Abstract
A negative extended-spectrum β-lactamase (ESBL) confirmation test result obtained after a positive ESBL screening test result using Clinical and Laboratory Standards Institute methods has been a common occurrence among isolates of Escherichia coli and Klebsiella pneumoniae in the SENTRY Antimicrobial Surveillance Program in the Asia-Pacific region. Among isolates collected between 1998 and 2004 this screen-positive, nonconfirmed profile (failed to show clavulanate synergy) was observed in 8.9% of 4,515 E. coli isolates and 20.3% of 2,303 K. pneumoniae isolates. We then selected 52 E. coli isolates and 68 K. pneumoniae isolates with a negative ESBL confirmation test, as well as comparable number of isolates with confirmed ESBL-positive tests, and examined them for the presence of TEM, SHV, plasmid-borne ampC, and CTX-M genes. We found that 62% of nonconfirming E. coli isolates and 75% of nonconfirming K. pneumoniae harbored a plasmid-borne AmpC enzyme of the CIT or DHA type. The majority of nonconfirming E. coli and K. pneumoniae from the Asia-Pacific region harbor important β-lactamases, and a positive screening test alone should be sufficient grounds to report resistance to extended-spectrum cephalosporins in this region.
Current Clinical and Laboratory Standards Institute (CLSI) standards recommend the use of a screening and confirmation test, in addition to standard susceptibility testing methods, to detect extended-spectrum β-lactamases (ESBLs) in the routine clinical laboratory among strains of Escherichia coli and Klebsiella pneumoniae (1, 6). This method has proven reliable over many years at detecting the great majority of conventional ESBLs, particularly of variants of the TEM and SHV enzyme classes. The CLSI method, however, does not address the significance of strains that are positive on the screening test but negative on the confirmation test. By default, the result of the standard susceptibility test (e.g., broth microdilution or disk diffusion) is applied to organisms with this ESBL test profile. However, the MIC distributions for these wild-type gram-negative species suggest that strains for which the MICs of any of the ESBL screening agents are >1 μg/ml are abnormal (8) and therefore likely to possess an acquired resistance mechanism. In particular, the emergence of plasmid-borne AmpC β-lactamases, which are not inhibited by clavulanic acid, in members of the Enterobacteriaceae (13) is likely to explain at least some of the strains that have a positive screening test but a negative confirmation test. It is important for both clinical and infection control reasons to detect strains harboring transmissible resistance mechanisms to extended-spectrum cephalosporins.
The present study examined isolates for which the MICs of extended-spectrum cephalosporins and/or aztreonam are >1 μg/ml, including strains that would be considered susceptible by routine susceptibility testing using CLSI broth microdilution. In particular, we focused on their overall frequency in a prospective collection of clinical isolates of E. coli and K. pneumoniae from the Asia-Pacific region and tested for the presence of older and newer β-lactamase genes.
MATERIALS AND METHODS
Isolates.
Isolates were collected in 17 clinical laboratories in nine countries in the Asia-Pacific region (including South Africa) between 1998 and 2003. Isolates came from blood, lower-respiratory-tract infections, skin or skin structure infections, urine, and intensive care specimens. All isolates were referred to a central laboratory (Women's and Children's Hospital, Adelaide, Australia) for testing.
Susceptibility testing.
Isolates were tested by using commercial validated broth microdilution panels (Trek Diagnostic Systems, East Grinstead, United Kingdom) against a wide range of antimicrobials, including ceftriaxone, ceftazidime, aztreonam, and cefoxitin. The MIC testing was done according to CLSI methods and interpretive breakpoint criteria (5, 6).
ESBL definitions, screening, and confirmation.
All isolates for which the MICs of either ceftriaxone, ceftazidime, or aztreonam were >1 μg/ml were considered to have a positive screening test for an ESBL and subjected to clavulanate confirmatory testing using the methods recommended by the CLSI (6), except that the substrates were extended to five agents: cefotaxime, ceftriaxone, ceftazidime, cefepime, and aztreonam (1). Isolates that demonstrated clavulanate enhancement with one or more of the substrates were designated as confirmed, while those that did not demonstrate enhancement were designated nonconfirmed.
Molecular tests.
Selected isolates with positive screen tests were subjected to molecular screening for β-lactamases using PCR tests as previously described for TEM (9), SHV (15), family-specific CTX-M (14), and plasmid-borne AmpC genes (12). The isolates were selected to provide a balance between the two species and between those that were confirmed or not confirmed as having an ESBL.
RESULTS
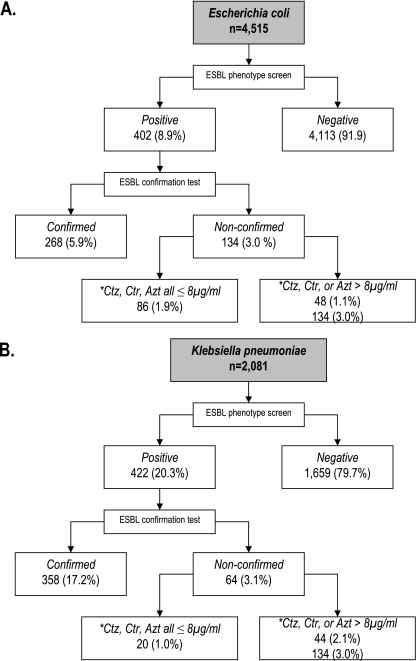
Of 4,515 isolates of E. coli, for 402 (8.9%) the MICs of ceftriaxone, ceftazidime, and/or aztreonam were >1 μg/ml (Fig. 1). Among the 2,081 K. pneumoniae, 422 (20.3%) fulfilled the same screening criteria. One-third of the screen-positive E. coli isolates (134 of 402 [33.3%]) and one-sixth of the screen-positive K. pneumoniae isolates (64 of 422 [15.2%]) could not be confirmed as ESBL positive using the conventional CLSI clavulanate confirmation test.
FIG. 1.
Breakdown of E. coli and K. pneumoniae isolates by results of ESBL screening and confirmation tests, and MICs to screening agents. Abbreviations: Ctz, ceftazidime; Ctr, ceftriaxone; and Azt, aztreonam.
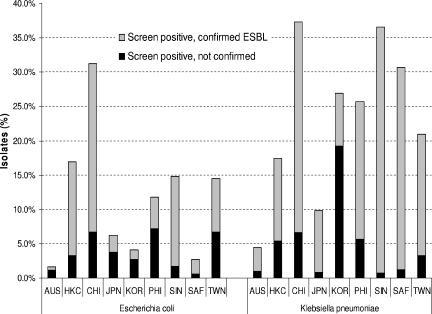
There was significant variation between participating countries with regard to both the prevalence of ESBL-positive strains and the prevalence of nonconfirmed strains on the ESBL confirmatory test (Fig. 2). Rates of nonconfirmed E. coli ranged from 0.6 to 7.2% of all strains between different countries; the range of rates for nonconfirmed K. pneumoniae was 0.8 to 19.2% among all strains. In the Philippines, nonconfirmed strains were predominant among the screen-positive E. coli isolates, whereas in Korea, nonconfirmed strains were predominant among both screen-positive E. coli and screen-positive K. pneumoniae isolates.
FIG. 2.
Prevalence by country of E. coli and K. pneumoniae with a positive ESBL screen that confirmed and did not confirm. AUS, Australia; HKC, Hong Kong; CHI, China; KOR, Korea; PHI, the Philippines; SIN, Singapore; SAF, South Africa; TWN, Taiwan. Numbers above the bars represent the total number of strains tested in each country.
The results of molecular testing on both ESBL-confirmed and nonconfirmed strains are shown in Table 1. As expected, known plasmid-borne AmpC enzymes predominated in the nonconfirmed isolates for both bacterial species, being found in 62% of nonconfirmed E. coli and 75% of nonconfirmed K. pneumoniae isolates. These enzymes were also found in ca. 10% each of the two species when the confirmatory test was positive. In each case these enzymes were found in combination with another β-lactamase, suggesting that the other β-lactamase was the dominant form of resistance. Only plasmid-borne AmpC enzymes of the CIT (Citrobacter) and DHA (Morganella) types were detected (Table 2).
TABLE 1.
β-Lactamase classes detected by PCR grouped by results of ESBL confirmation test on screening-positive strainsa
| β-Lactamase class(es) detected | Nonconfirmed ESBL test
|
Confirmed ESBL test
|
||||||
|---|---|---|---|---|---|---|---|---|
|
E. coli
|
K. pneumoniae
|
E. coli
|
K. pneumoniae
|
|||||
| n | % | n | % | n | % | n | % | |
| AmpC only | 3 | 5.8 | 9 | 17.6 | ||||
| TEM only | 11 | 21.2 | 7 | 10.4 | ||||
| SHV only | 1 | 1.9 | 5 | 9.8 | 4 | 6.0 | 17 | 25.0 |
| CTX-M only | 1 | 1.9 | 9 | 13.4 | 6 | 8.8 | ||
| TEM+SHV | 3 | 5.8 | 13 | 19.4 | 16 | 23.5 | ||
| TEM+CTX-M | 24 | 35.8 | ||||||
| SHV+CTX-M | 9 | 13.2 | ||||||
| AmpC+TEM | 26 | 50.0 | 1 | 2.0 | ||||
| AmpC+SHV | 1 | 1.9 | 19 | 37.3 | 1 | 1.5 | 6 | 8.8 |
| TEM+SHV+CTX | 3 | 5.9 | 1 | 1.5 | 13.2 | |||
| AmpC+TEM+SHV | 2 | 3.8 | 9 | 17.6 | 3 | 4.5 | ||
| AmpC+TEM+CTX | 2 | 3.0 | 1 | 1.5 | ||||
| AmpC+SHV+CTX-M | 1 | 1.5 | 1 | 1.5 | ||||
| AmpC+TEM+SHV+CTX-M | ||||||||
| PCR negative | 4 | 7.7 | 5 | 9.8 | 2 | 3.0 | 3 | 4.4 |
| Totals | 52 | 51 | 68 | 67 | ||||
| Any AmpC | 32 | 61.5 | 38 | 74.5 | 7 | 10.4 | 8 | 11.8 |
| Any TEM | 42 | 80.8 | 13 | 25.5 | 50 | 74.6 | 26 | 38.2 |
| Any SHV | 7 | 13.5 | 17 | 33.3 | 23 | 34.3 | 58 | 85.3 |
| Any CTX-M | 1 | 1.9 | 3 | 5.9 | 36 | 55.2 | 26 | 38.2 |
n, Number of isolates.
TABLE 2.
Prevalence of AmpC subgroups in AmpC-positive isolatesa
| Country | Prevalence (no. of isolates)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Nonconfirmed ESBL test
|
Confirmed ESBL test
|
|||||||
|
E. coli
|
K. pneumoniae
|
E. coli
|
K. pneumoniae
|
|||||
| CIT | DHA | CIT | DHA | CIT | DHA | CIT | DHA | |
| Australia | 1 | 1 | ||||||
| Hong Kong | 1 | 2 | 9 | 3 | 3 | |||
| Japan | 1 | |||||||
| Korea | 1 | 3 | 2 | |||||
| China | 2 | 2 | 1 | |||||
| The Philippines | 1 | 9 | 19 | 3 | ||||
| Singapore | 2 | |||||||
| South Africa | 1 | |||||||
| Taiwan | 12 | 1 | 2 | 2 | 1 | |||
| Total | 21 | 11 | 4 | 34 | 2 | 5 | 0 | 8 |
CIT and DHA, AmpC subgroups of CIT and DHA enzymes.
Genes encoding CTX-M-type enzymes were also found quite frequently in both bacterial species. In almost all cases (62 of 66 [94%]) the presence of these enzyme types resulted in a positive ESBL confirmatory test. As expected, the prevalence of TEM enzymes was high in E. coli (81% of screen-positive strains) since the molecular detection test captures almost all TEM genes, including the non-ESBL TEM-1 and -2. That these were largely TEM-1/2 was supported by the finding of a similar percentage (75%) of confirming strains of E. coli possessing a TEM gene. A TEM gene alone was the only β-lactamase gene detected in 11 of 52 (21%) of screen-positive nonconfirmed E. coli. They were isolated from seven different countries, and the majority were resistant to ampicillin, amoxicillin-clavulanate, cefazolin, and cefoxitin. In the absence of sequence data, it was not possible to interpret whether these were due to a problem with the confirmation method, the presence of inhibitor-resistant TEM enzymes, or the presence of another β-lactamase type not detected by the current PCR-based methods.
DISCUSSION
The SENTRY Program has previously shown that extended-spectrum β-lactamases are prevalent in many countries in the Asia-Pacific region and South Africa (1, 2, 10), having commenced monitoring in 1998. Over time we have noted an increasing proportion of strains that had elevated ceftriaxone, ceftazidime, and/or aztreonam MICs above those concentrations recommended for screening for ESBLs and which did not confirm as ESBLs using the CLSI-recommended method (5, 6). This more thorough examination of our isolates included first conducting confirmatory testing on all strains of E. coli and K. pneumoniae and testing all screen-positive strains for the presence of known β-lactamases of the ESBL and related types. In particular, we were interested to learn what proportion of the isolates of these species could not be confirmed as conventional ESBLs, and what proportion of this group might be explained by the presence of known plasmid-borne AmpC enzymes.
The proportion of nonconfirmed ESBLs was relatively high in the Asia-Pacific region and was particularly noticeable in isolates of E. coli. A substantial amount of nonconfirmation was associated with plasmid-borne AmpC enzymes of the CIT and DHA type. Our findings are clearly driven by the prevalence of different enzyme types in our region, and it was less clear how these results might reflect the situation in other regions of the world. However, the high detection rate of enzymes capable of inactivating third-generation cephalosporins in screen-positive, nonconfirmed strains should present a clear warning that the screening test itself was more meaningful than the confirmation test. In the Asia-Pacific region at least, a positive ESBL confirmation test merely suggests that the strain contains an enzyme or enzymes inhibited by clavulanate.
A number of mechanisms have been described that could account for the lack of clavulanate enhancement in the screen-positive nonconfirming E. coli isolates that possessed only TEM genes. These include TEM-1 hyperproduction, modification of outer membrane proteins, or the presence of inhibitor-resistant TEM enzymes (4). However, these mechanisms generally do not result in great increases in the MICs of extended-spectrum cephalosporins. We did not screen for OXA enzymes, which are usually clavulanate resistant and are only rarely found in E. coli or Klebsiella isolates (3, 11, 7). We also found a few isolates of both species, both confirming and nonconfirming, in which we were unable to detect any β- lactamase genes. This might be attributed to technical limitations with the present assays or perhaps to the existence of as-yet-undescribed β-lactamases, given the recent rapid expansion of β-lactamase types worldwide.
In summary, the majority of E. coli and K. pneumoniae isolates in the Asia-Pacific with a positive ESBL screening test and negative confirmation test harbor plasmid-borne β-lactamases that are of clinical and infection control importance. Hence, we would recommend that, in the Asia-Pacific region, laboratories should report all screen-positive isolates of these two species as resistant to extended-spectrum cephalosporins. Further work is necessary to determine whether there might be similar findings in other regions of the world and whether this should also apply to other members of the Enterobacteriaceae that lack natural chromosomal cephalosporinases. If this phenomenon can be shown to be similar around the world, a strong case could be made for reconsideration of the breakpoints for the extended-spectrum cephalosporins and aztreonam (6).
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Bell, J. M., J. D. Turnidge, R. N. Jones, et al. 2003. Prevalence of extended-spectrum β-lactamase-producing Enterobacter cloacae in the Asia-Pacific region: results from the SENTRY Antimicrobial Surveillance Program, 1998 to 2001. Antimicrob. Agents Chemother. 47:3989-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, J. M., J. D. Turnidge, A. C. Gales, M. A. Pfaller, R. N. Jones, and the SENTRY APAC group. 2002. Prevalence of extended-spectrum β-lactamase (ESBL)-producing clinical isolates in the Asia-pacific region and South Africa: regional results from SENTRY Antimicrobial Surveillance Program (1998-99). Diagn. Microbiol. Infect. Dis. 42:193-198. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, P. A. 2001. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev. 14:933-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaïbi, E. B., D. Sirot, G. Paul, and R. Labia. 1999. Inhibitor-resistant TEM β-lactamases: phenotypic, genetic and biochemical characteristics. J. Antimicrob. Chemother. 43:447-458. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 7th ed., M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.DiPersio, J. R., L. M. Deshpande, D. J. Biedenbach, M. A. Toleman, T. R. Walsh, and R. N. Jones. 2005. Evolution and dissemination of extended-spectrum β-lactamase-producing Klebsiella pneumoniae: epidemiology and molecular report from the SENTRY Antimicrobial Surveillance Program (1997-2003). Diagn. Microbiol. Infect. Dis. 51:1-7. [DOI] [PubMed] [Google Scholar]
- 8.European Committee on Antimicrobial Susceptibility Testing. 2006. Antimicrobial wild-type MIC distributions of microorganisms. http://217.70.33.99/Eucast2/.
- 9.Hanson, N. D., K. S. Thomson, E. S. Moland, C. C. Sanders, G. Berthold, and R. G. Penn. 1999. Molecular characterization of a multiply resistant Klebsiella pneumoniae encoding ESBLs and a plasmid-mediated AmpC. J. Antimicrob. Chemother. 44:377-380. [DOI] [PubMed] [Google Scholar]
- 10.Hirakata, Y., J. Matsuda, Y. Miyazaki, S. Kamihira, S. Kawakami, Y. Miyazawa, Y. Ono, N. Nakazaki, Y. Hirata, M. Inoue, J. D. Turnidge, J. M. Bell, R. N. Jones, S. Kohno, et al. 2005. Regional variation in the prevalence of extended-spectrum β-lactamase-producing clinical isolates in the Asia-Pacific region (SENTRY 1998-2002). Diagn. Microbiol. Infect. Dis. 52:323-329. [DOI] [PubMed] [Google Scholar]
- 11.Kaye, K. S., H. S. Gold, M. J. Schwaber, L. Venkataraman, Y. Qi, P. C. De Girolami, M. H. Samore, G. Anderson, J. K. Rasheed, and F. C. Tenover. 2004. Variety of β-lactamases produced by amoxicillin-clavulanate-resistant Escherichia coli isolated in the northeastern United States. Antimicrob. Agents Chemother. 48:1520-1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pérez-Pérez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippon, A., G. Arlet, and G. A. Jacoby. 2002. Plasmid-borne AmpC-type β-lactamases. Antimicrob. Agents Chemother. 46:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitout, J. D., D. A. Hossain, and N. D. Hanson. 2004. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J. Clin. Microbiol. 42:5715-5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasheed, J. K., C. Jay, B. Metchock, F. Berkowitz, L. Weigel, J. Crellin, C. Steward, B. Hill, A. A. Medeiros, and F. C. Tenover. 1997. Evolution of extended-spectrum β-lactam resistance (SHV8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob. Agents Chemother. 41:647-653. [DOI] [PMC free article] [PubMed] [Google Scholar]