Abstract
The development of reliable and rapid methods for the identification of patients colonized with vancomycin-resistant enterococci (VRE) is central to the containment of this agent within a hospital environment. To this end, we evaluated a prototype chromogenic agar medium (VRE-BMX; bioMérieux, Marcy l'Etoile, France) used to recover VRE from clinical specimens. This medium can also identify isolated colonies as either vancomycin-resistant Enterococcus faecium or Enterococcus faecalis, based on distinct colony colors. We compared the performance of VRE-BMX with bile esculin azide agar supplemented with vancomycin (BEAV). For this study, 147 stool samples were plated on each test medium and examined after 24 and 48 h of incubation. At 24 h, the sensitivity and specificity of each medium were as follows: BEAV, 90.9% and 89.9%, respectively; VRE-BMX, 96.4% and 96.6%, respectively. The positive predictive values (PPV) of VRE-BMX and BEAV at 24 h were 89.8% and 80.7%, respectively. VRE-BMX provided the identification of 10 isolates of vancomycin-resistant E. faecalis and 4 isolates of vancomycin-resistant E. faecium that were not recovered by BEAV. Further, VRE-BMX was capable of identifying patients colonized with both E. faecium and E. faecalis, a feature useful for infection control purposes that is not a function of BEAV. In terms of the recovery of vancomycin-resistant E. faecium and E. faecalis, the sensitivity and PPV were as follows: BEAV, 75.7% and 74.6%, respectively; VRE-BMX, 95.5% and 91.3%, respectively. In this initial evaluation, we found that VRE-BMX provided improved recovery of VRE from stool specimens, with the added advantage of being able to differentiate between vancomycin-resistant E. faecalis and E. faecium. Extending the incubation period beyond 24 h did not significantly improve the recovery of VRE and resulted in decreased specificity.
Enterococcus species are members of the normal intestinal flora and are the most common aerobic gram-positive cocci found in the large bowel of humans. The organisms have gained more notoriety, however, as nosocomial pathogens, now grouped as the third most common cause of bacteremia in the United States (5). Of the clinically important enterococci, E. faecium is more likely to acquire resistance to glycopeptides, while E. faecalis is linked more frequently to serious disease (12). The most common form of glycopeptide resistance is mediated by vanA, a transposon-mediated, inducible gene that leads to high-level resistance to vancomycin and teicoplanin. vanA can be plasmid mediated or chromosomal (10, 18). Glycopeptide resistance stemming from vanB is less common and confers resistance to vancomycin at moderate to high levels but not to teicoplanin (6, 12). Less commonly acquired vancomycin resistance genes include vanD, vanE, vanF, and vanG, which confer moderate to low-level resistance to glycopeptides.
The clinical impact of infection by vancomycin-resistant enterococci (VRE) has been examined in several studies (2, 3, 8, 9, 14-16, 19), with the most notable consequences being increases in mortality, length of hospital stay, and cost of hospitalization. While a clear link between colonization with glycopeptide-resistant Enterococcus and increased mortality has not been clearly established (12), antimicrobial therapy does promote selection and proliferation of VRE in the hospital environment (11, 12). Patients who become colonized with VRE can become asymptomatic carriers for months to years (12).
Until recently, detection of VRE colonization relied on culture techniques using selective/differential media (17). To date, the most effective medium identified for screening for VRE has been bile esculin azide agar supplemented with 6 μg of vancomycin/ml (BEAV) (11). While being reasonably sensitive, the use of selective culture techniques can be time consuming (the average recovery and identification time is 24 to 72 h) and labor intensive. Recent advances in the use of highly specific chromogenic substrates with sufficient sensitivity to identify VRE in a selective agar medium led to the development of VRE-BMX. The chromogenic substrates incorporated in VRE-BMX are targeted by enzymes specific for E. faecium or E. faecalis. Enzymatic degradation generates either purple (E. faecium) or blue-green (E. faecalis) by-products that distinguish the colony pigmentation of each species.
In this study, we compared the performance characteristics of VRE-BMX to BEAV for the detection and differentiation of VRE from stool samples obtained from hospitalized patients.
MATERIALS AND METHODS
Study enrollment and collection of clinical specimens.
Study subjects were selected from inpatients at Barnes-Jewish Hospital, a 1,442-bed tertiary care hospital in St. Louis, MO. Patients were considered for inclusion in the study if fecal specimens were submitted to the Barnes-Jewish microbiology laboratory for routine VRE screening. Collected specimens were transported and stored at 4°C for up to 24 h before being cultured. This study was approved by the Washington University Human Studies Committee (IRB no. 05-1091).
Media and culture conditions.
We compared the performance of an experimental chromogenic medium, VRE-BMX (bioMérieux, Marcy l'Etoile, France), with that of BEAV (Remel, Lenexa, KS). VRE-BMX and BEAV plates were directly inoculated by submerging a sterilized 10-μl inoculating loop in the specimen and streaking for isolation by the quadrant technique. This procedure was repeated for each medium type. Plates were incubated at 35°C in ambient air and examined for growth at 24 and 48 h. Colonies on VRE-BMX plates with purple or green pigmentation were presumptively identified as vancomycin-resistant E. faecium or E. faecalis, respectively (Fig. 1). The appearance of black colonies on BEAV plates was presumed to indicate positivity for VRE. Following observation of the colony morphologies, individual unique colonies were subcultured onto CPS ID 3 medium (bioMérieux), a chromogenic medium for presumptive identification of enterococci. Individual colonies were also subcultured to tryptic soy agar plates supplemented with 5% sheep blood (Remel) for confirmatory identification and antimicrobial susceptibility testing with the VITEK 2 automated identification and susceptibility system (bioMérieux). Additionally, the identification of isolated colonies and glycopeptide resistance were confirmed genotypically by PCR or PCR-restriction fragment length polymorphism (RFLP) analysis.
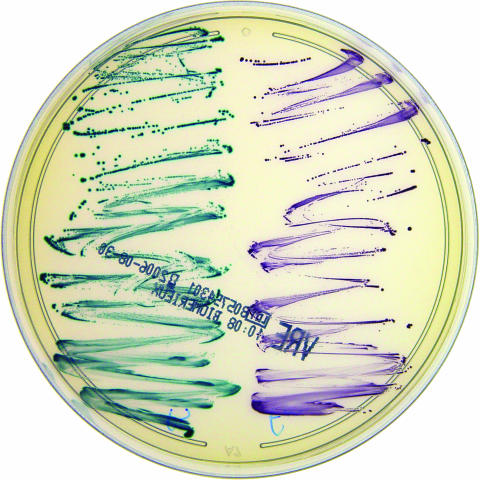
FIG. 1.
VRE-BMX medium plated with E. faecalis and E. faecium. This figure demonstrates the ability of VRE-BMX to identify E. faecium and E. faecalis based on the color of the colony; on VRE-BMX, colonies of E. faecalis appear blue-green and colonies of E. faecium appear purple. For the purposes of this study, fecal specimens were plated directly to VRE-BMX and BEAV and observed for growth at 24 and 48 h.
DNA isolation for PCR analysis.
Bacteria were grown overnight at 35°C on Columbia 5% sheep blood agar plates (bioMérieux). A bacterial suspension equivalent to a 0.5 McFarland standard was prepared, and cells were disrupted by mechanical lysis with glass beads of different sizes. After vortexing of the bacterial suspensions with beads for 2 min, samples were placed on ice and the supernatant was saved for amplification.
Multiplex PCR for detection of glycopeptide resistance genes and identification of E. faecium and E. faecalis.
Multiplex PCR was adapted from the method previously described by Depardieu et al. (7). Briefly, the multiplex PCR consisted of 25 μl of 2× reaction mix (QIAGEN Multiplex PCR master mix; QIAGEN, Courtaboeuf, France), 0.2 μM concentrations of each primer, 5 μl of 5× Q-solution (QIAGEN), and 5 μl of template DNA in a total volume of 50 μl. Samples were amplified as follows: an initial denaturation step at 95°C for 15 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 90 s, and elongation at 72°C for 1 min; and a final elongation at 72°C for 10 min. Amplified products were detected and quantified with a 2100 BioAnalyzer system (Agilent, Massy, France), according to the manufacturer's instructions.
Identification of enterococci by PCR-RFLP of the sodA gene.
A 429-bp fragment internal to the sodA gene was amplified with degenerate primers, as previously described by Poyart et al. (13). For RFLP analysis, 8 μl of PCR product was digested with 1 μl (10 units/μl) of MseI endonuclease (New England BioLabs, Ipswich, MA) in a total volume of 20 μl, according to the manufacturer's instructions. Digested products were then visualized by electrophoresis with a 2100 BioAnalyzer system (Agilent). RFLP patterns were analyzed by comparing the number and size of fragments between enterococcal species.
Statistical analysis.
To determine whether the results from each medium were significantly different, the significance of results was determined by MacNemar's test or by binomial distribution with the significance level fixed at 5% (if P was <5%, then the differences in performance were considered statistically significant).
RESULTS
Isolation of VRE from fecal samples.
Stool samples from 147 patients were cultured for VRE according to the protocol. Colonies on VRE-BMX were screened for a purple (vancomycin-resistant E. faecium) or blue-green (vancomycin-resistant E. faecalis) hue (Fig. 1). Purple or blue-green colonies were easily differentiated from other flora. BEAV plates were screened for colonies causing a blackening of the medium around the colony and were easily distinguished from surrounding contaminants. Following observation of the colony morphologies, suspect colonies of VRE were identified and tested for glycopeptide resistance with VITEK 2 and confirmed by molecular techniques, as described in Materials and Methods.
VRE-BMX performance characteristics.
Upon completion of the medium evaluation, an analysis was performed to determine the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) at 24 and 48 h. The sensitivity of each medium was calculated for detection of VRE as well as for the detection of vancomycin-resistant E. faecium and E. faecalis. After molecular confirmation, the sensitivity of VRE-BMX for the detection of VRE at 24 and 48 h was 96.4% and 94.8%, respectively (Table 1). The sensitivity of BEAV for the detection of VRE was 90.9% at 24 h and 91.4% at 48 h (Table 1). The differences in sensitivity between 24 and 48 h were not statistically significant.
TABLE 1.
Sensitivity and specificity analysis of VRE-BMX and BEAV for VRE at 24 and 48 h
| Incubation period (h) | Medium | No. of TPa | No. of FNb | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| 24 | VRE-BMX | 53 | 2 | 96.4 | 96.6 |
| BEAV | 50 | 5 | 90.9 | 89.9 | |
| 48 | VRE-BMX | 55 | 3 | 94.8 | 73.9 |
| BEAV | 53 | 5 | 91.4 | 77.2 |
A true positive (TP) is defined as a blue-green or purple colony on VRE-BMX or a gray to black colony on BEAV that was identified as VRE by VITEK 2 and confirmed by PCR.
A false negative (FN) is defined as an isolate that was confirmed as VRE on one medium but did not grow on another medium.
Further, the sensitivity of each medium was analyzed for the detection of Enterococcus species that carry clinically significant glycopeptide resistance determinants (vanA, vanB), most notably in vancomycin-resistant E. faecium and E. faecalis. The sensitivity of VRE-BMX for the detection of E. faecium was 94.4% at 24 h, and that of BEAV was 85.7% (Table 2). There was no significant change in sensitivity when the incubation period of either medium was extended to 48 h (Table 2). The sensitivity of VRE-BMX for the detection of E. faecalis colonization was 100% at both 24 and 48 h, while BEAV had a calculated sensitivity of 16.7 and 13.3% after 24 and 48 h of incubation, respectively (Table 2). The marked difference in the sensitivity of BEAV between vancomycin-resistant E. faecium and E. faecalis can be explained because VRE-BMX identified 11 patients that were colonized with both E. faecium and E. faecalis. While VRE-BMX can easily distinguish dual colonization because of the differential properties of the chromogenic substrates, BEAV has no such differential capabilities, making it impossible to differentiate vancomycin-resistant E. faecium from E. faecalis without further testing.
TABLE 2.
Sensitivity analysis of VRE-BMX and BEAV for E. faecium or E. faecalis at 24 and 48 h
| Incubation period (h) | Medium | Isolate | No. of TPa | No. of FNb | Sensitivity (%) |
|---|---|---|---|---|---|
| 24 | VRE-BMX | E. faecium | 51 | 3 | 94.4 |
| E. faecalis | 12 | 0 | 100.0 | ||
| BEAV | E. faecium | 48 | 8 | 85.7 | |
| E. faecalis | 2 | 10 | 16.7 | ||
| 48 | VRE-BMX | E. faecium | 54 | 3 | 94.7 |
| E. faecalis | 14 | 0 | 100.0 | ||
| BEAV | E. faecium | 51 | 6 | 89.5 | |
| E. faecalis | 2 | 13 | 13.3 |
A true positive (TP) is defined as a blue-green or purple colony on VRE-BMX or a gray to black colony on BEAV that was identified as VRE by VITEK 2 and confirmed by PCR.
A false negative (FN) is defined as an isolate that was confirmed as VRE on one medium but did not grow on another medium.
As with the sensitivity analysis, the specificity of VRE-BMX compared to BEAV was calculated at 24 and 48 h. The specificity analysis showed that VRE-BMX was 96.6% and 73.9% specific at 24 and 48 h, respectively (Table 1). The specificity of BEAV at 24 h was 89.9%, but it dropped to 77.2% after extended incubation (Table 1). The drop in specificity of each medium was due to the proliferation of contaminant flora that may cause false-positive results. These organisms included gram-negative bacilli and yeast on VRE-BMX and gram-positive bacilli, gram-positive cocci, and Enterococcus species other than E. faecalis or E. faecium on BEAV.
The PPV and NPV of both media are shown in Table 3. VRE-BMX produced a PPV of 92.7% for vancomycin-resistant E. faecium (vanA or vanB) and 75.0% for vancomycin-resistant E. faecalis (vanA or vanB) at 24 h. By comparison, the PPV of BEAV for all VRE was 76.9% at 24 h and did not change significantly when the incubation period was extended to 48 h.
TABLE 3.
PPV and NPV of VRE-BMX and BEAV for VRE at 24 and 48 h
| Incubation period (h) | Medium | No. of TPa | No. of FPb | No. of TNc | No. of FNd | PPV (%) | NPV (%) |
|---|---|---|---|---|---|---|---|
| 24 | VRE-BMX | 53 | 6 | 86 | 2 | 89.8 | 97.7 |
| BEAV | 50 | 12 | 80 | 5 | 80.7 | 94.1 | |
| 48 | VRE-BMX | 55 | 21 | 68 | 3 | 72.4 | 95.8 |
| BEAV | 53 | 18 | 71 | 5 | 74.7 | 93.4 |
A true positive (TP) is defined as a blue-green or purple colony on VRE-BMX or a gray to black colony on BEAV that was identified as VRE by VITEK 2 and confirmed by PCR.
A false positive (FP) is defined as an isolate that exhibited typical coloration on the respective medium but was not identified as VRE by VITEK 2 or confirmed by PCR.
A true negative (TN) is defined as the lack of a typically colored colony.
A false negative (FN) is defined as an isolate that was confirmed as VRE on one medium but did not grow on another medium.
Growth of contaminants.
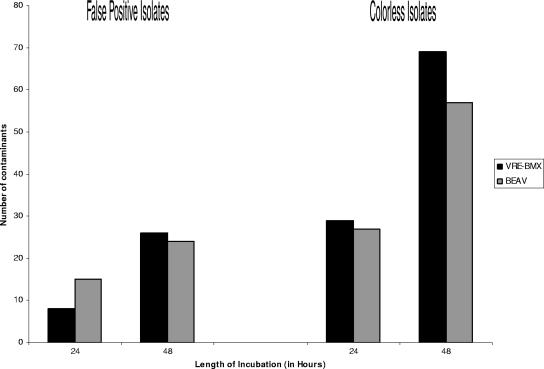
Breakthrough growth of contaminants on each medium was assessed at 24 and 48 h to determine the efficacy of the antimicrobial agents in VRE-BMX to suppress proliferation of normal stool flora (non-VRE). Contaminants were classified into two groups: (i) those that were considered potential false positives because they produced colonies with any hue of blue-green or purple on VRE-BMX that could be misinterpreted as vancomycin-resistant E. faecium or E. faecalis, or resulted in black colonies on BEAV, and (ii) colorless isolates. While BEAV grew 50% more contaminants at 24 h than did VRE-BMX, extending the incubation period to 48 h resulted in 24 and 26 cultures with breakthrough growth, respectively, on BEAV and VRE-BMX (Fig. 2). Isolates most likely to cause false-positive results on VRE-BMX included Candida spp. and gram-negative rods. Contaminants causing potential false-positive results on BEAV included Enterococcus spp. other than E. faecium and E. faecalis, gram-positive rods, and Streptococcus spp. Most breakthrough growth on VRE-BMX consisted of lightly green-colored colonies in the first quadrant of growth. However, contaminants on BEAV were found on all quadrants and were more likely to be Enterococcus spp. other than E. faecium and E. faecalis.
FIG. 2.
Growth of contaminants on each medium at 24 and 48 h. Contaminants are classified as potential false-positive isolates and colorless isolates. Isolates with any hue of blue-green or purple were classified as potential false positives on VRE-BMX; isolates with a black hue on BEAV were classified as potential false positives.
DISCUSSION
Recognizing the clinical impact of the VRE epidemic, the CDC drafted recommendations in 1995 to assist infection control personnel and hospital epidemiologists with the rapid spread of this organism within the hospital environment (4). Fundamental to these recommendations were strategies to contain cases or outbreaks of VRE and decrease the rates of transmission, including the isolation of VRE-infected or -colonized patients. The development of a sensitive method for detection of VRE colonization/infection, and preferably one that is rapid and simple to perform so as to facilitate screening of large numbers of patient samples with prompt isolation, is central to this goal.
The identification of VRE from colonized patients can be accomplished by screening cultures of stool or rectal swabs with differential and/or selective media. One such medium, BEAV, has the advantage of suppressing the normal fecal flora and allowing for growth of organisms carrying clinically significant glycopeptide resistance genes, such as vanA or vanB. However, the disadvantage of BEAV is that additional confirmatory tests are required to identify isolates and confirm glycopeptide resistance. Performing such tests on all of the colony morphotypes consistent with VRE can be time consuming and labor intense. In response to this problem, a number of manufacturers have developed novel methods of detecting VRE colonization that do not require confirmatory testing. These include molecular techniques, such as real-time PCR, and novel culture media.
While real-time gene amplification methods are currently being developed for the detection of agents such as VRE, they often add a level of cost and complexity that are not in line with the clinical relevance of the targeted organism. Though these methods offer the benefit of decreased turn-around time and have been shown to be significantly more sensitive than culture for many viruses, their utility for bacteria, especially organisms that comprise the normal gut flora, is more nebulous. For example, nucleic acid amplification techniques can identify antimicrobial resistance genes in the absence of a viable organism or when the resistance determinant is carried by an organism other than the targeted bacterium. In the latter example, the detection of vanA or vanB carried by an organism other than VRE in the presence of a pan-susceptible enterococcus could provide misleading information.
To address these issues, the ideal candidate for a VRE screen would be selective and differential: able to differentiate vancomycin-resistant Enterococcus faecalis from Enterococcus faecium without the additional step of identification and resistance confirmation. Further, the screen would have reasonable sensitivity for screening purposes and specificity to limit the need for additional testing.
The data presented in this article show that a chromogenic medium, VRE-BMX, from bioMérieux provides a viable alternative for screening of patients for gut colonization with VRE. This medium is able to identify and differentiate vancomycin-resistant E. faecium from E. faecalis while inhibiting growth of vancomycin-susceptible Enterococcus spp. Because this medium reliably identifies enterococci to species level and confirms glycopeptide resistance, it is not necessary to pursue additional biochemical analysis or determine antimicrobial susceptibilities in the clinical laboratory.
In this evaluation, BEAV culture combined with PCR was used as the “gold standard” for detection and confirmation of VRE from fecal specimens. After incubation of the plates for 24 and 48 h, the sensitivity, specificity, PPV, and NPV were calculated. In each of these analyses, we determined that VRE-BMX was at least a comparable, if not a superior, alternative to BEAV for the detection of VRE. Since BEAV does not identify Enterococcus to the species level and requires additional steps to obtain these data for infection control purposes, we conclude that VRE-BMX provides value-added qualities to a VRE screening agar. Our comparison showed that incubation of VRE-BMX beyond 24 h did not improve the sensitivity of the medium and actually reduced the specificity due to breakthrough growth of normal glycopeptide-resistant flora, such as Candida species. We noted that this was primarily observed in the first quadrant of the medium. When specimens were inoculated onto the medium, residual stool generally remained in the first quadrant. The excess specimen likely overwhelmed the inhibitory level of the vancomycin within the medium. To circumvent this problem, we would recommend first placing samples in a sterile diluent such as saline before plating and limiting the evaluation to 24 h of incubation.
Acknowledgments
W.M.D. is a consultant for bioMérieux, Inc., which provided support for the performance of this study.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Reference deleted.
- 2.Bhavnani, S. M., J. A. Drake, A. Forrest, J. A. Deinhart, R. N. Jones, D. J. Biedenbach, and C. H. Ballow. 2000. A nationwide, multicenter, case-control study comparing risk factors, treatment, and outcome for vancomycin-resistant and -susceptible enterococcal bacteremia. Diagn. Microbiol. Infect. Dis. 36:145-158. [DOI] [PubMed] [Google Scholar]
- 3.Carmeli, Y., G. Eliopoulos, E. Mozaffari, and M. Samore. 2002. Health and economic outcomes of vancomycin-resistant enterococci. Arch. Intern. Med. 28:2223-2228. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1995. Recommendations for preventing the spread of vancomycin resistance: recommendations of the Hospital Infection Control Practices Advisory Committee (HICPAC). Am. J. Infect. Control. 23:87-94. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1999. National nosocomial infections surveillance (NNIS) system report, data summary from January 1990 to May 1999. Am. J. Infect. Control. 27:520-532. [DOI] [PubMed] [Google Scholar]
- 6.Courvalin, P. 2005. Genetics of glycopeptide resistance in gram-positive pathogens. Int. J. Med. Microbiol. 294:479-486. [DOI] [PubMed] [Google Scholar]
- 7.Depardieu, F., B. Perichon, and P. Courvalin. 2004. Detection of the van alphabet and identification of enterococci and staphylococci at the species level by multiplex PCR. J. Clin. Microbiol. 42:5857-5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbutt, J. M., M. Ventrapragada, B. Littenberg, and L. M. Mundy. 2000. Association between resistance to vancomycin and death in cases of Enterococcus faecium bacteremia. Clin. Infect. Dis. 30:466-472. [DOI] [PubMed] [Google Scholar]
- 9.Lautenbach, E., W. B. Bilker, and P. J. Brennan. 1999. Enterococcal bacteremia: risk factors for vancomycin resistance and predictors of mortality. Infect. Control Hosp. Epidemiol. 20:318-323. [DOI] [PubMed] [Google Scholar]
- 10.Leclercq, R., E. Derlot, J. Duval, and P. Courvalin. 1988. Plasmid-mediated resistance to vancomycin and teicoplanin in Enterococcus faecium. N. Engl. J. Med. 319:157-161. [DOI] [PubMed] [Google Scholar]
- 11.Moellering, R. C. 1992. Emergence of Enterococcus as a significant pathogen. Clin. Infect. Dis. 14:1173-1178. [DOI] [PubMed] [Google Scholar]
- 12.Moellering, R. C. 2005. Enterococcus species, Streptococcus bovis, and Leuconostoc species, p. 2411-2421. In G. L Mandell, J. R. Bennett, and R. Dolin, Principles and practices of infectious diseases, 6th ed., vol 2. Elsevier Churchill Livingstone, Philadelphia, PA.
- 13.Poyart, C., G. Quesne, C. Boumaila, and P. Trieu-Cuot. 2001. Rapid and accurate species-level identification of coagulase-negative staphylococci by using the sodA gene as a target. J. Clin. Microbiol. 39:4296-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sloan, L. M., J. R. Uhl, E. A. Vetter, C. D. Schleck, W. S. Harmsen, J. Manahan, R. L. Thompson, J. E. Rosenblatt, and F. R. Cockerill III. 2004. Comparison of the Roche LightCycler vanA/vanB detection assay and culture for detection of vancomycin-resistant enterococci from perianal swabs. J. Clin. Microbiol. 42:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song, X., A. Srinivasan, D. Plaut, and T. M. Perl. 2003. Effect of nosocomial vancomycin-resistant enterococcal bacteremia on mortality, length of stay, and costs. Infect. Control Hosp. Epidemiol. 24:251-256. [DOI] [PubMed] [Google Scholar]
- 16.Stosor, V., L. R. Peterson, M. Postelnick, and G. A. Noskin. 1998. Enterococcus faecium bacteremia. Does vancomycin resistance make a difference? Arch. Intern. Med. 158:522-527. [DOI] [PubMed] [Google Scholar]
- 17.Teixeira, L. M., and R. M. Facklam. 2003. Enterococcus, p. 422-433. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed., vol 1. ASM Press, Washington, DC. [Google Scholar]
- 18.Uttley, A. H. C., C. H. Collins, J. Naidoo, and R. C. George. 1988. Vancomycin-resistant enterococci. Lancet i:57-58. [DOI] [PubMed] [Google Scholar]
- 19.Webb, M., L. W. Riley, and R. B. Roberts. 2001. Cost of hospitalisation for and risk factors associated with vancomycin-resistant Enterococcus faecium infection and colonization. Clin. Infect. Dis. 33:445-452. [DOI] [PubMed] [Google Scholar]