Abstract
Genotyping of Chlamydia trachomatis is limited by the low sequence variation in the genome, and no adequate method is available for analysis of the spread of chlamydial infections in the community. We have developed a multilocus sequence typing (MLST) system based on five target regions and compared it with analysis of ompA, the single gene most extensively used for genotyping. Sequence determination of 16 reference strains, comprising all major serotypes, serotypes A to L3, showed that the number of genetic variants in the five separate target regions ranged from 8 to 16. The genetic variation in 47 clinical C. trachomatis isolates of representative serotypes (14 serotype D, 12 serotype E, 11 serotype G, and 10 serotype K strains) was analyzed; and the MLST system detected 32 variants, whereas 12 variants were detected by using ompA analysis. Specimens of the predominant serotype, serotype E, were differentiated into seven genotypes by MLST but into only two by ompA analysis. The MLST system was applied to C. trachomatis specimens from a population of men who have sex with men and was able to differentiate 10 specimens of one predominant ompA genotype G variant into four distinct MLST variants. To conclude, our MLST system can be used to discriminate C. trachomatis strains and can be applied to high-resolution molecular epidemiology.
Chlamydia trachomatis can be subdivided by serological typing, based on the major outer membrane protein, into at least 15 serotypes. Serotypes A to C are associated with ocular trachoma, serotypes D to K preferably colonize the urogenital tract, and serotypes L1 to L3 cause lymphogranuloma venereum.
Serotyping of Chlamydia is laborious since it requires multiple passages in cell culture and the use of a large panel of monoclonal antibodies. Genotyping methods are more commonly used, and the major outer membrane protein gene, ompA, provides the best discriminatory capacity of the genes tested. Restriction fragment length polymorphism analysis of ompA is rapid, and its results show a high level of agreement with the results serotyping (9), while DNA sequencing of ompA has a higher resolution and can discriminate strains in clinically high-risk populations (2, 14). However, when it is applied to nonselected populations, the limited resolution of ompA sequencing restricts the amount of epidemiological information that can be obtained (6). This is especially true, given that the single serotype E comprises almost half of all urogenital chlamydial infections, and within this serotype, one genotypic variant appears to predominate (3, 4, 6). There is therefore an obvious need to develop better methods for evaluation of the molecular epidemiology of chlamydial infections. Furthermore, it has recently been shown that mutant strains evade systems commonly used for the detection of C. trachomatis (11). At present little is known about the spread of such changed chlamydial strains, but they are known to be prevalent in several regions of Sweden and are probably prevalent elsewhere. In this context, multilocus sequence typing (MLST) is an important tool for the investigation of whether several clones occur simultaneously and whether they have changed over time.
The aim of this study was to develop a high-resolution method for the discrimination of C. trachomatis strains. We explored the differences between the available genomes and tested a set of five candidate target regions for the design of an MLST system. Here we present data showing that the system developed has a high capacity to discriminate between strains of individual major outer membrane protein serovars; i.e., it is capable of identifying high intraserotype variation. The MLST system also demonstrated sequence variation when it was applied to clinical chlamydial specimens of common serotypes. This type of information can be used to gain epidemiological knowledge of C. trachomatis infections, and we present an example of how MLST can be applied in contact tracing. Furthermore, it may provide a better understanding of cell tropism and pathogenesis.
MATERIALS AND METHODS
Selection of target regions.
Complete or partial genomes of C. trachomatis were obtained from NCBI and the Sanger Institute (ftp://ftp.sanger.ac.uk/pub/pathogens/Chlamydia/) and comprised the genomes of strains A/Har-13, B/Jali, B/1A828, D/UW-3/Cx, and L2/unknown strain.
All genes from the raw genome sequences were extracted using Glimmer2 software (12). Homologous regions between multiple strains were identified and scanned for maximum nucleotide sequence difference with the help of a database-driven system called GENCOMP (Hans-Henrik Fuxelius and Siv G. E. Andersson, unpublished software). Homologous genes and intergenic regions were aligned with DIALIGN2 software (8) at the nucleotide level. dnapars (DNA parsimony) and dnadist (DNA distance) software from the PHYLIP package (J. Felsenstein, PHYLIP, phylogeny inference package, University of Washington, Seattle, 1993 [distributed by the author]) were applied to the aligned sequences and used to estimate nucleotide sequence divergence levels. Reference to dnadist in the following means that pairwise dnadist values exceeding a cutoff of 0.005 were counted for each cluster to give a scalar value for each cluster showing the highest pairwise variability. Candidate genes and intergenic regions were ranked by their scalar dnapars and dnadist values. Following visual inspection of the alignment, the sequences associated with maximal distance values were selected as candidates for the development of a genotyping system. Selected regions comprised genes coding for known and hypothetical proteins, along with intergenic regions. The regions were named after the gene (hypothetical or defined) dominating them: CT046 (hctB), CT058, CT144, CT172, and CT682 (pbpB).
Strain selection.
Reference strains comprising the 16 major serotypes, serotypes A to L3, of C. trachomatis were selected: A/Sa1, B/Iu1226, Ba/AP/2, C/TW3, D/IC-Cal8, D/UW-3/Cx, E/DK-20/ON, F/IC-Cal3, G/UW57/Cx, H/Wash, Ia/Iu-4168, J/UW36, K/UW31/Cx, L1/440, L2/434, and L3/404. A set of 47 isolates of serotypes D, E, G, and K from clinical specimens was collected over 6 months at Malmö University Hospital; and a set of 21 samples of serotypes D and G was collected from February 2004 to March 2005 at Venhälsan Gay Clinic, Karolinska University Hospital, Stockholm, Sweden (5). The samples of chlamydia used to demonstrate the application of MLST in contact tracing were from our earlier study (6).
DNA purification.
DNA from Chlamydia cultures and swab samples was purified by using a QIAamp DNA mini kit (QIAGEN, Hilden, Germany). DNA from urine samples was isolated by using a MagAttract DNA Mini M48 kit on a BioRobot M48 workstation (QIAGEN).
PCR amplification.
PCR was used to amplify the five different target regions of the genome of C. trachomatis. The primer pairs used amplified regions between 382 and 2,377 bp (Table 1). The reaction mixture contained 0.4 μM each primer, 0.2 mM deoxynucleoside triphosphates, 1.5 mM MgCl2, and 1.3 U Expand high-fidelity polymerase (Roche Applied Science, Mannheim, Germany). The same temperature program was used for most regions: initial denaturation for 2 min at 94°C, followed by 40 cycles of denaturation for 15 s at 94°C, annealing for 30 s at 60°C, and elongation for 1 min at 72°C. The elongation step was increased by 5 s per cycle after 10 cycles. The amplification was terminated with elongation for 7 min at 72°C. Different cycling conditions were required for pbpB, as this region is longer. The annealing temperature was initially 68°C, but it was decreased by 1°C per cycle during the first 10 cycles to 58°C. Elongation was for 80 s for the first 10 cycles, but it was then increased by 5 s per cycle for the remaining 30 cycles. The ompA gene was amplified as described by Lysén et al. (6).
TABLE 1.
Primers used for PCR
| Region | Primer name | Sequence | Length of PCR product (bp) |
|---|---|---|---|
| CT046 (hctB) | Hctb39F | 5′-CTCGAAGACAATCCAGTAGCAT-3′ | 686-904 |
| Hctb794R | 5′-CACCAGAAGCAGCTACACGT-3′ | ||
| CT058 | CT222F | 5′-CTTTTCTGAGGCTGAGTATGATTT-3′ | 1,499 |
| CT1678R | 5′-CCGATTCTTACTGGGAGGGT-3′ | ||
| CT144 | CT144:248F | 5′-ATGATTAACGTGATTTGGTTTCCTT-3′ | 799 |
| CT144:1046R | 5′-GCGCACCAAAACATAGGTACT-3′ | ||
| CT172 | Four268F | 5′-CCGTAGTAATGGGTGAGGGA-3′ | 378-685 |
| Four610R | 5′-CGTCATTGCTTGCTCGGCTT-3′ | ||
| CT682 (pbpB) | pbpB1F | 5′-TATATGAAAAGAAAACGACGCACC-3′ | 2,377 |
| pbpB2366R | 5′-TGGTCAGAAAGATGCTGCACA-3′ |
Sequencing.
The PCR products were purified by using an ExoSAP-IT purification kit (Amersham Biosciences). Sequencing PCR with the BigDye Terminator (version 3.1) cycle sequencing kit (Applied Biosystems, Foster City, CA) and purification were performed according to the instructions of the manufacturer, except that the annealing temperature was 60°C. The ompA gene was sequenced as described by Klint et al. (5). An ABI 3130 instrument (Applied Biosystems) was used for sample analysis, and the sequences obtained were examined by using the SeqScape (version 2.5) and BioEdit 7.0 sequence alignment editor (Ibis Therapeutics, Carlsbad, CA).
RESULTS
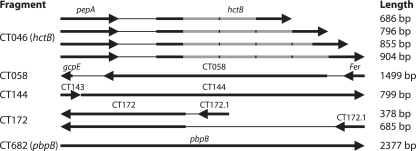
Using a computational approach, we examined the regional variability of six completely or partially sequenced genomes of C. trachomatis. The initial screen identified the eight most variable regions, which were sequenced and analyzed for their genetic variation, along with that of ompA, from 16 reference strains of serotypes A to L3. The five most variable regions thus identified comprised annotated genes (hctB and pbpB) as well as hypothetical genes (Table 2). The mutational variation included both nucleotide substitution and insertion-deletion (indel) differences and either was concentrated in one segment of the gene or was evenly distributed across the entire gene (Fig. 1). For example, tandem duplications of a repetitive element were observed in hctB, and an insertion-deletion difference was observed in CT172 (Fig. 1). The frequencies of substitutions and indels in these coding areas were substantially higher than those in the noncoding segments of the same set of genomes.
TABLE 2.
Genetic variation between and within serotypes
| Serotype (no. of isolates) | No. of variants for the following target regionsa:
|
|||||
|---|---|---|---|---|---|---|
| hctB | CT058 | CT144 | CT172 | pbpB | ompA | |
| A to L3 (reference strains) (16) | 11 | 11 | 8 | 11 | 16 | 16 |
| D (14) | 4 | 6 | 5 | 4 | 6 | 5 |
| E (12) | 5 | 2 | 3 | 2 | 3 | 2 |
| G (11) | 3 | 4 | 3 | 3 | 7 | 4 |
| K (10) | 2 | 3 | 1 | 2 | 2 | 1 |
Number of variants for each target region in 16 reference strains of serotypes A to L3 and in 47 clinical isolates of serotype D, E, G, or K.
FIG. 1.
Schematic view of the five MLST regions. Each region is named after the dominant gene. Lengths are approximate since there are minor deletions/insertions. The hctB region has a repetitive element (gray).
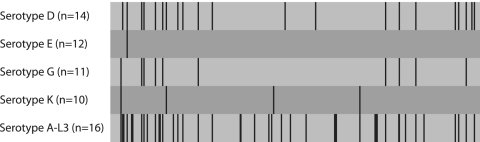
To make a closer study of sequence variations within serotypes, 47 clinical isolates of serotype D, E, G, or K were analyzed. They represented the most common types and the three serotype complexes B, C, and F/G. In Table 2, the number of variants for each target region is presented. Table 2 shows that strains of the most common serotype, serotype E, had a larger number of genetic variants in three MLST regions than in ompA. An example of the nucleotide variation is shown for the CT058 region (Fig. 2). Here the mutation rate in serotype E was similar to that in ompA. The same is true of serotype K strains, while strains of serotypes D and G had higher levels of sequence variation. The pbpB region had sequence variation that was higher than that in ompA in all the serotypes examined. Analysis of type K isolates demonstrated higher degrees of variation in four candidate regions compared with that in ompA.
FIG. 2.
Variable positions found in the CT058 target region when 16 reference strains of serotypes A to L3 and 47 clinical specimens of serotypes D, E, G, and K were analyzed. Some strains carry multiple variations.
Table 3 shows the number of variants for all MLST regions and also how ompA can contribute to strain discrimination. Overall, the number of variants for the MLST system was 32, whereas the number of variants was 12 when the information from ompA was used. The predominant serotype, serotype E, had two ompA variants, while in the MLST system, seven different genotypes were found. In the case of the isolates of serotype K examined, only one genetic variant was seen in ompA, while the MLST system was able to discriminate five. If the pbpB target region was extended by 1 kb, the number of serotype K variants increased to 7 and the combined MLST system (with ompA) detected 35 variants (data not shown). When pbpB was omitted from the analysis, the number of variants decreased to 29. This further demonstrates the discriminatory capacity of the pbpB region.
TABLE 3.
Discriminatory capacity of the MLST system
| Serotype (no. of isolates) | No. of variantsa
|
||
|---|---|---|---|
| MLST variants | MLST variants with ompA | ompA variants | |
| D (14) | 11 | 11 | 5 |
| E (12) | 7 | 8 | 2 |
| G (11) | 9 | 9 | 4 |
| K (10) | 5 | 5 | 1 |
| Total (47) | 32 | 33 | 12 |
Number of genetic variants when five target genes and ompA were analyzed in isolates of serotypes D, E, G, and K.
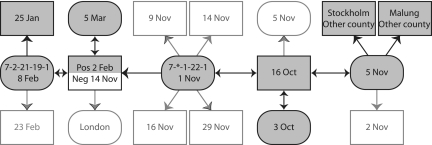
The usefulness of the MLST system was tested by analysis of a contact-tracing chain of chlamydia-infected persons. The chain comprised 18 individuals, of whom 8 were found to be infected with Chlamydia strains of serotype E with identical ompA sequences. MLST analysis was able to differentiate the strains into two distinct alleles, which were also separated in time (Fig. 3). This indicates a high discriminatory capacity that could be used to identify different chlamydial strains circulating in the community.
FIG. 3.
A contact-tracing chain of males (squares) and females (rounded), with sampling dates. Negative cases are indicated with white boxes. All positive cases were infected by serotype E and had identical ompA sequences. MLST was able to separate them, and the two variants are presented with different allele numbers in two samples. *, no sequence was obtained from the target region.
Analysis of chlamydial strain variation in men who have sex with men (MSM) is another area in which the MLST system was applied. As we have shown previously, certain genetic variants of ompA predominate in chlamydia-infected populations of MSM (5). We therefore wanted to investigate whether a similar picture emerged when the same strains were analyzed by MLST. Samples were collected over 1 year at a clinic for MSM where patients had had, on average, 4 partners (range, 1 to 85 partners) during the 3 months prior to examination for chlamydial infection.
A common ompA variant among MSM is EF017667, which comprised 98% of all serotype G strains in our previous study and which constituted 44% of all cases of chlamydial infection detected. Analysis of 10 such specimens by MLST differentiated them into four genotypes (Table 4). In addition, a single strain of serotype G with only one point mutation in ompA compared with the ompA sequence of variant EF017667 was examined. It was found to be markedly different in the MLST system, with divergent sequence variants in three of the five target regions.
TABLE 4.
MLST analysis of C. trachomatis specimens of predominant ompA serotypes in MSM
| Serotype | ompA variant (no. of specimens tested) | Designation for genetic variants detected in the following target regionsa:
|
No. of specimens | ||||
|---|---|---|---|---|---|---|---|
| hctB | CT058 | CT144 | CT172 | pbpB | |||
| G | EF017667 (10) | 20 | 21 | 5 | 4 | 6 | 6 |
| 20 | 8 | 5 | 4 | 6 | 1 | ||
| 20 | 21 | 13 | 4 | 6 | 1 | ||
| 10 | 8 | 5 | 4 | 6 | 2 | ||
| G | EF017668 (1) | 10 | 6 | 10 | 3 | 6 | 1 |
| D | EF017669 (9) | 5 | 20 | 5 | 4 | 8 | 9 |
| D | X62920 (D/IC-Cal8) (1) | 5 | 19 | 7 | 5 | 19 | 1 |
Each genetic variant of a target region is designated by an arbitrarily assigned number.
Another predominant variant, within serotype D (EF017669), comprised 89% of all cases of serotype D in our previous study with MSM. Nine of these specimens were examined by MLST, and their sequences were found to be identical in all five target regions. A single strain of another serotype D variant, which differed in only a single nucleotide position in ompA compared with the sequences of the nine specimens described above, was also analyzed by MLST. With that system, its sequence was found to differ in four of the target regions, and its sequence was clearly different from that of EF017669.
DISCUSSION
We present here a novel MLST system for the discrimination of C. trachomatis strains. The five hypervariable regions identified comprised three hypothetical genes and two genes annotated as hctB and pbpB, which code for a DNA- and a penicillin-binding protein, respectively. It is perhaps no coincidence that pbpB, like ompA, is a putative outer membrane protein potentially involved in the interaction with the host cell. The observed variability may be due to positive selection for amino acid sequence divergence for the purpose of escaping the host immune response. This is particularly likely, given that the level of sequence variation is much higher than that in noncoding regions of the genomes. The dnadist variability is, on average, 6.75 in coding DNA and 5.53 in noncoding DNA.
The C. trachomatis genome contains hypervariable regions called plasticity zones near the origin and terminus of replication. These are characterized by insertion-deletion differences and breakpoints in gene synteny (10). The CT172 locus is located within one of the plasticity zones, and CT144 is immediately next to it. This zone is characterized by a rapid rate of sequence evolution. The adjacent genes pbpB and ompA are centered in another area of the genome that is flanked by the conserved orthologs pkn5 (CT674) and CT695 and that also shows a high degree of sequence variability but no gene order structure differences. The hctB gene is located distantly from these in a shorter zone of six genes. The dnadist variability in these zones ranges from 13.2 (CT144 and CT172) to 17.9 (pbpB and ompA). Positive selection for high sequence variability makes these regions suitable for use for the typing of closely related clinical strains but not for the study of long-term evolutionary histories.
Initial analysis of the five regions separately in 16 reference serotype A to L3 strains indicated fewer genetic variants in those regions than in ompA, with the exception of pbpB, which showed the same number of polymorphisms. However, when the five MLST regions of clinical isolates of the predominant serotype, serotype E, were analyzed, four regions were found to have a similar or a higher number of genetic variants compared with the number for ompA. Although no covariation is expected, it is noteworthy that strains of serotype E have low levels of sequence variation (i.e., the number of nucleotide changes leading to distinct genetic variants is small) in other target regions compared with the numbers in strains of different serotypes. The pbpB target provided the highest intraserotype sequence variation and was more discriminatory than ompA for isolates of all four serotypes tested. This variation was based on the arbitrary selection of samples from 1 year in a laboratory that ran 27,000 annual tests and that detected 2,000 cases of chlamydial infection. Consequently, even higher levels of variation could be expected if sampling were to take place at different points in time and at different geographical sites.
To investigate the usefulness of MLST as a tool for molecular epidemiology in a real-world situation, it was used to test samples from a contact-tracing chain. This showed that MLST was superior to ompA analysis and was able to discriminate cases of chlamydial infection and link them to separate contact chains, whereas the ompA sequences were identical.
Analysis of MSM with chlamydial infections showed that cases with an identical and predominant serotype G variant of ompA were differentiated by MLST into four genotypes. In contrast, for infections of the predominant serotype D variant, all cases were also identical by MLST. Interestingly, this is in agreement with earlier findings of a lower relative frequency of shared ompA mutations in serotype G strains than in serotype D strains (7). Serotype G strains thus appear to be more variable than serotype D strains also in other regions of the genome.
Previous studies of different single genes have failed to find useful targets for a genotyping system of high resolution. This is explained by the genetic isolation of Chlamydia and the exceptionally low degree of horizontal DNA transfer during evolution (1, 13), leading to low sequence variation. When the results for the five candidate regions in our study were combined, the resolution of the MLST system was significantly higher than that obtained with ompA genotyping alone. Thus, within serotypes, between 50% and 82% of the strains tested had a unique MLST sequence pattern. Furthermore, ompA analysis only slightly enhanced the discrimination of C. trachomatis strains when it was added to analysis of the five candidate regions. However, in optimizing an MLST system, both sequence variation and sequence length are important. Omission of the longest target region, pbpB, considerably reduced the sequence information, but the resolution for genotyping was almost maintained. Further analysis based on more sequence information may indicate the optimal combination of target regions for use in MLST. The phylogenetics of these genetically variable regions may also provide information that will improve our understanding of regions important for pathogenesis.
In summary, our findings show that MLST can be used for the discrimination of C. trachomatis strains and can be applied to high-resolution molecular epidemiology. This provides a tool for understanding the spread of chlamydial infections in the community.
Acknowledgments
This work was supported by the National Institute of Public Health and the National Board of Health and Welfare, Sweden, and by Uppsala University Hospital, Uppsala (to B. Herrmann), and Uppsala University (to S. G. E. Andersson).
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Dalevi, D. A., N. Eriksen, K. Eriksson, and S. G. Andersson. 2002. Measuring genome divergence in bacteria: a case study using chlamydian data. J. Mol. Evol 55:24-36. [DOI] [PubMed] [Google Scholar]
- 2.Dean, D., E. Oudens, G. Bolan, N. Padian, and J. Schachter. 1995. Major outer membrane protein variants of Chlamydia trachomatis are associated with severe upper genital tract infections and histopathology in San Francisco. J. Infect. Dis. 172:1013-1022. [DOI] [PubMed] [Google Scholar]
- 3.Jonsdottir, K., M. Kristjansson, J. Hjaltalin Olafsson, and O. Steingrimsson. 2003. The molecular epidemiology of genital Chlamydia trachomatis in the greater Reykjavik area, Iceland. Sex. Transm. Dis. 30:249-256. [DOI] [PubMed] [Google Scholar]
- 4.Jurstrand, M., L. Falk, H. Fredlund, M. Lindberg, P. Olcen, S. Andersson, K. Persson, J. Albert, and A. Backman. 2001. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J. Clin. Microbiol. 39:3915-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klint, M., M. Lofdahl, C. Ek, A. Airell, T. Berglund, and B. Herrmann. 2006. Lymphogranuloma venereum prevalence in Sweden among men who have sex with men and characterization of Chlamydia trachomatis ompA genotypes. J. Clin. Microbiol. 44:4066-4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lysén, M., A. Osterlund, C. J. Rubin, T. Persson, I. Persson, and B. Herrmann. 2004. Characterization of ompA genotypes by sequence analysis of DNA from all detected cases of Chlamydia trachomatis infections during 1 year of contact tracing in a Swedish county. J. Clin. Microbiol. 42:1641-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Millman, K., C. M. Black, R. E. Johnson, W. E. Stamm, R. B. Jones, E. W. Hook, D. H. Martin, G. Bolan, S. Tavare, and D. Dean. 2004. Population-based genetic and evolutionary analysis of Chlamydia trachomatis urogenital strain variation in the United States. J. Bacteriol. 186:2457-2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morgenstern, B. 1999. DIALIGN 2: improvement of the segment-to-segment approach to multiple sequence alignment. Bioinformatics (Oxford, England) 15:211-218. [DOI] [PubMed] [Google Scholar]
- 9.Morre, S. A., J. M. Ossewaarde, J. Lan, G. J. van Doornum, J. M. Walboomers, D. M. MacLaren, C. J. Meijer, and A. J. van den Brule. 1998. Serotyping and genotyping of genital Chlamydia trachomatis isolates reveal variants of serovars Ba, G, and J as confirmed by omp1 nucleotide sequence analysis. J. Clin. Microbiol. 36:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ripa, T., and P. Nilsson. 2006. A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro. Surveill. 11:E061109.2. [DOI] [PubMed] [Google Scholar]
- 12.Salzberg, S. L., A. L. Delcher, S. Kasif, and O. White. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26:544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stephens, R. S. 2002. Chlamydiae in evolution: a billion years and counting, p. 3-12. In J. Schachter, G. Christiansen, I. N. Clarke, M. R. Hammerschlag, B. Kaltenboeck, C.-C. Kuo, R. G. Rank, G. L. Ridgway, P. Saikku, W. E. Stamm, R. S. Stephens, J. T. Summersgill, P. Timms, and P. B. Wyrick (ed.), Chlamydial infections. Proceedings of the 10th International Symposium on Human Chlamydial Infections. International Chlamydia Symposium, San Francisco, CA.
- 14.Sturm-Ramirez, K., H. Brumblay, K. Diop, A. Gueye-Ndiaye, J. L. Sankale, I. Thior, I. N′Doye, C. C. Hsieh, S. Mboup, and P. J. Kanki. 2000. Molecular epidemiology of genital Chlamydia trachomatis infection in high-risk women in Senegal, West Africa. J. Clin. Microbiol. 38:138-145. [DOI] [PMC free article] [PubMed] [Google Scholar]