species identification by phenotypic characterization, we conducted molecular identification of staphylococci by detection of the femB gene encoding for an enzyme important in cross-linking peptidoglycan in S. aureus (19). Direct sequencing of sodA and hsp60 genes was performed for the differentiation of SIG strains (24, 25, 38). Sequencing reactions were performed by using a Big Dye terminator (version 1.1) cycle sequencing kit (Applied Biosystems, Foster City, CA) with an ABI Prism 3100 genetic analyzer (Applied Biosystems). Multiple alignment was carried out by the CLUSTAL X program (46). The construction of the unrooted phylogenetic tree was performed by the neighbor-joining method (40).
Detection of mecA and SCCmec typing.
mecA was determined for all of the CPS to confirm methicillin-resistance, and SCCmec typing was performed for all mecA-positive CPS strains by amplification of the regions within SCCmec, the ccr region (three classes of ccr), and the mec region (IS1272, mecI and mecR1, and mecA) by PCR as described previously (17, 18, 30).
Detection of enterotoxins, exfoliative toxins, TSST-1 and the Panton-Valentine leukocidin (PVL) genes.
Localization of 11 exotoxin genes, encoding for staphylococcal enterotoxins SEA (sea), SEB (seb), SEC (sec), SED (sed), SEE (see), SEG (seg), SEH (seh), SEI (sei), SEJ (sei); exfoliative toxin A, B (ETA; eta, ETB; etb); TSST-1 (tst); Panton-Valentine leukocidin (lukS and lukF) were detected by PCR as reported previously (3, 28, 33).
PFGE analysis.
Chromosomal DNAs of the CPS strains digested with SmaI, AscI, and XmaI (New England Biolabs, Beverly, MA) were analyzed by pulsed-field gel electrophoresis (PFGE) as described previously (37) with minor modifications. The PFGE conditions for SmaI-, XmaI-, and AscI-digested DNAs were as follows: a switch time of 3.0 to 9.0 s and a run time of 9 h and a switch time of 8.0 to 45.0 s and a run time of 12 h (for SmaI and XmaI); a switch time of 5.0 to 40.0 s and a run time of 22 h (for AscI); included angle, 120°; and voltage, 6 V/cm. The buffer temperature was maintained at 11.3°C. The PFGE patterns were interpreted according to the criteria of Tenover et al. (45). Isolates showing six or fewer fragment differences were considered to be subtypes of a pulse type (45).
RESULTS
Surveillance of CPS from dogs and veterinary staff members.
Of 20 veterinary staff members, CPS strains were isolated from 6 (30.0%) and 3 of the 6 were MRCPS (Table 1) . Of the three MRCPS strains, two were MRSA. Another MRCPS strain was identified as S. intermedius phenotypically and categorized as SIG. This strain was methicillin-resistant SIG (MRSIG).
TABLE 1.
Carriage of methicillin-resistant staphylococci in veterinary staff members and dogs
| Group | No. tested | No. (%) of humans or dogs infected with:
|
|||||
|---|---|---|---|---|---|---|---|
| S. aureus | MRSA | SIG | MRSIG | CPS | MRCPSa | ||
| Veterinary staff | 20 | 5 (25.0) | 2 (10.0) | 1 (5.0) | 1 (5.0) | 6 (30.0) | 3 (15.0) |
| Dogs | |||||||
| Inpatient | 26 | 2 (7.7) | 1 (3.8) | 13 (50.0) | 11 (42.3) | 15 (57.7) | 12 (46.2) |
| Outpatient | 31 | 3 (9.7) | 0 (0) | 12 (38.7) | 6 (19.4) | 15 (48.4) | 6 (19.4) |
| Total | 57 | 5 (8.8) | 1 (1.75) | 25 (43.9) | 17 (29.8) | 30 (52.6) | 18 (31.6) |
MRCPS is equal to MRSA plus MRSIG.
In 57 dogs, CPS strains were isolated from 30 dogs (52.6%) were isolated, and 18 of the 30 strains were MRCPS. S. aureus strains were isolated from 5 dogs (8.8%), and SIG strains were isolated from 25 dogs (43.9%). A total of 20% (one of five) of S. aureus strains and 68% (17 of 25) of SIG strains showed methicillin resistance.
Species identification of MRSIG strains based on molecular phylogenetic analysis.
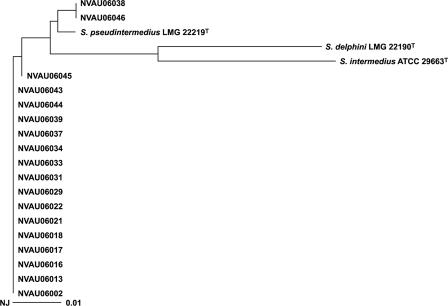
All of the 18 MRCPS strains other than MRSA in the present study were phenotypically identified as MRSIG. The sodA sequence-based analysis revealed that all of the 18 strains had only 96 to 97% sequence similarities with Staphylococcus delphini CIP 103732T and were quite different from S. intermedius CIP 8160T, with 91 to 92% similarities. Furthermore, there was only 93 to 94% similarity of hsp60 gene between these 18 strains with both S. delphini CIP 103732T and S. intermedius ATCC 29663T, respectively. These 18 strains belonged to the same cluster of sodA and hsp60 gene sequence, and the sequence similarities were greater than 99% with S. pseudintermedius LMG 22219 T for both the sodA gene (Fig. 1) and the hsp60 gene (Fig. 2). Thus, all 18 MRSIG strains were identified as S. pseudintermedius by phylogenetic analysis of the sodA and hsp60 genes.
FIG. 1.
Phylogenetic tree (unrooted) based on partial sodA gene sequences of the S. intermedius group isolated in the present study. The tree was constructed by the neighbor-joining method using CLUSTAL X.
FIG. 2.
Phylogenetic tree (unrooted) based on partial hsp60 gene sequences of the S. intermedius group isolated in the present study. The tree was constructed by the neighbor-joining method using CLUSTAL X.
Antimicrobial susceptibility profiles.
Among three MRSA strains isolated in the present study, two strains from a veterinary staff member and a dog were resistant to all β-lactams, minocycline, erythromycin, clindamycin, and levofloxacin but susceptible to vancomycin, trimethoprim-sulfamethoxazole, teicoplanin, and gentamicin. In contrast, one strain from a staff member showed susceptibility to all antimicrobial agents other than β-lactams.
On the other hand, all of methicillin-resistant S. pseudintermedius (MRSP) strains were divided into two antibiogram types based on oxacillin susceptibility (Table 2.). The majority of the MRSP strains (15 of 18 [83.3%]) showed high-level resistance to oxacillin, and the rest of them (3 of 18 [16.7%]; 1 from an inpatient dog and 2 from outpatient dogs) were low-level resistant (2-4 μg/ml) despite being mecA positive. These three strains were more susceptible to cefazolin, cefotiam, cefaclor, and fosfomycin than high-level oxacillin-resistant strains were; nevertheless, they were resistant to minocycline. All of the MRSP strains were resistant to erythromycin, clindamycin, trimethoprim-sulfamethoxazole, gentamicin (except for one strain that was intermediate), and levofloxacin. Most of the dogs with MRSP nasal carriage (14 of 17; 82.4%) had received antimicrobial agents as cefmetazole, cefaclor, enrofloxacin, or fosfomycin, within the previous 6 months.
TABLE 2.
MICs for antimicrobial agents against MRSP in a veterinary teaching hospital
| Antimicrobial agenta | MIC (μg/ml) of antimicrobial agent againstb:
|
|||||
|---|---|---|---|---|---|---|
| H-MRSP (n = 15)
|
L-MRSP (n = 3)
|
|||||
| Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | |
| OXA | >128 | >128 | >128 | 2-4 | 2 | 4 |
| AMC | 8->16 | >16 | >16 | 8->16 | 8 | >16 |
| CFZ | 4->32 | 4 | 8 | ≤0.5-2 | ≤0.5 | 2 |
| CTM | ≤0.5->32 | 4 | 8 | ≤0.5-2 | ≤0.5 | 2 |
| CEC | 32->32 | >32 | >32 | 2-32 | 2 | 32 |
| FMX | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| IPM | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 | ≤0.12 |
| GEN | 16->16 | 16 | >16 | 8-16 | 8 | 16 |
| ABK | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| MIN | ≤0.5 | ≤0.5 | ≤0.5 | 8-16 | 8 | 16 |
| ERY | >8 | >8 | >8 | >8 | >8 | >8 |
| CLI | >4 | >4 | >4 | >4 | >4 | >4 |
| VAN | ≤0.5-1 | 1 | 1 | ≤0.5-1 | ≤0.5 | 1 |
| TEC | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
| FOF | 32->32 | 32 | >32 | ≤8 | ≤8 | ≤8 |
| SXT | >76/4 | >76/4 | >76/4 | >76/4 | >76/4 | >76/4 |
| LVX | 8->8 | 8 | >8 | 8->8 | 8 | >8 |
OXA, oxacillin; AMC, ampicillin; CFZ, cefazolin; CTM, cefotiame; CEC, cefaclor; FMX, flomoxef; IPM, imipenem; GEN, gentamicin; ABK, arbekacin; MIN, minocycline; ERY, erythromycin; CLI, clindamycin; VAN, vancomycin; TEC, teicopranin; FOF, fosfomycin; SXT, sulfamethoxazole/trimethoprim (both values are given); LVX, levofloxacin.
H, high-level resistance to OXA (MIC of ≥128 μg/ml); L, low-level resistance to OXA (MIC of ≤64 μg/ml). n, Number of strains.
Molecular characterization of MRCPS strains.
Table 3 shows the genotypic characteristics of 21 MRCPS strains analyzed in the present study. Two MRSA strains with high-level resistance to oxacillin isolated from a staff member and a dog belonged to type II SCCmec and had tst, sec, seg, and sei genes. The PFGE patterns of these two strains (X1 and X2) were closely related to each other (Fig. 3). The remaining multidrug-susceptible MRSA strain belonged to type IV SCCmec and had no toxin genes. The PFGE pattern of this strain was different from those of the other two strains.
TABLE 3.
Molecular characterization of MRCPS
| Strain | Species | Groupa | Resistance to OXAb | SCCmec type | Toxin type | PFGE pattern
|
|
|---|---|---|---|---|---|---|---|
| SmaI | AscI | ||||||
| NVAU06003 | MRSA | Human | H | II | tst, sec, seg, sei | X1 | NDd |
| NVAU06030 | MRSA | Dog-i | H | II | tst, sec, seg, sei | X2 | ND |
| NVAU06032 | MRSA | Human | H | IV | None | Y | ND |
| NVAU06013 | MRSP | Human | H | III | None | A4 | a1 |
| NVAU06021 | MRSP | Dog-o | H | III | None | B2 | a4 |
| NVAU06029 | MRSP | Dog-o | H | III | None | C2 | a1 |
| NVAU06002 | MRSP | Dog-o | H | III | None | D | a1 |
| NVAU06018 | MRSP | Dog-o | H | III | None | D | a1 |
| NVAU06037 | MRSP | Dog-o | H | NTc | None | E | a3 |
| NVAU06046 | MRSP | Dog-o | L | NT | None | H | a6 |
| NVAU06016 | MRSP | Dog-i | H | III | None | A2 | a1 |
| NVAU06017 | MRSP | Dog-i | H | III | None | A3 | a1 |
| NVAU06022 | MRSP | Dog-i | H | III | None | B3 | a3 |
| NVAU06031 | MRSP | Dog-i | H | III | None | C1 | a1 |
| NVAU06039 | MRSP | Dog-i | H | III | None | C3 | a1 |
| NVAU06045 | MRSP | Dog-i | L | V | None | F | a5 |
| NVAU06043 | MRSP | Dog-i | H | NT | None | A1 | a1 |
| NVAU06044 | MRSP | Dog-i | H | NT | None | A1 | a1 |
| NVAU06034 | MRSP | Dog-I | H | NT | None | B1 | a1 |
| NVAU06033 | MRSP | Dog-i | H | NT | None | B4 | a2 |
| NVAU06038 | MRSP | Dog-i | L | NT | None | G | a6 |
Dog-o and Dog-i refer to outpatient and inpatient dogs, respectively.
H, high-level resistance to OXA (MIC of ≥128 μg/ml); L, low-level resistance to OXA (MIC of 0.5 to 64 μg/ml).
NT, nontypeable.
ND, not done.
FIG. 3.
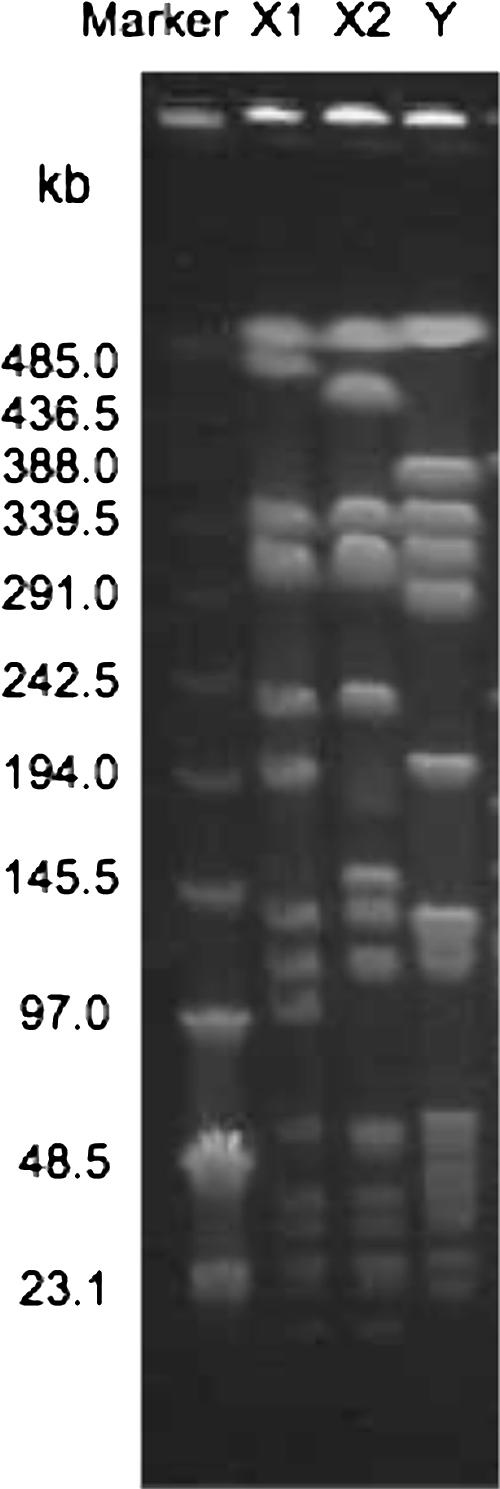
PFGE patterns of SmaI-digested MRSA strains in the veterinary hospital. Low-molecular-weight marker DNA λ is shown on the left side. The three lanes on the right display results obtained with strains. Each lane shows a different PFGE type.
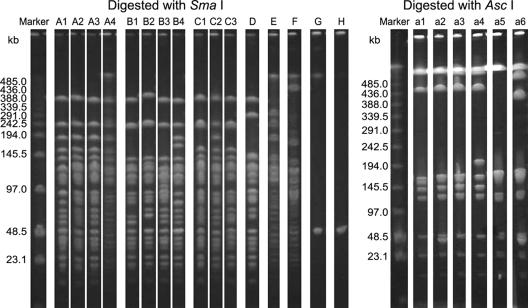
For MRSP strains, 10 of 18 strains (55.6%) carried a type III SCCmec element, one strain (5.6%) had a type V SCCmec element, and 7 strains (38.9%) were nontypeable. PFGE analysis of AscI-digested DNA was less discriminative than that of SmaI-digested DNA because all of the MRSP strains were considered to be subtypes within the same cluster (Fig. 4). The PFGE patterns of SmaI-digested DNA from the MRSP strains were differentiated into eight clones (A to H). Clones A to C were divided into several subtypes as A1 to A4, B1 to B4, and C1 to C3, respectively (Fig. 4). Only two pairs of strains shared patterns identical to each other (A1 and D). The genomic DNAs from strain NVAU06046 and NVAU06038 could not be digested by SmaI. The neoschizomer XmaI, which cuts the same recognition sequence of SmaI but at a different position, also could not digest the two DNAs (data not shown). Although the number of bands in their PFGE profiles was too small to match the criteria (45), each PFGE type was designated type G and type H tentatively. Clones with PFGE types A to E showed high-level resistance to oxacillin. PFGE types A and B were predominant in inpatient dogs (7 of 11 [63.6%]). One MRSP strain, NVAU06013, isolated from a veterinary staff member, also belonged to clone A with type III SCCmec. Both PFGE types B and C were detected in inpatient and outpatient dogs. In contrast, PFGE types D to H were only isolated from outpatient dogs, and PFGE types F and G were only found in inpatient dogs.
FIG. 4.
PFGE patterns of MRSP strains digested with SmaI and AscI in the veterinary hospital. Low-molecular-weight marker DNA λ is shown on the left. The lanes on the right side display results obtained with strains. Each lane shows a different PFGE type.
PFGE type H strain with nontypeable SCCmec, PFGE type F strain with type V SCCmec, and PFGE type G strain with nontypeable SCCmec showed low-level resistance to oxacillin. No toxin genes were detected in the MRSP strains.
DISCUSSION
To our knowledge, this is the first report on the prevalence of MRSP in dogs and humans in a veterinary institution. In dogs, the majority of CPS strains from nasal, oral, or ear commensal flora or pyoderma specimens have been reported to be S. intermedius (7, 39). In the present study, there was a low rate of S. aureus carriage in dogs (only 5 of 57 [8.8%]) compared to that observed in humans; only one MRSA strain was detected from a dog with MRSA infection. In dogs, S. intermedius is considered to be more predominant than S. aureus (7, 39), but we could not find any MRSI in the present study. Surprisingly, all of the MRSIG strains were identified as S. pseudintermedius using sodA or hsp60 sequencing. Meyer and Schleifer described how different biotypes of S. intermedius strains showed only 50 to 65% DNA homology using DNA-DNA reassociation tests, indicating that they belonged to different species (31). Other authors also reported that S. intermedius isolates from different sources could be genotypically or phenotypically differentiated (6, 13, 14). In the phylogenetic point of view, these results suggest that S. pseudintermedius is not a novel emerging species among dogs but might have been recognized as one of the different biotypes of S. intermedius.
SIG strains did not seem to be found frequently in humans (only one MRSP was isolated in the present study). This human MRSP strain showed susceptibility patterns and genotypes (PFGE pattern and type III SCCmec) similar to those of dog-derived MRSP strains. This suggests dog-to-human transmission. Similar reports have been published previously (16, 21, 44).
The prevalence of MRSIG isolates from healthy or sick dogs has still been uncommon in previous reports (15, 23, 47, 49); however, the rate of methicillin resistance among SIG isolates observed in the present study was higher (17 of 25 [68%]) than those determined in previous reports. The use of cephalosporins, carbapenems, and fluoroquinolones is a well-known risk factor for acquisition of MRSA in humans (51). Since broad-spectrum antimicrobials were used for 14 of 17 dogs with MRSP in the present study, antimicrobial selective pressure might have contributed to this prevalence of methicillin resistance.
There was no type I, II, or IV SCCmec in MRSP strains. In Japan, the majority of human healthcare-associated MRSA strains carry type II SCCmec, and type III SCCmec is detected in major HA-MRSA strains from South America, East Europe, and Asian countries other than Japan and South Korea (10, 35). No type III SCCmec has been reported in Japanese MRSA strains (10). From the evolutionary point of view, the difference of SCCmec element between MRSA and MRSP suggests that the acquisition and spread of mecA gene in S. aureus and S. pseudintermedius occurred via independent transmission routes.
A minority of MRSP strains (3 of 18) with low-level oxacillin-resistance showed different PFGE patterns from those of any high-level oxacillin-resistant strains. Kania et al. reported that 22 (40%) of 55 methicillin-susceptible S. intermedius strains determined by the disk diffusion method had the mecA gene (20). Furthermore, Bemis et al. found that the breakpoint of oxacillin for S. aureus was too low for the detection of oxacillin resistance in S. intermedius and S. schleiferi strains (4). These results suggest that low-level oxacillin resistance in S. intermedius and S. schleiferi strains might be underestimated or unrecognized in the veterinary laboratories. Heterogeneous resistance is well known in many species of mecA-positive staphylococci, but the mechanism of the different resistance level has not been elucidated yet. Future studies are needed to clarify this mechanism.
In our study, all staphylococcal enterotoxin genes, toxic shock syndrome toxin 1, Pantone-Valentine leukocidin, and exfoliative toxin genes were not detected in the MRSP strains. Becker et al., using the same PCR system that was used in the present study, reported that 33 of 281 (11.3%) S. intermedius strains had SEC genes (2). The reason for this discrepancy is uncertain, but it may be due to the difference in species; ours were identified as S. pseudintermedius, a species distinct from S. intermedius. Further testing of the enterotoxigenic potential among S. intermedius, S. delphini, and S. pseudintermedius should be carried out.
The genomic DNAs from strains NVAU06046 and NVAU06038 could not be digested by SmaI and XmaI; however, AscI-digested DNAs of both strains yielded banding patterns denoted as A6. This result might be due to an unknown restriction and/or methylation system in the recognition sequence of the SmaI and XmaI site. Although Bens et al. reported similar findings (5), the DNA modification enzyme in our strains was different from the ones sited in that report due to the protection of both SmaI and XmaI digestion.
The PFGE diversities of MRSP strains suggest that they were not only acquired in this veterinary teaching hospital but also acquired in primary veterinary clinics in the community. Although other comparable data of MRSP strains are not available, Middleton reported that MRSA isolates in seven veterinary teaching hospitals showed large genetic diversities (32).
MRSA strains in the veterinary environment have been described in several reports (27, 29, 36, 41, 42, 48, 49). In the present study, three MRSA strains isolated from two staff members and a dog were also isolated. They had profiles similar to the MRSA strains that have been recently predominant in healthcare-associated or community-acquired MRSA in Japan (22, 34, 37). Loeffler et al. also reported that most MRSA isolates from the veterinary hospital setting were related to human epidemic MRSA clone EMRSA-15 in the United Kingdom (29). Molecular epidemiology suggests that MRSA isolates from dogs are derived from humans.
In conclusion, the prevalence of not MRSI but MRSP carriage in dogs in this veterinary teaching hospital was unexpected since S. pseudintermedius is a new species established in 2005 (11). Further investigations to reclassify canine CPS based on genetic identification such as sodA or hsp60 sequencing should be carried out.
Acknowledgments
This study was partially supported by a Grant-in-Aid for 21st Century COE Research from The Ministry of Education, Science, Sports, Culture, and Technology of Japan.
We thank M. Bonkobara, T. Yougo, and T. Washizu for their help in collecting specimens.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Becker, K., D. Harmsen, A. Mellmann, C. Meier, P. Schumann, G. Peters, and C. von Eiff. 2004. Development and evaluation of a quality-controlled ribosomal sequence database for 16S ribosomal DNA-based identification of Staphylococcus species. J. Clin. Microbiol. 42:4988-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker, K., B. Keller, C. von Eiff, M. Brück, G. Lubrittz, J. Etienne, and G. Peters. 2001. Enterotoxigenic potential of Staphylococcus intermedius. Appl. Environ. Microbiol. 67:5551-5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, K., R. Roth, and G. Peters. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J. Clin. Microbiol. 36:2548-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bemis, D. A., R. D. Jones, L. E. Hiatt, E. D. Ofori, B. W. Rohrbach, L. A. Frank, and S. A. Kania. 2006. Comparison of tests to detect oxacillin resistance in Staphylococcus intermedius, Staphylococcus schleiferi, and Staphylococcus aureus isolates from canine hosts. J. Clin. Microbiol. 44:3374-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bens, C. C. P. M., A. Voss, and C. H. W. Klaassen. 2006. Presence of a novel DNA methylation enzyme in methicillin-resistant Staphylococcus aureus isolates associated with pig farming leads to uninterpretable results in standards pulsed-field gel electrophoresis analysis. J. Clin. Microbiol. 44:1875-1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bes, M., S. L. Saidi, F. Becharnia, H. Meugnier, F. Vandenesch, J. Etienne, and J. Freney. 2002. Population diversity of Staphylococcus intermedius isolates from various host species: typing by 16S-23S intergenic ribosomal DNA spacer polymorphism analysis. J. Clin. Microbiol. 40:2275-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beiberstein, E. L., S. S. Jang, and D. C. Hirsh. 1984. Species distribution of coagulase-positive staphylococci in animals. J. Clin. Microbiol. 19:610-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers, H. F. 2001. The changing epidemiology of Staphylococcus aureus. Emerg. Infect. Dis. 7:178-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. Document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Chongtrakool, P., T. Ito, X. X. Ma, Y. Kondo, S. Trakulsomboon, C. Tiensasitorn, M. Jamklang, T. Chavalit, J.-H. Song, and K. Hiramatsu. 2006. Staphylococcal cassette chromosome mec (SCCmec) typing of methicillin-resistant Staphylococcus aureus strains isolated in 11 Asian countries: a proposal for a new nomenclature for SCCmec elements. Antimicrob. Agents Chemother. 50:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Devriese, L. A., M. Vancanneyt, M. Baele, M. Vaneechoutte, E. de Graef, C. Snauwaert, I. Cleenwerck, P. Dawyndt, J. Swings, A. Decostere, and F. Haesebrouck. 2005. Staphylococcus pseudintermedius sp. nov., a coagulase-positive species from animals. Int. J. Syst. Evol. Microbiol. 55:1569-1573. [DOI] [PubMed] [Google Scholar]
- 12.Freney, J., W. E. Kloos, V. Hajek, and J. A. Webster. 1999. Recommended minimal standards for description of new staphylococcal species. Int. J. Syst. Bacteriol. 49:489-502. [DOI] [PubMed] [Google Scholar]
- 13.Futagawa-Saito, K., M. Suzuki, M. Ohsawa, S. Ohshima, N. Sakurai, W. Ba-Thein, and T. Fukuyasu. 2004. Identification and prevalence of an enterotoxin-related gene, se-int, in Staphylococcus intermedius isolates from dogs and pigeons. 2004. J. Appl. Microbiol. 96:1361-1366. [DOI] [PubMed] [Google Scholar]
- 14.Futagawa-Saito, K., S. Sugiyama, S. Karube, N. Sakurai, W. Ba-Thein, and T. Fukuyasu. 2004. Prevalence and characterization of leukotoxin-producing Staphylococcus intermedius in isolates from dogs and pigeons. J. Clin. Microbiol. 42:5324-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gortel, K., K. L. Campbell, L. Kakoma, T. Whittem, D. J. Schaeffer, and R. M. Weisiger. 1999. Methicillin resistance among staphylococci isolated from dogs. Am. J. Vet. Res. 60:1526-1530. [PubMed] [Google Scholar]
- 16.Guardabassi, L., M. E. Loeber, and A. Jacobson. 2004. Transmission of multiple-resistant Staphylococcus intermedius between dogs affected by deep pyoderma and their owners. Vet. Microbiol. 98:23-27. [DOI] [PubMed] [Google Scholar]
- 17.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jonas, D., H. Grundmann, D. Hartung, F. D. Daschner, and K. J. Towner. 1999. Evaluation of the mecA, femB duplex polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18:643-647. [DOI] [PubMed] [Google Scholar]
- 20.Kania, S. A., N. L. Williamson, L. A. Frank, R. P. Wilkes, R. D. Jones, and D. A. Bemis. 2004. Methicillin resistance of staphylococci isolated from the skin of dogs with pyoderma. Am. J. Vet. Res. 65:1265-1268. [DOI] [PubMed] [Google Scholar]
- 21.Kikuchi, K., T. Karasawa, C. Piao, I. Itoda, H. Hidai, H. Yamaura, K. Totsuka, T. Morikawa, and M. Takayama. 2004. Molecular confirmation of transmission route of Staphylococcus intermedius in mastoid cavity infection from dog saliva. J. Infect. Chemother. 10:46-48. [DOI] [PubMed] [Google Scholar]
- 22.Kikuchi, K., N. Takahashi, C. Piao, K. Totsuka, H. Nishida, and T. Uchiyama. 2003. Molecular epidemiology of methicillin-resistant Staphylococcus aureus strains causing neonatal toxic shock syndrome-like exanthematous disease in neonatal and perinatal wards. J. Clin. Microbiol. 41:3001-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, T. J., Y. R. Na, and J. I. Lee. 2005. Investigation into the basis of chloramphenicol and tetracycline resistance in Staphylococcus intermedius isolates from cases of pyoderma in dogs. J. Vet. Med. B 52:119-124. [DOI] [PubMed] [Google Scholar]
- 24.Kwok, A. Y., and A. W. Chow. 2003. Phylogenetic study of Staphylococcus and Macrococcus species based on partial hsp60 gene sequences. Int. J. Syst. Evol. Microbiol. 53:87-92. [DOI] [PubMed] [Google Scholar]
- 25.Kwok, A. Y., S. C. Su, R. P. Reynolds, S. J. Bay, S. J. Av-Gay, Y. Dovichi, and A. W. Chow. 1999. Species identification and phylogenetic relationships based on partial HSP60 gene sequences within the genus Staphylococcus. Int. J. Syst. Bacteriol. 49:1181-1192. [DOI] [PubMed] [Google Scholar]
- 26.Layer, F., B. Ghebremedhin, K.-A. Moder, W. König, and B. König. 2006. Comparative study using various methods for identification of Staphylococcus species in clinical specimens. J. Clin. Microbiol. 44:2824-2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, J. H. 2003. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolated from major food animals and their potential transmission to humans. Appl. Environ. Microbiol. 69:6489-6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lina, G., Y. Piémont, F. Godail-Gamot, M. Bes, M.-O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 29.Loeffler, A., A. K. Boag, J. Sung, J. A. Lindsay, L. Guardabassi, A. Dalsgaard, H. Smith, K. B. Stevens, and D. H. Lloyd. 2005. Prevalence of methicillin-resistant Staphylococcus aureus among staff and pets in a small animal referral hospital in the UK. J. Antimicrob. Chemother. 56:692-697. [DOI] [PubMed] [Google Scholar]
- 30.Ma, X. X., T. Ito, C. Tiensasitorn, M. Jamklang, P. Chongtrakool, S. Boyle-Vavra, R. S. Daum, and K. Hiramatsu. 2002. Novel type of staphylococcal cassette chromosome mec identified in community-acquired methicillin-resistant Staphylococcus aureus strains. Antimicrob. Agents Chemother. 46:1147-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer, S. A., and K. H. Schleifer. 1978. Deoxyribonucleic acid reassociation in the classification of coagulase-positive staphylococci. Arch. Microbiol. 117:183-188. [DOI] [PubMed] [Google Scholar]
- 32.Middleton, J. R., W. H. Fales, C. D. Luby, J. L. Oaks, S. Sanchez, J. M. Kinyon, C. C. Wu, C. W. Maddox, R. D. Walsh, and F. Hartmann. 2005. Surveillance of Staphylococcus aureus in veterinary teaching hospitals. J. Clin. Microbiol. 43:2916-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monday, S. R., and G. A. Bohach. 1999. Use of multiplex PCR to detect classical and newly described pyrogenic toxin genes in staphylococcal isolates. J. Clin. Microbiol. 37:3411-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliveira, D. C., A. Tomasz, and H. de Lencastre. 2002. Secrets of success of a human pathogen: molecular evolution of pandemic clones of methicillin-resistant Staphylococcus aureus. Lancet Infect. Dis. 2:180-189. [DOI] [PubMed] [Google Scholar]
- 36.O'Mahony, R., Y. Abbott, F. C. Leonard, B. K. Markey, P. J. Quinn, P. J. Pollock, S. Fanning, and A. S. Rossney. 2005. Methicillin-resistant Staphylococcus aureus (MRSA) isolated from animals and veterinary personnel in Ireland. Vet. Microbiol. 109:285-296. [DOI] [PubMed] [Google Scholar]
- 37.Piao, C., T. Karasawa, K. Totsuka, T. Uchiyama, and K. Kikuchi. 2005. Prospective surveillance of community-onset and healthcare-associated methicillin-resistant Staphylococcus aureus isolated from a university-affiliated hospital in Japan. Microbiol. Immunol. 49:959-970. [DOI] [PubMed] [Google Scholar]
- 38.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rich, M. 2005. Staphylococci in animals: prevalence, identification and antimicrobial susceptibility, with an emphasis on methicillin-resistant Staphylococcus aureus. Br. J. Biomed. Sci. 62:98-105. [DOI] [PubMed] [Google Scholar]
- 40.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:846-849. [DOI] [PubMed] [Google Scholar]
- 41.Seguin, J. C., R. D. Walker, J. P. Caron, W. E. Kloos, C. G. George, R. J. Hollis, R. N. Jones, and M. A. Pfaller. 1999. Methicillin-resistant Staphylococcus aureus outbreak in a veterinary teaching hospital: potential human-to-animal transmission. J. Clin. Microbiol. 37:1459-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strommenger, B., C. Kehrenberg, C. Kettlitz, C. Cuny, J. Verspohl, W. Witte, and S. Schwarz. 2006. Molecular characterization of methicillin-resistant Staphylococcus aureus strains from pet animals and their relationship to human isolates. J. Antimicrob. Chemother. 57:461-465. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi, T., I. Satoh, and N. Kikuchi. 1999. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int. J. Syst. Bacteriol. 49:725-728. [DOI] [PubMed] [Google Scholar]
- 44.Tanner, M. A., C. L. Everett, and D. C. Youvan. 2000. Molecular phylogenetic evidence for noninvasive zoonotic transmission of Staphylococcus intermedius from a canine pet to a human. J. Clin. Microbiol. 38:1628-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Duijkeren, E., A. T. A. Box, M. E. O. C. Heck, W. J. B. Wannet, and A. C. Fluit. 2004. Methicillin-resistant staphylococci isolated from animals. Vet. Microbiol. 103:91-97. [DOI] [PubMed] [Google Scholar]
- 48.van Duijkeren, E., M. J. H. M. Wolfhagen, A. T. A. Box, M. E. O. C. Heck, W. J. B. Wanet, and A. C. Fluit. 2004. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 10:2235-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vengust, M., M. E. C. Anderson, J. Rousseau, and J. S. Weese. 2006. Methicillin-resistant staphylococcal colonization in clinically normal dogs and horses in the community. Lett. Appl. Microbiol. 43:602-606. [DOI] [PubMed] [Google Scholar]
- 50.Weese, J. S., M. Archanbault, B. M. Willey, H. Dick, P. Hearn, B. N. Kreiswirth, B. Said-Salim, A. McGeer, Y. Likhoshvay, J. F. Prescott, and D. E. Low. 2005. Methicillin-resistant Staphylococcus aureus in horses and horse personnel, 2000-2002. Emerg. Infect. Dis. 11:430-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Westh, H., C. S. Zinn, V. T. Rosdahl, and the SARISA study group. 2004. An international multicenter study of antimicrobial consumption and resistance in Staphylococcus aureus isolates from 15 hospitals in 14 countries. Microb. Drug Resist. 10:169-176. [DOI] [PubMed] [Google Scholar]