Abstract
A large outbreak with an aminoglycoside-resistant Enterobacter cloacae (AREC) clone occurred at the University Medical Center Utrecht beginning in 2001 and continued up through the time that this study was completed. This clone (genotype I) contains a conjugative R plasmid carrying the qnrA1, blaCTX-M-9, and aadB genes, encoding resistance to quinolones, extended-spectrum β-lactamases, and aminoglycosides, respectively. The aim of this study was to determine whether this clone was more transmissible than other AREC strains. Therefore, the dissemination of this genotype and of other E. cloacae strains was studied. In addition, infection control measures taken were evaluated. Pulsed-field gel electrophoresis analysis divided the 191 AREC strains into 42 different genotypes, of which 5 (12%) involved at least three patients. Aside from this outbreak (133 patients), only two other small outbreaks occurred, showing that the infection control measures were successful for all strains but one. Among 324 aminoglycoside-susceptible E. cloacae strains, 34/166 (20%) genotypes were identified from at least three patients, but only 4 involved small outbreaks. The outbreak strain was also detected in 11 of 15 other Dutch hospitals and caused outbreaks in at least 4. Evaluation of infection control measures showed that the outbreak strain disseminated throughout the hospital despite adequate implementation of internationally accepted guidelines on the control of multidrug-resistant Enterobacteriaceae (MRE). In conclusion, some MRE strains are better able to spread than others, and these strains may not be controlled by the current infection control guidelines. Strategies to identify such strains in an early phase and adapted guidelines for such “superbugs” are needed to prevent these clones from becoming endemic.
Worldwide, the prevalence of multidrug-resistant Enterobacteriaceae (MRE) in nosocomial settings is rising, and outbreaks are frequently reported (6, 7, 9, 10, 14, 17, 22, 29, 32, 33). Although in the past most reports described outbreaks that were confined to one ward or hospital, in recent years outbreaks have increasingly been reported to involve multiple health care facilities (e.g., other hospitals, nursing homes, and rehabilitation centers) (1, 4, 5, 12, 15, 36, 37). Infections with MRE lead to higher morbidity and mortality among affected patients and increase the hospital costs substantially (16, 18). Furthermore, most of these strains carry conjugative R plasmids that are easily transferred to other species, increasing the antimicrobial resistance problems in health care facilities (21, 22, 26, 28). In The Netherlands, national guidelines for the infection control of MRE have been developed by the Dutch Working Party on Infection Prevention (WIP) in accordance with international infection control guidelines, the guidelines of the Hospital Infection Control Practices Advisory Committee, and the guidelines of the Centers for Disease Control and Prevention (hygienic measures A1 to A3 and B1 to B5 in Table 1) (11, 13, 19, 30). By Dutch law, each hospital must have an infection prevention policy according the guidelines of the WIP, and the implementation of these guidelines is monitored by Dutch inspection of health care services. As a result, there is high compliance with these guidelines nationwide. At the University Medical Center Utrecht (UMCU), Utrecht, The Netherlands, the Division of Hospital Hygiene and Infection Prevention (HHIP) working group is responsible for the implementation of these guidelines. This strategy has been highly successful in preventing the spread of methicillin-resistant Staphylococcus aureus and vancomycin-resistant enterococci (VRE) in The Netherlands in previous years (2, 24, 31, 34, 35).
TABLE 1.
Summary of infection control measures for MRE at the UMCU
| Type of intervention | Designation | Descriptiona |
|---|---|---|
| Standard infection prevention policy | A1 | Contact isolation (gloves + gowns) of cases (patients with MRE)b |
| A2 | Active surveillance of patients with an increased risk for MRE (e.g., transfer from a hospital abroad)b | |
| A3 | Informing receiving institution about presence of MRE upon transfer of a patient with MRE to another health care institutionc | |
| Policy during an outbreak with MREb | B1 | Formation of a multidisciplinary team to develop an outbreak policy |
| B2 | Contact isolation of patients in single rooms | |
| B3 | Cohorting of patients; cohorting at ICU/MCU into two cohorts (contact patients and new admitted patients) | |
| B4 | Periodic surveillance of patients on wards involved | |
| B5 | Identification of possible (environmental) reservoirs | |
| Additional measures during the outbreak described in this study | C1 | For contact patient cohort, contact isolation until discharge from ICU/MCU; after transfer to the wards, stopping contact isolation upon three consecutive negative screening cultures |
| C2 | Observations to evaluate compliance to the implemented contact isolations | |
| C3 | Disinfection of room with chlorine solution (300 ppm) and change of bed curtains after discharge of a patient | |
| C4 | Labeling of cases in the hospital information system to install contact isolation in single rooms at readmission | |
| C5 | Genotyping of at least one AREC isolate per patient until outbreak strain was detected | |
| C6.1 | Weekly screening for MRE on the ICUs/MCUs involved until 4 wks after discharge of the last patient | |
| C6.2 | Monthly screening for MRE at all ICUs/MCUs | |
| C7 | Periodic information program for the health care workers | |
| C8 | For newly admitted patient cohort, preemptive contact isolation at the ICUs/MCUs until 2 wks after discharge of the last patient; after transfer to the wards, stopping contact isolation upon three consecutive negative screening cultures | |
| C9 | Daily disinfection of all bed sites with alcohol and of floors and sinks with a chlorine solution for the ICUs/MCUs involved | |
| C10 | Implementation of a restrictive antibiotic policy for ICUs/MCUs involved as described previously (39); based on the antibiogram of the outbreak clone, recommendation of carbapenems with or without amikacin as empirical therapy for infections with Enterobacteriaceae and proven infections with the outbreak strain |
In January 2003, it became clear that despite the implementation of the guidelines, a large outbreak with an aminoglycoside-resistant Enterobacter cloacae (AREC) strain (genotype I) had been occurring at the UMCU since January 2001 (20, 28). This clone contained a conjugative R plasmid carrying at least the resistance genes qnrA1, blaCTX-M-9, aadB, and sulI, encoding resistance to quinolones, extended-spectrum β-lactamases (ESBLs), aminoglycosides (gentamicin and tobramycin), and sulfamethoxazole, respectively (28). Implementation of the recommended guidelines extended with some far-reaching control measures had not controlled the spread of this MRE clone up through the time that this study was completed. The aim of this study was to determine whether this clone was more transmissible than other AREC strains. Therefore, the dissemination of this outbreak strain as well as of other E. cloacae isolates was studied. In addition, infection control measures were evaluated. Furthermore, it was investigated whether this strain had emerged in other Dutch hospitals as well.
MATERIALS AND METHODS
Setting.
The UMCU is a 1,042-bed tertiary care hospital with all medical disciplines represented. Each discipline has its own ward(s) with its own staff. There are four adult intensive care units (ICUs), of which three (neurology/neurosurgery, surgery, and thoracic surgery) are located next to each other; the fourth one (medical ICU) is located on a different floor in a different part of the building. The children's hospital is located next to the adult hospital. Approximately 27,000 patients are admitted to the hospital each year.
Infection control policy.
At the UMCU, the HHIP is responsible for the development and implementation of the infection control policy. In Table 1, the infection control policy for resistant Enterobacteriaceae in the UMCU is summarized. From 2001 until the recognition of the outbreak at the end of January 2003, the standard infection prevention policy was applied to all patients with aminoglycoside- or carbapenem-resistant or ESBL-positive Enterobacteriaceae (Table 1, measures designated A). After the recognition of the outbreak, additional measures were taken either hospital-wide or only at the wards of the two divisions (surgery and neurology/neurosurgery) predominantly involved (Table 1, measures designated B and C).
Description of the outbreak.
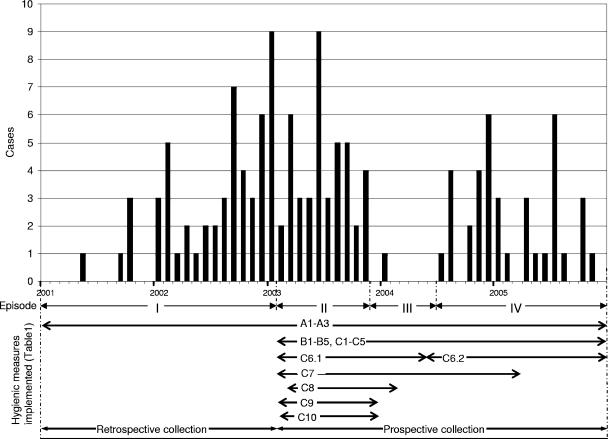
In retrospect, the outbreak with the genotype I strain could be divided in four episodes (Fig. 1) as described below.
FIG. 1.
Monthly incidence of E. cloacae genotype I cases and the four episodes (I to IV) into which the outbreak was divided. The arrows indicate the periods of time in which the different hygienic measures described in Table 1 were implemented.
(i) Episode I (January 2001 to January 2003).
At the end of January 2003, genotyping results revealed a large outbreak of one clonal lineage (genotype I) involving 53 patients (20).
(ii) Episode II (February 2003 to November 2003).
In February 2003, several additional hygienic measures were taken at the neurology, neurosurgery, and surgery wards along with the associated ICUs (Table 1). Environmental screening, performed to trace the possible source of the outbreak, had revealed the presence of the outbreak strain in the cleaning water used for the floors and sinks. Disinfection procedures with a chlorine solution (300 ppm) were introduced. Despite the measures taken, 42 new cases were identified at different wards: 31 at the neurosurgery or surgery wards and 11 patients on other wards, including the other two ICUs. The vast majority of these patients could be epidemiologically linked to one another by their overlapping hospital stays. The total number of patients nursed in isolation for (suspected) carriership and/or infected patients with MRE (Table 1) increased from 2001 to 2003 from 526 to 1,123 patients.
(iii) Episode III (December 2003 to June 2004).
As of December 2003, the outbreak seemed under control. Consequently, the preemptive contact isolation precautions taken for all new admitted patients at ICUs and medium care units were stopped. In the 7 months from December 2003 to June 2004, only one case was identified as a result of the weekly surveillance.
(iv) Episode IV (July 2004 to December 2005).
In July, a new case was identified by a positive screening culture obtained within 48 h of the admission of a patient after this patient was transferred from another Dutch hospital. Subsequently, 20 new cases that were epidemiologically linked to this index patient were identified. Finally, from February 2005 to the end of 2005, another 16 cases were identified: 13 at or linked to the neurosurgery or surgery wards and 3 at unrelated wards.
Microbiological investigation.
At the UMCU, clinical specimens were cultured according Clinical and Laboratory Standards Institute (CLSI) guidelines. Identification and susceptibility testing were performed by use of the Phoenix 100 automated microbiology system with version V3.22 software (Becton Dickinson Biosciences, Sparks, MD) and interpreted according CLSI guidelines (27). Intestinal screening was performed by culturing rectal swab samples in a selective Mueller-Hinton bouillon containing 8 μg/ml tobramycin, 10 μg/ml vancomycin, and 5 μg/ml nystatine. Genotyping was performed by pulsed-field gel electrophoresis (PFGE) using XbaI in a modified protocol of the PFGE method described previously (3, 21). Cluster analysis was performed by using BioNumerics, version 2.5 (Applied Maths, Kortrijk, Belgium) (20).
Groups of isolates included in this study. (i) Group 1.
Group 1 consisted of all AREC strains and aminoglycoside-susceptible Enterobacter cloacae (ASEC) strains stored in our laboratory from 2001 through 2005. The storage of all invasive isolates as well as that of the first aminoglycoside-resistant Enterobacteriaceae isolate of each patient is a routine procedure in our laboratory (20). From 2003 to 2005, incidentally noninvasive E. cloacae isolates from patients that were suspected to have hospital-acquired infections were stored. For some patients, more than one isolate was available, but only one isolate of each PFGE type per patient was included.
(ii) Group 2.
After the detection of the outbreak strain in cultures taken from a patient within 48 h after transfer from another Dutch hospital, the extent to which the strain had disseminated nationwide was investigated. Therefore, 15 clinical microbiology laboratories throughout The Netherlands representing 15 hospitals, including 5 university hospitals, were asked to send AREC strains isolated in 2005 or 2006, of which one isolate per patient was included in group 2.
Typing and characterization of Dutch isolates.
The isolates obtained from other microbiological laboratories were typed by PFGE (3, 21). Isolates belonging to genotype I were further characterized by PCR for the presence of the qnrA1 gene encoding reduced susceptibility to quinolones as described before (28). The presence of the qnrA1 gene was considered indicative of the presence of the 180-kb multiresistance plasmid pQC (28).
Definitions.
A cluster was defined as a group of isolates obtained from at least three patients with PFGE patterns that showed at least 80% similarity, as was defined earlier for the outbreak genotype (20). An outbreak was defined as a cluster in which two subsequent isolates were obtained less than 2 months after the first isolate was obtained.
RESULTS
Genotyping.
In total, 515 E. cloacae isolates from 480 patients were genotyped: 191 AREC strains obtained from 179 patients and 324 ASEC strains from 320 patients (from 19 patients, both AREC and ASEC were isolated). Of the 515 isolates, 405 (79%) were isolated from clinical specimens and 110 (21%) from surveillance cultures. The clinical isolates caused the following infections: 60 bloodstream infections (8 AREC strains; 52 ASEC strains), 114 invasive infections (25 AREC strains; 89 ASEC strains) (36 intra-abdominal infections, 23 other surgical infections, 28 orthopedic infections, 11 neurosurgical infections [7 cases of meningitis], 3 cases of pleural empyema, 3 cases of peritoneal dialysis-related peritonitis, 10 other), 72 wound infections (26 AREC strains; 46 ASEC strains), 117 respiratory tract infections or colonizations (48 AREC strains; 69 ASEC strains), and 42 urinary tract infections or colonizations (21 AREC strains; 21 ASEC strains).
Cluster analysis divided the 191 AREC strains into 42 different genotypes, of which 5 (12%) were represented by at least three patients. Genotype I, representing the outbreak strain, was isolated from 133 patients. The other four genotypes were obtained from the following numbers of patients: six patients (genotype 176), five patients (genotype 33), and two times three patients (genotypes 35 and 96). If cluster analysis was restricted to the 128 clinical isolates, 24 different genotypes were identified, of which 2 (8%) were represented by at least 3 patients: genotype I (97 patients) and genotype 33 (4 patients). However, outbreaks were observed only for clusters 33 and 176, with four and six patients, respectively, that had entirely overlapping times of hospitalization.
Cluster analysis divided the 324 ASEC strains into 166 different genotypes, of which 34 (20%) were represented by at least 3 patients (1, 2, 1, 4, 9, and 17 different genotypes detected from 13, 9, 8, 6, 4, and 3 patients, respectively). However, only four outbreaks with only three patients each were recorded.
Interestingly, the most prevalent genotype among the ASEC strains was genotype Ia (13 patients). This genotype is that of the outbreak strain lacking the R plasmid and was isolated for the first time from a blood culture in 1998. In the first three periods of the outbreak, genotype Ia was found only in patients who harbored the resistant genotype I as well. However, in episode IV, genotype Ia was isolated independently of genotype I from six of the seven patients. Three of these patients were hospitalized at different wards involved in the outbreak. Surprisingly, the other three were isolated at two different ICUs at the children's hospital, where genotype I had never been isolated.
To summarize, the aminoglycoside-resistant genotype I was by far the most common strain. This remains true even when only clinical isolates are taken into account. Among ASEC strains, genotype Ia represented the largest cluster as well.
Comparison of susceptibility patterns.
To determine whether the increased transmission rate of genotype I was associated with increased drug resistance, the susceptibility patterns for these isolates were compared with the patterns for the aminoglycoside-resistant isolates belonging to other genotypes. The quinolone resistance rate was higher (89% versus 26%; P < 0.0001) and the cotrimoxazole resistance rate was lower (11% versus 40%; P < 0.0001) in the genotype I isolates than in the AREC strains of other genotypes, while no significant difference regarding susceptibility to expanded-spectrum and broad-spectrum cephalosporins was detected. The other AREC genotypes (genotypes 33 and 176) causing small outbreaks were ciprofloxacin susceptible.
The group of ASEC isolates was significantly more susceptible for the antibiotics tested (chi-square test; P < 0.0001 for all antibiotics tested) than was the group of genotype I isolates or the group of AREC strains with other genotypes.
Multicenter outbreak.
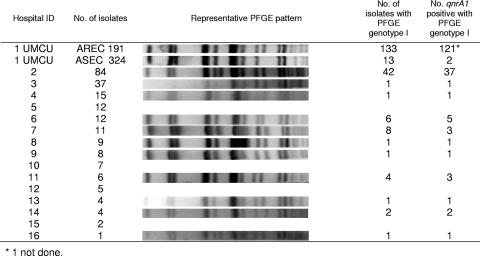
In total, 217 AREC strains obtained from 15 hospitals (range, 1 to 84 [mean, 15; median, 8]) from across The Netherlands were genotyped. At these facilities, the national recommended infection control practices were in place. Cluster analysis showed that in addition to being found at the UMCU, genotype I resided in 11 other hospitals, a nursing home, and a rehabilitation center, involving 68 patients in total (Fig. 2). Nearly all isolates were positive for qnrA1. The presence of genotype I isolates in two hospitals and in the last two health care facilities could be traced back to transfers of cases from one of the other hospitals involved. In four other hospitals, genotype I isolates caused outbreaks as well, involving at least 42, 8, 5, and 4 patients, respectively. Two of these outbreaks were still not under control up through the time that this study was completed. In addition, one other AREC strain (genotype 35) was identified as a “multicenter” strain, being present in four hospitals and involving at least 3, 1, 1, and 1 patient(s), respectively.
FIG. 2.
PFGE patterns for genotype I isolates from 11 of 15 Dutch hospitals and the UMCU and the presence or absence of the qnrA1 gene.
DISCUSSION
The results of this study show that an MRE strain disseminated throughout the hospital despite adequate implementation of the internationally accepted guidelines on the control of MRE. Only after the implementation of additional extended, sometimes far-reaching, control measures could the spread of this MRE clone be stopped initially. Unfortunately, a few months later the strain was reintroduced into the hospital by the transfer of a patient from another hospital. The far-reaching control measures were not reinstalled, and the strain has become endemic at the UMCU.
Although compliance to the infection control measures was not prospectively monitored, strong circumstantial evidence is provided that the measures were adequately implemented. In the period of the outbreak, only two very small outbreaks with other AREC genotypes occurred. Another finding indicative of an adequate compliance to the guidelines was the doubling of the total number of patients that were nursed in isolation for carriership for MRE in this period. In addition, for ASEC strains only four small outbreaks, each involving three patients, were observed, which supports the notion that standard infection control measures were adequately implemented. Routine hospital-wide surveillance of multidrug-resistant Enterobacteriaceae, VRE, and methicillin-resistant Staphylococcus aureus and Pseudomonas aeruginosa at the ICUs did not show any relevant outbreaks (data not shown).
These data indicate that genotype I had, in comparison to other multidrug-resistant strains, an extra capacity to disseminate. This hypothesis is supported by the observation that the outbreak strain was found at other Dutch health care institutions and caused outbreaks in at least four other hospitals. Furthermore, previous reports indicate that strains with an increased epidemic character exist. The spread of genotype I to different health care facilities resembles the spreads of a Klebsiella pneumoniae clone in New York City, NY, of different K. pneumoniae strains in France and Scotland, of an Enterobacter aerogenes clone in France and Spain, of an Escherichia coli clone in Taiwan, and of an ESBL-producing E. coli clone that was detected not only in six health care facilities but also in the community (1, 4, 5, 15, 23, 36, 37). All these outbreak strains resemble each other in that they were multidrug resistant. They were all were ESBL positive, and nearly all were resistant to aminoglycosides and ciprofloxacin. The question is whether this increased transmission rate was purely due to a selective advantage of their multidrug resistance phenotype or whether additional genetic properties played a role as well. The results of this study show that multidrug resistance alone is not enough to cause a strain to become highly epidemic. In the same hospital setting, other multidrug-resistant strains did not disseminate; among ASEC strains, genotype Ia had the highest dissemination rate, and this susceptible variant of the outbreak clone has been endemic within our hospital since 1989. Therefore, in addition to the resistance phenotype, other features improving the adaptation of the strain to the hospital environment may play a role as well. These features may result from mutations mediated by stress response pathways that are induced by a stressful environment. Recently, it has been shown that quinolone resistance requires the induction of such stress response pathways, which facilitate mutations in general (8). Since genotype I was quinolone resistant, it can be hypothesized that the induction of these responses resulted in other features improving the survival capacity of the strain in addition to quinolone resistance. Further research is needed to identify these features.
To our knowledge, this is the first time that internationally (and nationally) recommended guidelines for the control of MRE were evaluated in a study. The results showed that the recommended infection control measures were successful for all strains but one. The lack of success for genotype I indicates that an alternative strategy for highly transmissible strains is needed. If such an outbreak strain may be identified in an early stage of the outbreak, infection control measures may become tailor-made, reducing the need for extensive measures to only a limited number of resistant strains for a limited period of time. Such an approach was very successful during a VRE outbreak in our hospital in which stringent infection control measures were taken only for a specific clone (25).
In conclusion, the results of this study show that some MRE strains are more transmissible than others and that these strains may not be controlled by the current infection control guidelines. Therefore, strategies to identify such strains in an early phase as well as implementation of adapted guidelines for such “superbugs” are needed.
Acknowledgments
We thank the microbiology departments of the other Dutch hospitals for sending E. cloacae isolates and Judith P. M. Vlooswijk, Kim P. Jalink, and Annemieke Overbeek for providing assistance.
Footnotes
Published ahead of print on 21 February 2007.
REFERENCES
- 1.Arlet, G., M. Rouveau, I. Casin, P. J. Bouvet, P. H. Lagrange, and A. Philippon. 1994. Molecular epidemiology of Klebsiella pneumoniae strains that produce SHV-4 beta-lactamase and which were isolated in 14 French hospitals. J. Clin. Microbiol. 32:2553-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armeanu, E., and M. J. M. Bonten. 2005. Control of vancomycin-resistant enterococci: one size fits all? Clin. Infect. Dis. 41:210-216. [DOI] [PubMed] [Google Scholar]
- 3.Barrett, T. J., H. Lior, J. H. Green, R. Khakhria, J. G. Wells, B. P. Bell, K. D. Greene, J. Lewis, and P. M. Griffin. 1994. Laboratory investigation of a multistate food-borne outbreak of Escherichia coli O157:H7 by using pulsed-field gel electrophoresis and phage typing. J. Clin. Microbiol. 32:3013-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, P. A., S. Bratu, C. Urban, M. Visalli, N. Mariano, D. Landman, J. J. Rahal, S. Brooks, S. Cebular, and J. Quale. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 beta-lactamases in New York City. Clin. Infect. Dis. 39:55-60. [DOI] [PubMed] [Google Scholar]
- 5.Bratu, S., D. Landman, R. Haag, R. Recco, A. Eramo, M. Alam, and J. Quale. 2005. Rapid spread of carbapenem-resistant Klebsiella pneumoniae in New York City: a new threat to our antibiotic armamentarium. Arch. Intern. Med. 165:1430-1435. [DOI] [PubMed] [Google Scholar]
- 6.Canton, R., T. M. Coque, and F. Baquero. 2003. Multi-resistant Gram-negative bacilli: from epidemics to endemics. Curr. Opin. Infect. Dis. 16:315-325. [DOI] [PubMed] [Google Scholar]
- 7.Canton, R., A. Oliver, T. M. Coque, M. del Carmen Varela, J. C. Perez-Diaz, and F. Baquero. 2002. Epidemiology of extended-spectrum beta-lactamase-producing Enterobacter isolates in a Spanish hospital during a 12-year period. J. Clin. Microbiol. 40:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirz, R. T., J. K. Chin, D. R. Andes, V. de Crecy-Lagard, W. A. Craig, and F. E. Romesberg. 2005. Inhibition of mutation and combating the evolution of antibiotic resistance. PLoS Biol. 3:e176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte, A., F. Boavida, F. Grosso, M. Correia, L. M. Lito, J. Melo Cristino, and M. J. Salgado. 2003. Outbreak of GES-1 beta-lactamase-producing multidrug-resistant Klebsiella pneumoniae in a university hospital in Lisbon, Portugal. Antimicrob. Agents Chemother. 47:1481-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dubouix, A., C. Roques, C. Segonds, M. J. Jeannot, S. Malavaud, S. Daude, G. Chabanon, and N. Marty. 2005. Epidemiological investigation of a Serratia liquefaciens outbreak in a neurosurgery department. J. Hosp. Infect. 60:8-13. [DOI] [PubMed] [Google Scholar]
- 11.Dutch Working Party on Infection Prevention. 2005. Maatregelen tegen overdracht van bijzondere-resistente micro-organsimen (BRMO). WIP guidelines. www.wip.nl/free_content/Richtlijnen/11BRMO.pdf.
- 12.Galdbart, J.-O., F. Lemann, D. Ainouz, P. Feron, N. Lambert-Zechovsky, and C. Branger. 2000. TEM-24 extended-spectrum beta-lactamase-producing Enterobacter aerogenes: long-term clonal dissemination in French hospitals. Clin. Microbiol. Infect. 6:316-323. [DOI] [PubMed] [Google Scholar]
- 13.Garner, J. S., and the Hospital Infection Control Practices Advisory Committee. 1996. Guideline for isolation precautions in hospitals. Infect. Control Hosp. Epidemiol. 17:53-80. [DOI] [PubMed] [Google Scholar]
- 14.Giraud-Morin, C., and T. Fosse. 2003. A seven-year survey of Klebsiella pneumoniae producing TEM-24 extended-spectrum beta-lactamase in Nice University Hospital (1994-2000). J. Hosp. Infect. 54:25-31. [DOI] [PubMed] [Google Scholar]
- 15.Hobson, R. P., F. M. MacKenzie, and I. M. Gould. 1996. An outbreak of multiply-resistant Klebsiella pneumoniae in the Grampian region of Scotland. J. Hosp. Infect. 33:249-262. [DOI] [PubMed] [Google Scholar]
- 16.Hyle, E. P., A. D. Lipworth, T. E. Zaoutis, I. Nachamkin, W. B. Bilker, and E. Lautenbach. 2005. Impact of inadequate initial antimicrobial therapy on mortality in infections due to extended-spectrum beta-lactamase-producing Enterobacteriaceae: variability by site of infection. Arch. Intern. Med. 165:1375-1380. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, X., Y. Ni, Y. Jiang, F. Yuan, L. Han, M. Li, H. Liu, L. Yang, and Y. Lu. 2005. Outbreak of infection caused by Enterobacter cloacae producing the novel VEB-3 beta-lactamase in China. J. Clin. Microbiol. 43:826-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang, C.-I., S.-H. Kim, W. B. Park, K.-D. Lee, H.-B. Kim, M. Oh, E.-C. Kim, and K.-W. Choe. 2004. Bloodstream infections caused by Enterobacter species: predictors of 30-day mortality rate and impact of broad-spectrum cephalosporin resistance on outcome. Clin. Infect. Dis. 39:812-818. [DOI] [PubMed] [Google Scholar]
- 19.Kluytmans-Vandenbergh, M. F. Q., J. A. J. W. Kluytmans, and A. Voss. 2005. Dutch guideline for preventing nosocomial transmission of highly resistant microorganisms (HRMO). Infection 33:309-313. [DOI] [PubMed] [Google Scholar]
- 20.Leverstein-van Hall, M. A., H. E. M. Blok, A. Paauw, A. C. Fluit, A. Troelstra, E. M. Mascini, M. J. M. Bonten, and J. Verhoef. 2006. Extensive hospital-wide spread of a multidrug-resistant Enterobacter cloacae clone, with late detection due to a variable antibiogram and frequent patient transfer. J. Clin. Microbiol. 44:518-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leverstein-van Hall, M. A., A. T. A. Box, H. E. M. Blok, A. Paauw, A. C. Fluit, and J. Verhoef. 2002. Evidence of extensive interspecies transfer of integron-mediated antimicrobial resistance genes among multidrug-resistant Enterobacteriaceae in a clinical setting. J. Infect. Dis. 186:49-56. [DOI] [PubMed] [Google Scholar]
- 22.Liu, C.-P., N.-Y. Wang, C.-M. Lee, L.-C. Weng, H.-K. Tseng, C.-W. Liu, C.-S. Chiang, and F.-Y. Huang. 2004. Nosocomial and community-acquired Enterobacter cloacae bloodstream infection: risk factors for and prevalence of SHV-12 in multiresistant isolates in a medical centre. J. Hosp. Infect. 58:63-77. [DOI] [PubMed] [Google Scholar]
- 23.Mammeri, H., G. Laurans, M. Eveillard, S. Castelain, and F. Eb. 2001. Coexistence of SHV-4- and TEM-24-producing Enterobacter aerogenes strains before a large outbreak of TEM-24-producing strains in a French hospital. J. Clin. Microbiol. 39:2184-2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mascini, E. M., and M. J. M. Bonten. 2005. Vancomycin-resistant enterococci: consequences for therapy and infection control. Clin. Microbiol. Infect. 11(Suppl. 4):43-56. [DOI] [PubMed] [Google Scholar]
- 25.Mascini, E. M., A. Troelstra, M. Beitsma, H. E. M. Blok, K. P. Jalink, T. E. M. Hopmans, A. C. Fluit, R. J. Hené, R. J. L. Willems, J. Verhoef, and M. J. M. Bonten. 2006. Genotyping and preemptive isolation to control an outbreak of vancomycin-resistant Enterococcus faecium. Clin. Infect. Dis. 42:739-746. [DOI] [PubMed] [Google Scholar]
- 26.Naiemi, N. A., B. Duim, P. H. M. Savelkoul, L. Spanjaard, E. de Jonge, A. Bart, C. M. Vandenbroucke-Grauls, and M. D. de Jong. 2005. Widespread transfer of resistance genes between bacterial species in an intensive care unit: implications for hospital epidemiology. J. Clin. Microbiol. 43:4862-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.NCCLS/CLSI. 2003. Performance standards for antimicrobial susceptibility testing: 14th international supplement. NCCLS document M100-S14. NCCLS, Wayne, PA.
- 28.Paauw, A., A. C. Fluit, J. Verhoef, and M. A. Leverstein-van Hall. 2006. Enterobacter cloacae outbreak and emergence of quinolone resistance gene in Dutch hospital. Emerg. Infect. Dis. 12:807-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peña, C., M. Pujol, C. Ardanuy, A. Ricart, R. Pallares, J. Liñares, J. Ariza, and F. Gudiol. 1998. Epidemiology and successful control of a large outbreak due to Klebsiella pneumoniae producing extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 42:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirwitz, S., and the Hospital Infection Control Practices Advisory Committee. 1997. HICPAC guidelines for isolation precautions. Am. J. Infect. Control 25:287-288. [DOI] [PubMed] [Google Scholar]
- 31.Sprenger, M. J. M., J. E. Degener, B. J. Kullberg, J. W. M. van der Meer, D. J. Mevius, and E. E. Stobberingh. 1999. European strategy for control of resistance to antibiotics. Ned. Tijdschr. Geneeskd. 143:1322-1325. (In Dutch.) [PubMed] [Google Scholar]
- 32.Talon, D., P. Menget, M. Thouverez, G. Thiriez, H. Gbaguidi Haore, C. Fromentin, A. Muller, and X. Bertrand. 2004. Emergence of Enterobacter cloacae as a common pathogen in neonatal units: pulsed-field gel electrophoresis analysis. J. Hosp. Infect. 57:119-125. [DOI] [PubMed] [Google Scholar]
- 33.van der Zwet, W. C., N. van Riessen, P. W. M. Bergervoet, J. R. van der Laan, P. H. M. Savelkoul, and F. W. Sebens. 2005. Outbreak of multi-resistant Escherichia coli on a surgical ward: course, measures and consequences for future admissions of contaminated patients. Ned. Tijdschr. Geneeskd. 149:2281-2286. (In Dutch.) [PubMed] [Google Scholar]
- 34.Verhoef, J., D. Beaujean, H. Blok, A. Baars, A. Meyler, and C. van der Werken, and A. Weersink. 1999. A Dutch approach to methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 18:461-466. [DOI] [PubMed] [Google Scholar]
- 35.Vriens, M., H. Blok, A. Fluit, A. Troelstra, C. van der Werken, and J. Verhoef. 2002. Costs associated with a strict policy to eradicate methicillin-resistant Staphylococcus aureus in a Dutch university medical center: a 10-year survey. Eur. J. Clin. Microbiol. Infect. Dis. 21:782-786. [DOI] [PubMed] [Google Scholar]
- 36.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum beta-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]
- 37.Yu, W. L., R. N. Jones, R. J. Hollis, S. A. Messer, D. J. Biedenbach, L. M. Deshpande, and M. A. Pfaller. 2002. Molecular epidemiology of extended-spectrum beta-lactamase-producing, fluoroquinolone-resistant isolates of Klebsiella pneumoniae in Taiwan. J. Clin. Microbiol. 40:4666-4669. [DOI] [PMC free article] [PubMed] [Google Scholar]