Abstract
In 1997, we developed a PCR assay for the detection of herpes simplex virus (HSV) DNA. Recently, we determined an optimal positivity criterion based on research specimens and a dilution study. We found that a cutoff of 50 HSV DNA copies/ml of swab specimen, a level 10-fold lower than our previous cutoff, minimizes misclassification.
We developed a validated method to detect herpes simplex virus (HSV) DNA in genital swabs (3) and have extensively utilized this assay in a variety of investigations as well as the clinical management of patients. We previously used a cutoff of 10 copies of HSV DNA per 20 μl (or 500 copies per ml) to determine positivity of the specimen (2, 5). This cutoff was defined because of concern over imperfect sensitivity at low concentrations and also over potential false positives due to environmental contamination or small amounts of DNA leaking from one sample to another (6).
Over the last 5 to 6 years, we have accumulated a large number of genital samples in HSV type 2 (HSV-2)-seropositive and HSV-seronegative participants and wanted to use this database to optimize the assay's sensitivity and specificity. Additionally, we have performed a series of dilution experiments to define the sensitivity of the PCR assay at low copy numbers. Using these experiments, we more accurately computed the test sensitivity, specificity, and misclassification rates.
We sought to determine the optimal cutoff for HSV shedding detection by PCR by comparing misclassification rates. A result is misclassified if the participant was shedding but is not detected (false negative) or if the participant was not shedding but tested positive by PCR (false positive). Misclassification rates require estimates of (i) sensitivity, (ii) specificity, (iii) shedding rate, and (iv) shedding quantity. These quantities were obtained as described below. The details of the assay were reported previously (8). Samples from each subject were not run sequentially by collection time.
Determination of sensitivity.
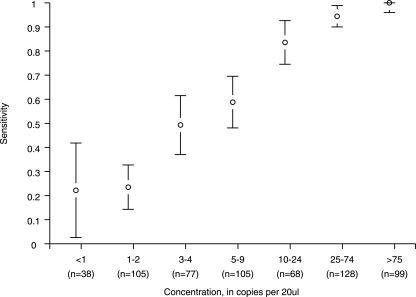
Multiple samples at various concentrations of HSV-2 were created by sequentially diluting a single sample of a known amount. The sensitivity of the assay was computed as the proportion of samples with detectable virus at each concentration. Fifteen HSV-2 concentrations were created, and similar concentrations were combined to provide seven ranges of from <1 to >75 copies per 20 μl. At least 38 repetitions in each range were made, with over 500 repetitions in total. Figure 1 shows each dilution and the estimated sensitivity based on the proportion of samples that were PCR positive. As expected, the sensitivity of detection increased at higher copy number.
FIG. 1.
Detection probabilities (quantity-specific sensitivity) of HSV-2-positive samples by HSV-2 concentration. The 95% confidence intervals are indicated with error bars, and the numbers of samples used to estimate sensitivity are shown below the horizontal axis.
Shedding rate and quantity among HSV-2-seropositive participants.
Samples from HSV-2-seropositive participants were taken from a wide variety of clinical studies in which daily swabs of genital secretions were collected, usually for 30 to 60 consecutive days per subject (1, 2, 4, 5, 7, 9, 10). We estimated the shedding rate as days with detectable virus by PCR out of days when genital swabs were taken; the swabbed areas included genital skin (penile shaft and buttocks) or mucosa (vulva, cervix, and anus). We also summarized, on days when shedding occurred, the shedding quantity and the corresponding frequencies. Three hundred twenty participants, 165 (52%) of whom were also HSV-1 seropositive, contributed 20,162 PCR samples of genital secretions that were assayed for HSV DNA by PCR. HSV DNA was detected in 3,992 samples (19.8%), 708 (18% of positive samples and 3% of samples overall) of which were below 10 copies per 20 μl (low positives). A total of 67% of the shedding occurred at >75 copies per 20 μl.
Determination of specificity.
We analyzed genital swabs from participants known to be HSV-1 and HSV-2 seronegative by Western blotting. We enrolled both pregnant women at the time of labor and nonpregnant participants who participated in daily home swabbing sessions. Two samples (0.6%) obtained from 350 pregnant HSV-seronegative women were found to be positive by PCR. Among six other HSV-seronegative participants who contributed 915 days during home swabbing sessions, 7 days (0.8%) from three subjects were positive by PCR. Typing results demonstrated HSV-1 in all three subjects. Most of these positive results were of low copy number. Thus, the specificity of the assay was above 99% (95% confidence interval within 98.4% to 100%) for all cutoffs considered.
Computation of misclassification.
We used the following formula, where s is the shedding rate, ϕ is the specificity, and θk and pk are the sensitivity of PCR and the observed frequency of shedding in copy level range k, respectively (both s and the observed pk are estimated from the shedding studies of HSV-2-seropositive participants, θk was from the dilution study, and ϕ was from HSV-seronegative participants): misclassified = false positives + false negatives = (1 − ϕ) × (1 − s) + Σ (1 − θk) × pk × s.
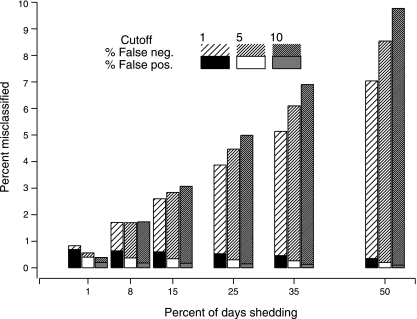
Figure 2 depicts the estimated false-positive, false-negative, and total misclassification rates, with shedding rates between 1% and 50%, using three cutoffs: ≥1, ≥5, and ≥10 copies per 20 μl. False-positive results comprised only a minority of misclassified results. At shedding rates greater than 8%, the lowest cutoff resulted in the lowest total misclassification rate.
FIG. 2.
Misclassification rates by shedding rate and cutoff for determination of positivity. Total bar height indicates overall misclassification, with shading distinguishing false negatives (neg.) and false positives (pos.).
We also examined the timing of low positives (below 10 copies per 20 μl) in relation to episodes of shedding, where an episode is any consecutive shedding period at copy levels greater than 10 copies per 20 μl. We tested whether low positives occurred randomly (as would be expected with contamination) or in close proximity to episodes. Of the 708 samples in which HSV DNA was detected at 1 to 10 copy numbers, 54% occurred during or within 2 days of a shedding episode, compared to 13% of 16,170 totally negative samples (P < 0.0001 by hypergeometric test), confirming that low positives are associated with confirmed HSV shedding episodes. A total of 15% of low positives also occurred coincidently with lesions, compared to 5% of completely negative samples (P < 0.0001).
Quantitative PCR for HSV DNA appears to be highly accurate. The test probably detects all shedding when the actual quantity is greater than 75 copies per 20 μl (3.75 × 103 copies/ml) (67% of received samples) and correctly identifies more than 99% of negative samples. Given the high specificity of the assay, and the occurrence of low positives in substantial numbers (3% of samples) and in temporal association with episodes and lesions, a detection of even low copy numbers of HSV DNA (1 copy/20 μl or 50 copies/ml) is almost invariably a true positive.
However, a cutoff determination also requires a consideration of the clinical laboratory context. For example, it may be deemed more risky to provide a false-positive diagnosis of genital HSV in an adult, and hence, we would prefer the cutoff with maximum specificity, whereas for detecting HSV encephalitis or neonatal herpes, the consequences of a false-negative assay are high.
Acknowledgments
This work was funded by NIH grant AI-30731-13 and by the Bill and Melinda Gates Foundation, grant 26469.
Colleagues Ted Gooley and Katherine Guthrie at the Fred Hutchinson Cancer Research Center provided valuable advice.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Corey, L., A. Wald, R. Patel, S. L. Sacks, S. K. Tyring, T. Warren, J. M. Douglas, Jr., J. Paavonen, R. A. Morrow, K. R. Beutner, L. S. Stratchounsky, G. Mertz, O. N. Keene, H. A. Watson, D. Tait, and M. Vargas-Cortes. 2004. Once-daily valacyclovir to reduce the risk of transmission of genital herpes. N. Engl. J. Med. 350:11-20. [DOI] [PubMed] [Google Scholar]
- 2.Gupta, R., A. Wald, E. Krantz, S. Selke, T. Warren, M. Vargas-Cortes, G. Miller, and L. Corey. 2004. Valacyclovir and acyclovir for suppression of shedding of herpes simplex virus in the genital tract. J. Infect. Dis. 190:1374-1381. [DOI] [PubMed] [Google Scholar]
- 3.Hobson, A., A. Wald, N. Wright, and L. Corey. 1997. Evaluation of a quantitative competitive PCR assay for measuring herpes simplex virus DNA content in genital tract secretions. J. Clin. Microbiol. 35:548-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krone, M. R., S. R. Tabet, M. Paradise, A. Wald, L. Corey, and C. L. Celum. 1998. Herpes simplex virus shedding among human immunodeficiency virus-negative men who have sex with men: site and frequency of shedding. J. Infect. Dis. 178:978-982. [DOI] [PubMed] [Google Scholar]
- 5.Posavad, C. M., A. Wald, S. Kuntz, M. L. Huang, S. Selke, E. Krantz, and L. Corey. 2004. Frequent reactivation of herpes simplex virus among HIV-1-infected patients treated with highly active antiretroviral therapy. J. Infect. Dis. 190:693-696. [DOI] [PubMed] [Google Scholar]
- 6.Ryncarz, A. J., J. Goddard, A. Wald, M. L. Huang, B. Roizman, and L. Corey. 1999. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J. Clin. Microbiol. 37:1941-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schacker, T., J. Zeh, H. L. Hu, E. Hill, and L. Corey. 1998. Frequency of symptomatic and asymptomatic herpes simplex virus type 2 reactivations among human immunodeficiency virus-infected men. J. Infect. Dis. 178:1616-1622. [DOI] [PubMed] [Google Scholar]
- 8.Wald, A., M. L. Huang, D. Carrell, S. Selke, and L. Corey. 2003. Polymerase chain reaction for detection of herpes simplex virus (HSV) DNA on mucosal surfaces: comparison with HSV isolation in cell culture. J. Infect. Dis. 188:1345-1351. [DOI] [PubMed] [Google Scholar]
- 9.Wald, A., J. Zeh, S. Selke, T. Warren, R. Ashley, and L. Corey. 2002. Genital shedding of herpes simplex virus among men. J. Infect. Dis. 186(Suppl. 1):S34-S39. [DOI] [PubMed] [Google Scholar]
- 10.Wald, A., J. Zeh, S. Selke, T. Warren, A. J. Ryncarz, R. Ashley, J. N. Krieger, and L. Corey. 2000. Reactivation of genital herpes simplex virus type 2 infection in asymptomatic seropositive persons. N. Engl. J. Med. 342:844-850. [DOI] [PubMed] [Google Scholar]