Abstract
This study presents the data of an evaluation of the automated Nuclisens easyMAG and EasyQ systems versus the Roche AmpliPrep-AMPLICOR combination for testing of high-volume human immunodeficiency virus (HIV) load. This represents a follow-up of a previous study investigating the performance of the real-time Nuclisens assay using the semiautomated NucliSENS miniMAG extraction procedure. Three hundred eighteen patient samples were analyzed using both methods. The easyMAG-EasyQ HIV type 1 system has a higher sensitivity and broader dynamic range than the Cobas AmpliPrep-AMPLICOR system when the standard Roche assay is used alone, 25 to 3,000,000 IU/ml versus 400 to 750,000 HIV RNA copies/ml, respectively. There was significant correlation between the assays (0.93; P < 0.0001), with good accuracy (percent similarity mean μ = 96%), good precision (percent similarity standard deviation = 4.97%), and overall good agreement with a low percent similarity coefficient of variation of 5.17 to 6.11%. Bland-Altman analysis revealed that the AMPLICOR assay generated higher values than the EasyQ combination, with 95% of results within clinically acceptable limits. The throughput of samples was greatly improved using the easyMAG-EasyQ system, allowing 144 samples to be processed within 6 h. The potential for contamination has been dramatically reduced using the automated extraction system. Additional negative controls have been added to the kit to monitor for contamination based on the South African experience. This assay thus presents a real option for monitoring HIV load assays in high-volume testing environments.
Plasma viral load monitoring is considered standard care for human immunodeficiency virus (HIV)-infected patients on antiretroviral (ARV) therapy in the developed world (http://AIDSinfo.nih.gov). Numerous countries in Africa have elected to base their treatment follow-up on CD4 monitoring alone. South Africa is unique in its approach in that HIV viral load monitoring is used to determine changes to second-line drug regimens (3). We have previously reported on the performance of an assay selected for use on the National ARV rollout program: the NucliSENS EasyQ HIV type 1 (HIV-1) assay version 1.1 from bioMérieux (5). This first evaluation conducted at the National Health Laboratory Service compared the results of this assay using a semiautomated preparation step called the NucliSENS miniMag to the Roche AMPLICOR Monitor version 1.5 assay. This extraction procedure reflects a combination of boom chemistry and magnetic silica particles. The back end is a real-time nucleic acid sequence-based amplification assay using molecular beacon-based detection technology (6). In this study, plasma samples from 284 seropositive individuals at different stages of infection were analyzed. The results showed that HIV RNA values quantitated by the NucliSENS EasyQ assay (i) correlated significantly with those of the AMPLICOR Monitor value (r = 0.874, P < 0.00001) and (ii) were reproducible. A perceived drawback of the NucliSENS EasyQ combination was that the procedure was found to be quite labor intensive at the NucliSENS miniMAG extraction step and hence probably more prone to variation because of the strong technician interaction. This aspect, as discussed by Stevens et al. (5) might also be related to the relatively high invalidity and contamination rates that were initially observed, especially if combined with high testing volume pressure, as in this case.
To address the NucliSENS miniMAG ease-of-use issues, bioMérieux recently introduced the NucliSENS easyMAG platform for nucleic acid isolation. This instrument has exactly the same extraction chemistry but automates the labor-intensive NucliSENS miniMAG washing steps, thereby significantly reducing hands-on time. The NucliSENS easyMAG-NucliSENS EasyQ HIV-1 bioMérieux combination v1.1 (Biomerieux, Boxtel, The Netherlands) was compared to the Roche combination COBAS AmpliPrep-COBAS AMPLICOR HIV-1 Monitor (Roche Diagnostics, Branchburg, NJ) and described in this study. Throughput capacities, ease-of-use, reliability, and robustness of both systems in a clinical laboratory setting were also evaluated.
MATERIALS AND METHODS
Sample collection.
Samples were selected from patients attending HIV treatment centers at two Johannesburg regional hospitals (Helen Joseph and Johannesburg Hospital) as part of the South African National ARV rollout program. Five-milliliter EDTA blood samples were collected and centrifuged, and the plasma was submitted to the laboratory for routine HIV-1 viral load analysis. Samples collected at Helen Joseph were placed in NucliSENS lysis buffer prior to transportation. Samples received at the Johannesburg site were placed in lysis buffer at the Johannesburg reference laboratory. Samples were selected randomly in the laboratory based on an available minimum plasma volume of 3.3 ml. A total of 318 patient specimens were analyzed using both methods. All values were reported according to the limits set by each assay: (i) for the Roche AMPLICOR standard assay, the linear range is 400 RNA copies/ml to 750,000 RNA copies/ml; (ii) for the EasyQ assay, the linear range is 25 IU/ml to 3,000,000 IU/ml. This analysis was conducted using ethics clearance number M00-01-07 from the University of the Witwatersrand Human Ethics Committee.
Instrumentation and analysis.
The NucliSENS easyMAG and the NucliSENS EasyQ analyzer were installed in a separate dedicated laboratory for the duration of the study. The AmpliPrep and AMPLICOR Roche analyzers already installed and maintained in the main PCR accredited laboratory were used for the comparative analysis. All assays were conducted according to the manufacturer's instructions and good laboratory practice standards.
For the Roche assay, the input volume was 0.35 ml. Results below the detection limit of the standard Roche assay were not evaluated further using the ultrasensitive version of the kit. Plasma for the NucliSENS assay was transferred to lysis buffer according to the manufacturer's protocol. Extractions for the NucliSENS assay were performed using a 1-ml input volume with an internal calibrator added to each sample prior to extraction. Purified HIV-1 RNA and internal calibrator RNA were coamplified, and amplicon formation was measured in real time on the EasyQ analyzer. In cases where undetectable levels of viral load were obtained in both assays, they were considered concordant. For the NucliSENS assay, the only deviations to the manufacturer's instructions were the addition of negative and positive control samples to the run, which at the time, were not routinely supplied by the manufacturer.
Control samples.
High-positive controls (control 1, 25,000 IU/ml), low-positive controls (control 2, 2,500 IU/ml), and negative controls (control 3) were obtained from the VQC Laboratories (Acrometrix, Alkmaar, The Netherlands) and used as reference material in each run. The number of controls per easyMAG run was 8 (control 1, n = 2; control 2, n = 2; control 3, n = 4). Similar runs were prepared with high- and low-positive controls and negative controls at identical positions for the Roche platform.
Linearity with specimens known to contain HIV-1 RNA.
Dilution series of known HIV-1-positive samples were prepared by diluting the samples in steps of ∼0.5 logs in HIV-negative pooled plasma. Each dilution was tested in quadruplicate. The quantitative log result was plotted against the dilution factor, and reproducibility was measured by the percent coefficient of variation (%CV) at each dilution range. The R2 value was also reported for the linear regression equation of the relationship between quantitative viral result and dilution factor. Linearity studies were only performed using the NucliSENS easyMAG and NucliSENS EasyQ HIV-1 combination.
Statistical models applied.
A detailed statistical analysis included the following: (i) correlation studies (Pearson and Spearman correlation coefficients), (ii) the Bland-Altman (1) difference plot for bias and agreement, including limits of agreement and confidence intervals, and (iii) the percent similarity model (4) for measuring agreement, including accuracy (percent similarity mean) and precision (percent similarity standard deviation [SD]) and overall agreement (percent similarity CV). The Bland-Altman model measures the difference between two methods (a − b). This model is represented by scatter plots of the difference between the methods on the vertical axis and the absolute value of the reference on the horizontal axis. The average absolute value is not used, as this evaluation is to determine whether the new method can replace the existing method and reported patient results are given as absolutes not averages between two methods. The percent similarity model applies the formula (a + b)/2/a × 100, where a is the reference method and b is the new method. The percent similarity values between data pairs are then represented in a histogram format overlaid with a normal curve. The peak distance (mean percent similarity) from 100% shows the accuracy between the two methods, and the spread (SD) of the curve shows the precision between two methods. The overall agreement between the two methods is then represented by a single unit, the percent similarity CV (SD/mean) which summarizes both accuracy and precision into one unit. A low percent similarity CV shows good agreement between methods. All statistical analysis was performed on log-transformed data, after converting AMPLICOR results in RNA copies/ml into IU/ml as described by Stevens et al. in 2005 (copies/ml × 0.51 = IU/ml) (5). For purposes of the statistical analyses, the COBAS AmpliPrep-AMPLICOR assay was considered the reference method against which the easyMAG-NucliSENS EasyQ HIV-1 v1.1 combination was evaluated.
RESULTS
A total of 318 clinical samples were available for analysis. Fifty-three samples could not be included in the analysis for the following reasons: (i) insufficient volume for reanalysis on AMPLICOR assay (n = 13), (ii) sample clotting on AMPLICOR analyzer (n = 3), (iii) no or invalid EasyQ results (n = 35), and (iv) no result available on both systems (n = 2). The 35 EasyQ samples (11.6% of the total number of samples) suffered from sample gelation and could therefore not be processed or led to invalid results flagged by the easyMAG-EasyQ HIV-1 combination. Upon retesting another plasma specimen (taken at the same time point from the same patient), a valid result was obtained for 20 samples.
Thus, statistical analyses were performed on the results of the 265 samples for which both a valid NucliSENS EasyQ HIV-1 and AMPLICOR result were available.
Results of QC controls.
The quality control (QC) samples were used to calculate the intervariability of each assay as well as the background error. Table 1 outlines the control results from both assays. The CVs for both assays increased at the lower control (2) range and overall showed slightly more variability in Roche AMPLICOR, with lower average values than in the EasyQ HIV-1 assay. Four of 50 control (2) samples were not detected in the Amplicor standard assay (8%).
TABLE 1.
Results of control samples incorporated in easyMAG-NucliSENS EasyQ HIV-1 combination (EasyQ) and in AmpliPrep-AMPLICOR (Roche) runs
| Control no. (result) and parameter | Result for assay:
|
|
|---|---|---|
| EasyQ (log IU/ml) (n = 39) | AMPLICOR (log IU/ml) (n = 50) | |
| 1 (4.40 log IU/ml) | ||
| n | 39 | 50 |
| SD | 0.13 | 0.19 |
| CV (%) | 3.1 | 5.0 |
| Mean | 4.21 | 3.85 |
| Sensitivity (%) | 100 | 100 |
| 2 (3.40 log IU/ml) | ||
| n | 39 | 46 |
| SD | 0.17 | 0.19 |
| CV (%) | 4.8 | 6.9 |
| Mean | 3.41 | 2.69 |
| Sensitivity (%) | 100 | 92 |
| 3 (negative), specificity (%) | 100a | 100b |
Specificity for 39 reportable results (<25 IU/ml, <LDL).
Specificity for 49 reportable results (<400 copies/ml). One isolate had a recorded Roche result of 104 copies/ml, which as a reportable result is <400 copies/ml. The number generated (continuous value) in place of the typical <400 copies/ml (discrete value), however, may indicate contamination and has been investigated.
Summary statistics of patient sample distribution and data trimming.
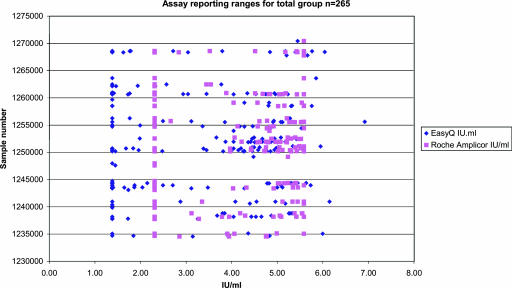
The scatter plot in Fig. 1 summarizes the spread of reportable results for both assays and clearly shows the broader range of the EasyQ and the limits of the Roche AMPLICOR standard assay reported results within this range. The data set describes that 50.2% of the data set (n = 133) reads <400 copies/ml on the AMPLICOR assay (<2.3 log IU/ml) and that 39.2% (n = 104) of the data read <25 IU/ml on the EasyQ HIV-1 assay (1.4 log IU/ml); 9.1% of the data (n = 24) provided results greater than the upper detection limit (UDL) in the AMPLICOR assay. For EasyQ, only 1 sample was above the UDL of the assay. Interestingly, only 5 samples were above the UDL of the Roche assay.
FIG. 1.
A scatter plot of the Roche AMPLICOR and EasyQ results reported in log IU/ml, showing the range in reported results and limits to each assay. The horizontal axis shows log IU/ml, and the vertical axis shows sample numbers in order of preparation for the total number of samples analyzed (n = 256).
The presence of negative (<400 copies/ml and <25 IU/ml) and “overrange” values (>750,000 copies/ml and 3,000,000 IU/ml) for the Roche and EasyQ sample results, respectively, complicates the statistical analysis. To address this, in one approach, the data in the lower and upper ranges were used in the analysis as if having a quantitative result representing the corresponding limit of detection (400 and 750,000 copies/ml, respectively, for the AMPLICOR assay and 24 IU/ml for EasyQ HIV-1). Alternatively, the results with no quantifiable result can be removed from the analysis. The corresponding data set from which the data were beyond the lower and upper limits of the AMPLICOR assay (<400, >750,000 copies/ml) were removed is referred to as the trimmed data. One test result was present for which the EasyQ HIV-1 assay yielded a negative result (<25 IU/ml), whereas Roche Amplicor yielded a clearly positive result (276,000 AMPLICOR copies/ml; 140,760 AMPLICOR IU/ml), and this was considered an outlier. The analyses will also be reported after removal of this outlying observation. Two results were >400 IU/ml in the NucliSENS EasyQ assay and <400 copies/ml in the AMPLICOR assay.
The statistical analysis is performed on both the total group (n = 265) and the trimmed group (n = 108). The summary statistics for the total and trimmed group are shown in Table 2. Therefore, part of the analysis was performed on three data sets: (i) total data set including those below the detection limit (designated LDL by the analyzer) and overrange results (n = 265), (ii) trimmed group, LDL results and overrange results are excluded (n = 108), and (iii) trimmed group, of which the one outlier was excluded (n = 107).
TABLE 2.
Data summary for the total sample group (n = 265) and the trimmed group (n = 108)
| Group (n) | Mean (IU/mla) | Range (IU/mla) |
|---|---|---|
| Total (256) | ||
| EasyQ | 94,244 | 24-830,000 |
| EasyQ log | 3.0 | 1.38-6.92 |
| Roche AMPLICOR | 162,175 | 400-750,000 |
| Roche AMPLICOR converted to IU | 82,709 | 204-382,500 |
| Roche AMPLICOR IU log | 3.6 | 2.3-5.58 |
| Trimmed (108) | ||
| EasyQ | 61,303 | 24-430,000 |
| EasyQ log | 4.36 | 1.38-5.63 |
| Roche AMPLICOR | 230,772 | 901-692,000 |
| Roche AMPLICOR converted to IU | 117,694 | 460-352,920 |
| Roche AMPLICOR IU log | 4.77 | 2.66-5.55 |
Results are for Roche AMPLICOR are in copies/ml; all other results are in IU/ml.
Direct assay correlation.
The correlation coefficients provide information on the linear dependency of the two assay results expressed on a logarithmic scale. Spearman's correlation is determined from ranks (nonparametric with weaker assumptions) and is the preferred measure in the case of unquantifiable test results. The Spearman's and Pearson's correlation coefficients are listed in Table 3. Both models show significant correlation between the EasyQ HIV-1 assay (IU/ml) and the Roche AMPLICOR (IU/ml) for this range of data, indicating that high quantitation values in one assay are accompanied by high results in the other. The correlation between the two assays decreased upon data trimming, but the correlations remain statistically highly significant. The outlying value is apparently influential, though the overall conclusion does not change.
TABLE 3.
Pearson and Spearman correlation coefficients
| Data set | n | Spearman correlation
|
Pearson correlation
|
||
|---|---|---|---|---|---|
| Value | Pa | Value | Pa | ||
| Total | 256 | 0.930 | <0.0001 | 0.961 | <0.0001 |
| Trimmed | 108 | 0.730 | <0.0001 | 0.697 | <0.0001 |
| Trimmed without outlier | 107 | 0.751 | <0.0001 | 0.777 | <0.0001 |
The P values test the null hypothesis that the true correlation equals zero.
Measure of agreement between the assays.
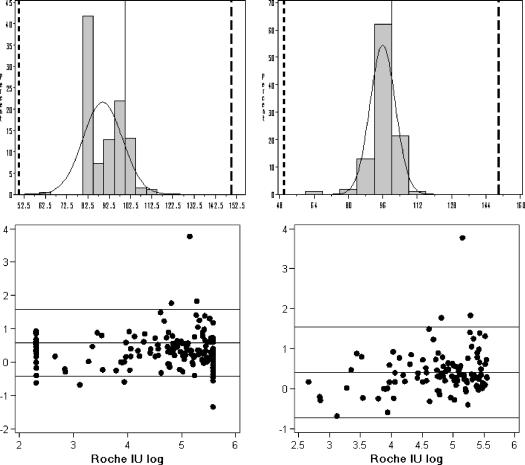
The correlation models showed a significant linear relation between the two amplification assays. Whether the easyMAG-EasyQ HIV-1 combination can replace the AMPLICOR system in this clinical setting without new baseline testing or recalculation of the original results obtained with the Roche system can only be determined by the Bland-Altman and percent similarity models measuring agreement between the two assays. The percent similarity histograms are illustrated in Fig. 2 followed by the Bland-Altman plots for the total and trimmed data. The relevant model statistics are summarized in Table 4.
FIG. 2.
Percent similarity histograms and Bland-Altman difference scatter plots for the total group (n = 256, left) and the trimmed group (n = 108, right). The histograms plot the frequency of the percent similarity values between the AMPLICOR and EasyQ results and show the number of similar results in each percent similarity interval. The normal curves are included, which visually show the accuracy between the methods (closeness of the peak to the 100% similarity line) and the precision (spread around the mean). The 100% similarity and 50 and 100% reference lines are also included. The summary statistics for these models are presented in Table 4.
TABLE 4.
Summary statistics of the observed differences in quantitation results (log Roche AMPLICOR [IU] − log EasyQ) for the three data subsets
| Data set | n | Difference
|
Limits of agreement | % Similarity
|
|||
|---|---|---|---|---|---|---|---|
| Mean (confidence interval) | SD | Mean | SD | CV | |||
| Total | 265 | 0.581 (0.52, 0.64) | 0.506 | −0.43, 1.59 | 89.4 | 9.2 | 10.3 |
| Trimmed | 108 | 0.411 (0.3, 0.52) | 0.565 | −0.72, 1.54 | 95.9 | 5.86 | 6.11 |
| Trimmed (without outlier) | 107 | 0.380 (0.29, 0.47) | 0.463 | −0.55, 1.31 | 96.2 | 4.97 | 5.17 |
This percent similarity analysis shows the EasyQ HIV-1 assay has good accuracy (%μ = 96%) relative to the AMPLICOR IU/ml method and good precision (% SD = 4.97%). Overall, the assays showed good acceptable agreement with a low percent similarity CV of 5.17 to 6.11%. The observation that the mean similarity is less than 100% again indicates that the AMPLICOR results are, on average, higher than with EasyQ HIV-1. This too is reflected by the Bland-Altman analysis with a positive bias that shows that the AMPLICOR values read higher than the EasyQ HIV-1 values. A greater than 0.3 log difference is acceptable laboratory variability in an assay, and a >log 1.0 difference would be considered clinically significant. The width between the limits of agreement is 1.86 log (based on n = 107). If the mean difference is taken into account, then more than 95% of the differences in quantitation results between the two assays are less than 1 log, which is considered within acceptable limits of clinical significance. The samples with the largest observed differences in test results were not clustered according to sample number, and were all distributed in different runs. The Bland-Altman plots in Fig. 2 also show, in general, that AMPLICOR results yield greater values in the higher range (>log 4.5 IU/ml), whereas the EasyQ yields greater values in the lower range (<log3.5 IU/ml).
Linearity with specimens known to contain HIV-1 RNA.
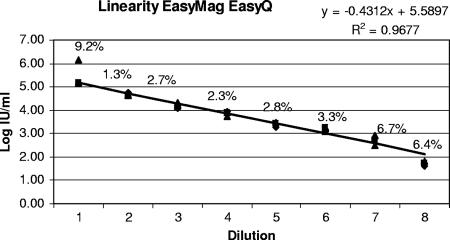
The linearity in quantitation using the EasyQ HIV-1 assay was analyzed by testing a dilution series of two high-positive samples derived from two HIV-1-infected individuals. Dilution series of samples were prepared by diluting the samples with steps of 0.5 logs. A good linearity was observed for both samples at an R2 of >0.96, with the greatest variability in the low range ± log 2.0 (CV = 6.7%.) and high ranges log 5.0 (CV = 9.2%). Figure 3 represents the linear regression for one sample including the %CV over the range in dilution.
FIG. 3.
Linear regression of one of the samples prepared in quadruplicate dilutions over log ranges to show linearity of the easyMAG EasyQ method. The horizontal axis shows the dilution number, and the vertical axis shows log intervals (IU/ml). The equation of the line is presented as well as the R2, which shows that 96.7% of the data are represented by this equation. The %CV values for the quadruplicate results at each dilution are also given and show that the greatest variability occurs in the high and low ranges.
Qualitative assessment of performance: ease of use.
In this study, significant emphasis was placed on the technical ease with which the assay could be conducted. Technicians at the Johannesburg hospital laboratory were trained, and the general conclusion was that the extraction method for the assay was found to be relatively easy to conduct with extremely user-friendly software. Staff did not require more than 2 to 3 days training prior to implementation. This was a vast improvement on the miniMAG extraction methodology previously used for the assay. Six runs on the easyMAG and 3 runs EasyQ HIV-1 can be completed fairly easily (by one technician) in an 8-h working day (translating into a throughput of 144 samples). For the reference method, the COBAS AmpliPrep-AMPLICOR system, the processing of 144 samples takes approximately 24 h when one AmpliPrep and three COBAS Amplicor instruments are used. It should also be noted that in the South African context where all equipment is leased, the cost of reagents is significantly less expensive for the NucliSENS assay.
DISCUSSION
Several publications are available describing the performance of the NucliSENS EasyQ analyzer: (i) comparison of NucliSENS EasyQ to NucliSENS HIV-1 QT assay (7), (ii) multicenter study comparing results to three other commercially available assays, COBAS Amplicor Monitor HIV-1 v1.5 (Roche), Versant HIV-1 RNA assay (Bayer), and Nuclisens HIV-1 QT (biomérieux) (2), and (iii) publication referenced in the introduction describing the NucliSENS mini-MAG versus EasyMAG combination (6). This paper represents the first formal description of the easyMAG-EasyQ combination and compares its performance to that of the Roche AmpliPrep-AMPLICOR combination.
In this study a significant correlation existed between the log values of the easyMAG-EasyQ HIV-1 and COBAS AmpliPrep-AMPLICOR methods. This showed a linear relationship between the AMPLICOR and EasyQ HIV-1 methods in spite of removing the influencing values from the total group: Spearman, r = 0.93 for the total group (n = 265); Spearman, r = 0.73 for the trimmed group (n = 108), and Spearman, r = 0.75 for the trimmed group without outlier (n = 107). The high similarity in quantitation between the assays suggests that switching from one assay to the other can be done without new baseline testing. This was confirmed by the additional analysis using methods of agreement, the percent similarity model and Bland-Altman analysis: (i) percent similarity model which revealed a mean observed similarity of 96% and a mean percent difference (MPD) (±SD) of 3.8 ± 5.0%, with overall good agreement (low CV) with the AMPLICOR assay (% similarity CV = 5.2%); (ii) for Bland-Altman, the limits of agreement (1.31, −0.55) and the mean difference (0.38) are within the boundaries of clinically acceptable differences between reportable results of the assays.
The easyMAG-EasyQ HIV-1 system has a higher sensitivity and broader dynamic range than the COBAS AmpliPrep-AMPLICOR system when the standard Roche assay is used alone, with 25 to 3,000,000 IU/ml versus 400 to 750,000 copies/ml, respectively. This system thus obviates the need for processing with a second assay, the Roche Ultrasensitive assay. In the future, this two-test strategy can be replaced by the Roche AmpliPrep-Roche Taqman combination, which has a linear range of 40 to 10E6 RNA copies/ml and is currently under evaluation in this laboratory.
Fifty percent of the samples tested in this study had values below the LDL of the AMPLICOR assay (400 copies/ml), which shows the impact of ARV therapy on the patients being referred to the laboratory but influences the statistical analysis. Forty percent of the samples tested in the study had negative results in the EasyQ HIV-1 assay (LDL, 25 IU/ml). All 24 samples (9.1%) with a UDL result in the AMPLICOR assay could be quantified by EasyQ HIV-1 assay; in these cases, a high viral load result was always obtained with EasyQ HIV-1. In spite of sensitivity controls in each AMPLICOR run, 8% of the low-positive QC control samples containing 2,500 IU/ml were not detected by the AmpliPrep-AMPLICOR system, while the same control sample was detected with 100% sensitivity (n = 50) by the easyMAG-EasyQ HIV-1 combination. No explanation was found for the AMPLICOR reduced sensitivity on this control material. Both control 1 and 2, however, yielded, on average, lower values with the AMPLICOR assay than the EasyQ. This was similarly noted in the Bland-Altman scatter plots with the clinical samples in Fig. 2 where the EasyQ assay yields higher values than the AMPLICOR in the lower quantitative results. This may relate to differences between the two systems' nucleic amplification and detection technologies.
The throughput of samples was greatly improved using the easyMAG-EasyQ HIV-1 system. One hundred forty-four samples could easily be processed within 6 h using one easyMAG and one EasyQ instrument. Using one AmpliPrep and three COBAS AMPLICOR instruments, processing of the same number of samples takes 24 h. Concerns that require ongoing evaluation include the frequent occurrence of gelation of the samples during transport in the lysis buffer provided by biomérieux, leading to the higher number of invalid samples flagged by the EasyQ analyzer. The potential for contamination has been dramatically reduced with the automated extraction system, but as with any real-time analyzer, significant caution needs to be used to prevent contamination at the back end amplification step. It is for this reason that additional negative controls have been included in the system based on the South African laboratory experience.
The assay thus presents a very real option for HIV-1 viral load monitoring in high-volume environments due to its robust nature and the ability to produce rapid, reproducible results.
Footnotes
Published ahead of print on 31 January 2007.
REFERENCES
- 1.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307-310. [PubMed] [Google Scholar]
- 2.de Mendoza, C., M. Koppelman, B. Montes, V. Ferre, V. Soriano, H. Cuypers, M. Segondy, and T. Oosterlaken. 2005. Multicenter evaluation of the NucliSens EasyQ HIV-1 v1.1 assay for the quantitative detection of HIV-1 RNA in plasma. J. Virol. Methods 127:54-59. [DOI] [PubMed] [Google Scholar]
- 3.National Department of Health. 2004. National antiretroviral treatment guidelines. National Department of Health, Johannesburg, South Africa. http://www.doh.gov.za/docs/factsheets/guidelines/artguide04-f.html.
- 4.Scott, L. E., J. S. Galpin, and D. K. Glencross. 2003. Multiple method comparison: statistical model using percentage similarity. Cytometry B Clin. Cytom. 54:46-53. [DOI] [PubMed] [Google Scholar]
- 5.Stevens, W., T. Wiggill, P. Horsfield, L. Coetzee, and L. E. Scott. 2005. Evaluation of the NucliSens EasyQ assay in HIV-1-infected individuals in South Africa. J. Virol. Methods 124:105-110. [DOI] [PubMed] [Google Scholar]
- 6.Weusten, J. J., W. M. Carpay, T. A. Oosterlaken, M. C. van Zuijlen, and P. A. van de Wiel. 2002. Principles of quantitation of viral loads using nucleic acid sequence-based amplification in combination with homogeneous detection using molecular beacons. Nucleic Acids Res. 30:e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao, J., Z. Liu, L. S. Ko, G. Pan, and Y. Jiang. 2005. Quantitative detection of HIV-1 RNA using NucliSens EasyQ HIV-1 assay. J. Virol. Methods 129:40-46. [DOI] [PubMed] [Google Scholar]