Abstract
Two highly discriminatory fingerprinting assays, short tandem repeat typing and amplified fragment length polymorphism (AFLP), were compared to determine the genetic relatedness between 55 isolates of Aspergillus fumigatus obtained from 15 different patients suffering from proven invasive aspergillosis. Both techniques showed that interpatient isolates belonged to different genotypes and that intrapatient isolates from deep sites were all of the same genotype. By contrast, multiple genotypes were found among isolates originating from respiratory samples. Both techniques have specific advantages and disadvantages. AFLP is more universally applicable, but short tandem repeat analysis offers better discriminatory power and should be the preferred method for standardizing typing of clinical isolates of Aspergillus fumigatus.
Invasive aspergillosis (IA) is a life-threatening disease of patients who are immunocompromised by hematopoietic stem cell or solid organ transplantation, intensive chemotherapy, or treatment with corticosteroids (11). Aspergillus fumigatus is most frequently involved in causing this life-threatening disease. IA is a growing problem in hospitals because of the inexorable rise in the number of severely immunocompromised patients (12, 14). A better understanding of the epidemiology of this disease requires molecular techniques to investigate interstrain relatedness and strain dissemination (12).
Fingerprinting methods with high discriminatory power have to be applied to discriminate between unrelated isolates because of the wide genetic variability of A. fumigatus, (5) and several typing techniques have been described previously. Pattern-based techniques, such as random amplified polymorphic DNA (RAPD) analysis (2, 13), restriction fragment length polymorphism (RFLP) analysis (12, 19), and amplified fragment length polymorphism (AFLP) (20), have been employed but suffer from poor interlaboratory reproducibility and subjective interpretation of the fingerprinting data. Typing methods based on short tandem repeats (STRs), such as microsatellite length polymorphism (3) and STRAf typing (8), yield unambiguous typing data, as do those based on sequence-based typing schemes, such as multilocus sequence typing. These methods may be preferable to other typing methods, provided that they possess sufficient discriminatory power, and may have the characteristics necessary for the standardization of A. fumigatus typing.
STRAf typing is a recently described highly discriminatory and potentially reproducible fingerprinting assay for A. fumigatus (8, 9), but its utility in epidemiological surveys has not yet been established. In this study, we compared AFLP and STRAf typing to study the genetic relatedness between isolates of A. fumigatus from respiratory and tissue samples that had been obtained from 15 patients with proven IA (1). Previous studies have shown that the genotypes of isolates from intrapatient samples of deep sites were identical (12), so these samples could be considered internal controls. We therefore set out to evaluate the advantages and disadvantages of both techniques in terms of their discriminatory power and applicability as well as technical aspects and computerized data processing.
MATERIALS AND METHODS
Isolates.
Fifty-five isolates of A. fumigatus were obtained from ante- and postmortem specimens of 15 patients with proven IA who had been nursed in the hematology ward of the Radboud University Nijmegen Medical Center in the period from 1992 to 1998 (Table 1). Isolates were stored as spore suspensions in regular microbial freezing broth containing 12.5% (vol/vol) glycerol at −80°C. The isolates were revived by scraping off part of the sample, plating on Sabouraud's agar, and cultivation at 30°C. Fungal isolates were identified at the time of collection and again after revival by their macroscopic or microscopic appearance and their ability to grow at 48°C. Every isolate displayed normal growth characteristics after revival.
TABLE 1.
Origin of A. fumigatus isolates from patients with IA
| Patient | Origin | Date of isolation (day-mo-yr) | AFLP type | STRAf type | Sample |
|---|---|---|---|---|---|
| A | Bronchial secretion | 22-07-1992 | 1 | 1 | A1 |
| Tissue | 25-07-1992 | 1 | 1 | A2 | |
| B | Bronchial secretion | 26-08-1992 | 2 | 2 | B1 |
| Bronchial secretion | 28-08-1992 | 2 | 2 | B2 | |
| Pleural abscess | 31-08-1992 | 2 | 2 | B3 | |
| C | Bronchial secretion | 15-01-1993 | 3 | 3 | C1 |
| Lung biopsy sample | 19-01-1993 | 3 | 3 | C2 | |
| BAL fluid | 19-01-1993 | 3 | 3 | C3 | |
| D | Left lung | 28-12-1993 | 4 | 4 | D1 |
| Right kidney | 28-12-1993 | 4 | 4 | D2 | |
| Left kidney | 28-12-1993 | 4 | 4 | D3 | |
| E | Feces | 01-06-1994 | 5 | 5 | E1 |
| Pharynx swab | 06-06-1994 | 5 | 5 | E2 | |
| Feces | 09-06-1994 | 5 | 5 | E3 | |
| Pharynx swab | 13-06-1994 | 5 | 5 | E4 | |
| Sputum | 20-06-1994 | 5 | 5 | E5 | |
| Lung | 13-07-1994 | 5 | 5 | E6 | |
| F | Pharynx swab | 04-08-1994 | 6 | 6 | F1 |
| Right lung | 18-08-1994 | 6 | 6 | F2 | |
| Left kidney | 18-08-1994 | 6 | 6 | F3 | |
| Left lung | 18-08-1994 | 6 | 6 | F4 | |
| G | BAL fluid | 28-10-1994 | 7 | 7 | G1 |
| Lung | 04-11-1994 | 8 | 8 | G2 | |
| H | Sputum | 18-04-1995 | 9 | 9 | H1 |
| Left lung | 02-05-1995 | 10 | 10 | H2 | |
| I | Pus | 29-08-1995 | 11 | 11 | I1 |
| Feces | 31-08-1995 | 11 | 11 | I2 | |
| Pharynx swab | 04-09-1995 | 11 | 11 | I3 | |
| Feces | 14-09-1995 | 11 | 11 | I4 | |
| J | BAL fluid | 16-10-1995 | 12 | 12 | J1 |
| Right lung | 25-10-1995 | 12 | 12 | J2 | |
| Left lung | 25-10-1995 | 12 | 12 | J3 | |
| K | Feces | 29-03-1996 | 13 | 13 | K1 |
| Left lung | 01-04-1996 | 13 | 13 | K2 | |
| Right kidney | 01-04-1996 | 13 | 13 | K3 | |
| Heart | 01-04-1996 | 13 | 13 | K4 | |
| Left kidney | 01-04-1996 | 13 | 13 | K5 | |
| Right lung | 01-04-1996 | 13 | 13 | K6 | |
| Spleen | 01-04-1996 | 13 | 13 | K7 | |
| Liver | 01-04-1996 | 13 | 13 | K8 | |
| L | Bronchial secretion | 02-01-1997 | 14 | 14 | L1 |
| Bronchial secretion | 03-01-1997 | 14 | 14 | L2 | |
| Bronchial secretion | 09-01-1997 | 14 | 14 | L3 | |
| Right lung | 13-01-1997 | 14 | 14 | L4 | |
| M | Lung biopsy sample | 26-03-1997 | 15 | 15 | M1 |
| BAL fluid | 26-03-1997 | 16 | 16 | M2 | |
| BAL fluid | 26-03-1997 | 17 | 17 | M3 | |
| Lung biopsy sample | 26-03-1997 | 18 | 15 | M4 | |
| N | Pharynx swab | 11-12-1997 | 13 | 18 | N1 |
| Left lung | 17-12-1997 | 13 | 18 | N2 | |
| Spleen | 17-12-1997 | 13 | 18 | N3 | |
| Liver | 17-12-1997 | 13 | 18 | N4 | |
| O | BAL fluid | 19-01-1998 | 19 | 19 | O1 |
| BAL fluid | 19-01-1998 | 20 | 20 | O2 | |
| Left lung | 23-01-1998 | 19 | 19 | O3 |
DNA isolation.
Isolates were grown on Sabouraud's agar plates at 30°C until sporulation occurred. Spores were first mechanically lysed using the following procedures. A prewetted cotton swab was saturated with conidia from the sporulating culture. These spores were then suspended in a vial of MagNA Lyser green beads containing 350 μl lysis buffer and 50 μl proteinase K (all from Roche Diagnostics, Almere, The Netherlands). Lysis was performed in a MagNA Lyser instrument (Roche Diagnostics) for 30 s at 6,500 rpm. Next, the DNA was extracted and purified using a MagNA Pure LC instrument (Roche Diagnostics) in combination with MagNA Pure DNA isolation kit III according to the manufacturer's instructions. DNA yield and purity were estimated by UV absorbance measurements.
STRAf typing.
Nine STR markers specific for A. fumigatus were analyzed using the STRAf primers and reaction conditions as previously described (8). Briefly, three multiplex PCRs each amplified three STR markers. The obtained PCR products were separated by size and detected on a MegaBACE 500 platform equipped with a 48-capillary array (GE Healthcare, Diegem, Belgium). The assignment of repeat numbers in each marker was performed using Fragment Profiler 1.2 software (GE Healthcare).
AFLP.
Approximately 50 ng of genomic DNA was subjected to a combined restriction-ligation procedure containing 50 pmol of HpyCH4 IV adapter, 50 pmol MseI adapter, 2 U of HpyCH4 IV (New England Biolabs, Beverly, MA), 2 U of MseI (New England Biolabs), and 1 U of T4 DNA ligase (Promega, Leiden, The Netherlands) in a total volume of 20 μl of 1× reaction buffer for 1 h at 20°C. Next, the mixture was diluted five times with 10 mM Tris-HCl (pH 8.3) buffer. Adapters were made by mixing equimolar amounts of complementary oligonucleotides (5′-CTCGTAGACTGCGTACC-3′ and 5′-CGGGTACGCAGTC-3′ for HpyCH4 IV; 5′-GACGATGAGTCCTGAC-3′ and 5′-TAGTCAGGACTCAT-3′ for MseI) and heating to 95°C, subsequently cooling slowly to ambient temperature. One microliter of the diluted restriction-ligation mixture was used for amplification in a volume of 25 μl under the following conditions: 1 μM HpyCH4 IV primer with one selective residue (underlined) (5′-Flu-GTAGACTGCGTACCCGTC-3′), 1 μM MseI primer with four selective residues (underlined) (5′-GATGAGTCCTGACTAATGAA-3′), 0.2 mM of each deoxynucleoside triphosphate, and 1 U of Taq DNA polymerase (Roche Diagnostics) in 1× reaction buffer containing 1.5 mM MgCl2.
Amplification was done as follows. After an initial denaturation step for 4 min at 94°C in the first 20 cycles, a touchdown procedure was applied: 15 s of denaturation at 94°C, 15 s of annealing at 66°C, with the temperature for each successive cycle lowered by 0.5°C, and 1 min of extension at 72°C. Cycling was then continued for a further 30 cycles with an annealing temperature of 56°C. After completion of the cycles, an incubation at 72°C for 10 min was performed before the reaction mixtures were cooled to room temperature. The amplicons were then combined with the ET400-R size standard (GE Healthcare) and analyzed on a MegaBACE 500 automated DNA platform (GE Healthcare) according to the manufacturer's instructions.
Data analysis.
All data were analyzed with the BioNumerics 4.5 unweighted-pair group method using average linkage clustering (Applied Maths, Sint-Martens- Latem, Belgium), employing the multistate categorical similarity coefficient for STR data and the Pearson correlation coefficient for DNA fragments between 60 and 250 bases for AFLP analysis.
RESULTS
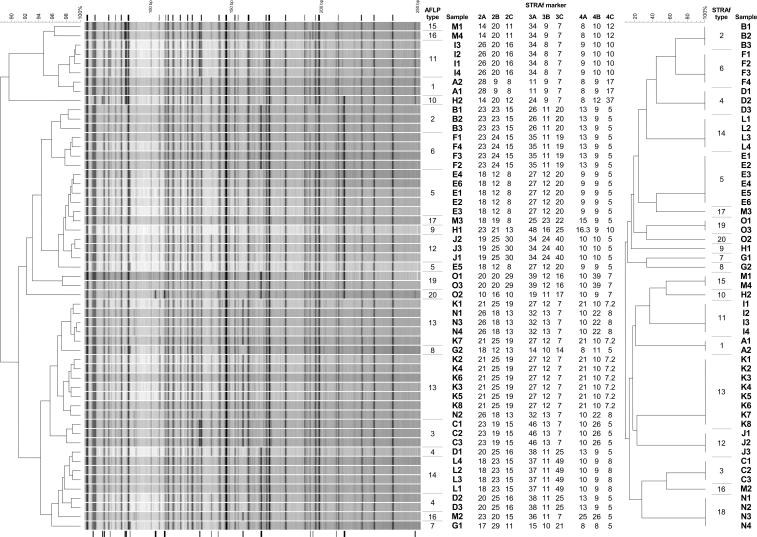
All AFLP fingerprints obtained with MseI and HpyCH4 IV contained multiple bands in the range of 50 to 250 bp (Fig. 1), and strong and weak bands were visible. When all samples were compared to each other, the fingerprints consisted of some 30 invariable bands present in all samples and around 20 variable bands which were present only in certain samples. Based on the variable bands, the different fingerprints can be distinguished from each other. In total, 20 visually recognizable different patterns were obtained. The calculated similarity for visually identical patterns ranges from 97 to 99%. The calculated similarity for patterns with visually recognizable differences ranges from 89 to 99%. Originally, the collection consisted of 56 isolates. However, one isolate yielded a fingerprint without resemblance to any of the other isolates. The reidentification of this isolate by internal transcribed spacer sequencing showed that this particular isolate proved to be Neosartorya fischeri (results not shown). This isolate was excluded from further analysis.
FIG. 1.
Results of both AFLP and STRAf typing on a collection of 55 clinical A. fumigatus isolates. The dendrogram on the left is based on the AFLP fingerprints. The indicator lines above the traces represent invariable bands, whereas the indicator lines below the traces represent variable bands in the AFLP fingerprints. After each fingerprint are the typing results for the nine markers of the STRAf panel. The dendrogram on the right is based on the STRAf results. Types were assigned arbitrarily, starting with the oldest isolate. The scale bars indicate the percentages of similarity. Sample numbers correspond to those in Table 1.
Using STRAf typing, all A. fumigatus isolates yielded a numerical typing result (Fig. 1). A total of 20 different genotypes were obtained with this technique. The N. fischeri isolate did not yield PCR products with the STRAf assay.
In general, there is an excellent agreement between the two typing methods, with only two tissue isolates (M1 and M4) of patient M that were differentiated by AFLP but not by STRAf typing, which yielded the same genotype. AFLP yielded the same genotype for all isolates of patients K and N, but these could be clearly distinguished from each other using STRAf typing. When all intrapatient isolates were compared to each other, samples of nonrespiratory origin belonged to the same genotype. By contrast, multiple genotypes were detected in isolates originating from respiratory samples, such as sputum and bronchoalveolar lavage (BAL) fluid samples, from the same patient. For instance, all eight isolates from the lung, kidney, heart, spleen, and liver of patient K shared the same STRAf genotype. An identical unique genotype was found in isolates from BAL fluid and lung samples obtained from patient O, whereas another BAL fluid sample yielded a different genotype. A similar observation was made for patients G, H, and M, as interpatient isolates belonged to different genotypes as determined by STRAf typing.
DISCUSSION
We compared two high-resolution PCR fingerprinting assays for analyzing the epidemiological relationships between multiple A. fumigatus isolates from patients with proven IA. Both AFLP analysis and STRAf analysis provided high-resolution data allowing discrimination between clinical isolates from different patients and between different genotypes in multiple samples from a patient. In general, interpatient isolates belonged to different genotypes whereas intrapatient isolates were of the same genotype. Cross-infections of respiratory origin or common-source infections were highly unlikely since the patients were nursed at different times in the ward.
The use of molecular epidemiological tools such as RAPD, multilocus enzyme electrophoresis, and sequence-specific DNA primer analyses (10) has led to suggestions that A. fumigatus isolates from several IA patients nursed in the same ward may originate from a single environmental source. However, these techniques lack discriminatory power and are not considered by us to be sufficiently reproducible to employ for this study. Rather, our results confirm the results reported by Girardin et al. (12) and Denning et al. (7), who investigated the genetic diversity of patients with IA by using RFLP and REA analyses, respectively. Our results are consistent with a scenario in which the respiratory tract may be colonized by several different genotypes, as inhalation of fungal conidia is assumed to be the primary means of acquiring IA (11), but a single genotype gains supremacy and invades the surrounding tissue, subsequently disseminating to the lung tissue or other organs. Thus, an isolate obtained from respiratory samples may not necessarily represent the same genotype as that found in deep sites, as appeared to be the case for patients G, H, M, and O. This also confirms that individuals may well be constantly exposed to a large variety of different Aspergillus genotypes from the environment (5, 15).
Both AFLP and the STRAf assay are seen as high-resolution and robust molecular fingerprinting assays for A. fumigatus isolates (8, 9, 20). As shown here, both techniques are suitable for unraveling the genetic relationship between clinical A. fumigatus isolates, with good correlation between the results of the two assays. Yet, compared to each other, both assays have their advantages and disadvantages. A clear advantage of the STRAf assay over AFLP is its ability to identify mixtures of strains. Mixed genotypes are recognized instantaneously by the presence of multiple peaks in each of the nine markers analyzed (8). Mixed AFLP genotypes are extremely difficult, if not impossible, to detect. Thus, when employing AFLP analysis for epidemiological analysis, it is important that pure cultures be obtained and analyzed. Assuming that individuals are, in fact, likely to be colonized by more than one genotype, it is important to subculture and analyze each colony separately when several are obtained from a respiratory sample in order to prevent false conclusions from being drawn (4, 6).
DNA fragments are amplified in AFLP from random locations throughout the entire genome and, although in a single AFLP experiment only a very small fraction of the entire genome is analyzed (<0.1%), the amount analyzed by the STRAf assay is even smaller. In addition, unlike the AFLP assay, the STRAf approach is highly biased toward nine arbitrarily selected loci, so any genomic change outside these loci will remain undetected, as was the case for isolates M1 and M4. The AFLP technique is basically applicable to any organism's genome without the need for prior sequence information. However, depending on the genome size and GC content, there may be a need for optimizing the choice of restriction enzymes and selective residues. By contrast, the STRAf assay is specific for A. fumigatus and requires specific sequence information for the detection of STR loci and flanking sequences to be used as primer binding sites. However, both techniques do permit improperly identified isolates to be recognized. The nonvariable bands that are obtained with A. fumigatus isolates may represent species-specific markers. Upon analysis of isolates from species other than A. fumigatus, most of these invariable bands will not show up and a banding pattern will be obtained that is usually less than 30% similar to the fingerprint of A. fumigatus isolates (as was the case for the N. fischeri isolate). Likewise, using the STRAf approach, improperly identified isolates can be recognized by the absence of typical amplification products, provided that technical issues that may lead to PCR failure can be ruled out.
In order to compare the different AFLP fingerprints to each other and to determine the genetic relatedness between multiple isolates, one has to rely on specialized computer software. AFLP fingerprints are usually analyzed using a pattern-based algorithm. However, repeat analyses may show small differences in the AFLP fingerprint due to small variations occurring during restriction/ligation, PCR amplification, and fragment analysis that may affect the final peak intensity. For the same reasons, the long-term reproducibility and interlaboratory comparisons of AFLP fingerprints may be quite challenging. Thus, identical samples are seldom identical using a pattern-based approach, although they may appear to be so by visual examination. In the pattern-based dendrogram shown in Fig. 1, visually identical fingerprints were calculated to be 97 to 99% similar (i.e., all samples of patient K). However, in the same analysis, samples with visually recognizable differences were also calculated to be up to 99% similar (i.e., samples D1 and L4) (Fig. 1). Consequently, a gray zone was obtained during interpretation of the results of a pattern-based approach in which the wrong conclusions may be drawn when relying solely on the computer-based dendrogram. This requires a visual inspection of the fingerprints to be included in the final interpretation. Similar limitations apply to computer-based analysis of other pattern-based fingerprinting methods, such as RAPD and RFLP analyses. By contrast, due to the more exact nature of STRAf typing, intralaboratory reproducibility is virtually 100% and the typing result is unambiguous.
Due to the large number of available restriction enzymes, an almost indefinite number of restriction enzyme combinations can be used to generate an AFLP fingerprint. The success of a particular combination is dictated primarily by the size and composition (i.e., GC content and the presence of multicopy elements) of the genome to be investigated. The number of fragments to be obtained can be modulated by changing the number of selective residues. In previous studies, other restriction enzyme combinations such as EcoRI and MseI (16, 20) and EcoRI and BfaI (17) have been used to generate AFLP fingerprints for Aspergillus spp. To rule out the possibility that the results of this study may have been influenced by the specific use of the combination of HpyCH4 IV and MseI, all isolates were also tested with the combination of EcoRI and MseI. Although obviously completely different fingerprints were obtained, the same interstrain relatedness was observed (data not shown).
A problem with any typing method is the subjective interpretation of a “genotype.” The assignment of a genotype is user dependent using AFLP, whereas the genotype resulting from STRAf typing is unambiguous. Nonetheless, two different but highly similar fingerprints could originate from two closely related isolates. Unfortunately, at present there are no established guidelines for defining a genotype for these methods (such as the criteria of Tenover et al. for the interpretation of pulsed-field gel electrophoresis fingerprints [18]). However, since there is an excellent correlation between the groupings of genotypes by using AFLP and STRAf typing, we believe that the differences between the fingerprints of the isolates in this study, although minor, are substantial enough to validate our assignment of genotypes. For instance, isolates E3 and M3 differed by only one band in the AFLP fingerprint but by five markers in the STRAf assay. This means that even a single band difference between two AFLP fingerprints could be interpreted as constituting different genotypes. Likewise, with the STRAf assay, any two different genotypes differ by a minimum of three markers. For instance, isolates from patients B and F differed by one repeat in marker 2B, nine repeats in marker 3A, and one repeat in marker 3C. In the AFLP fingerprint, there were at least two visually recognizable differences between these isolates. At present, it is unclear whether fewer than three differences in an STRAf genotype should also be interpreted as different genotypes, so more closely related isolates need to be analyzed.
In conclusion, both AFLP and STRAf typing are excellent techniques for analyzing the interstrain relatedness of A. fumigatus isolates. Each of the two techniques has its specific advantages and disadvantages. Therefore, the choice should be based primarily on the specific goal for analyzing collections of isolates. STRAf typing offers the most benefits for the standardization of typing A. fumigatus isolates.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Aufauvre-Brown, A., J. Cohen, and D. W. Holden. 1992. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J. Clin. Microbiol. 30:2991-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bart-Delabesse, E., J.-F. Humbert, E. Delabesse, and S. Bretagne. 1998. Microsatellite markers for typing Aspergillus fumigatus isolates. J. Clin. Microbiol. 36:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertout, S., F. Renaud, R. Barton, F. Symoens, J. Burnod, M. A. Piens, B. Lebeau, M. A. Viviani, F. Chapuis, J. M. Bastide, R. Grillot, and M. Mallie. 2001. Genetic polymorphism of Aspergillus fumigatus in clinical samples from patients with invasive aspergillosis: investigation using multiple typing methods. J. Clin. Microbiol. 39:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chazalet, V., J. P. Debeaupuis, J. Sarfati, J. Lortholary, P. Ribaud, P. Shah, M. Cornet, H. Vu Thien, E. Gluckman, G. Brucker, and J. P. Latgé. 1998. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J. Clin. Microbiol. 36:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cimon, B., F. Symoens, R. Zouhair, D. Chabasse, N. Nolard, A. Defontaine, and J. P. Bouchara. 2001. Molecular epidemiology of airway colonisation by Aspergillus fumigatus in cystic fibrosis patients. J. Med. Microbiol. 50:367-374. [DOI] [PubMed] [Google Scholar]
- 7.Denning, D. W., K. V. Clemons, L. H. Hanson, and D. A. Stevens. 1990. Restriction endonuclease analysis of total cellular DNA of Aspergillus fumigatus isolates of geographically and epidemiologically diverse origin. J. Infect. Dis. 162:1151-1158. [DOI] [PubMed] [Google Scholar]
- 8.de Valk, H. A., J. F. G. M. Meis, I. M. Curfs, K. Muehlethaler, J. W. Mouton, and C. H. W. Klaassen. 2005. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J. Clin. Microbiol. 43:4112-4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Valk, H. A., J. F. G. M. Meis, and C. H. W. Klaassen. 2007. Microsatellite based typing of Aspergillus fumigatus: strengths, pitfalls and solutions. J. Microbiol. Methods 69:268-272. [DOI] [PubMed] [Google Scholar]
- 10.Ellis, M., M. Richardson, and B. de Pauw. 2000. Epidemiology. Hosp. Med. 61:605-609. [DOI] [PubMed] [Google Scholar]
- 11.Fridkin, S. K., and W. R. Jarvis. 1996. Epidemiology of nosocomial fungal infections. Clin. Microbiol. Rev. 9:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girardin, H., J. Sarfati, F. Traore, J. Dupouy Camet, F. Derouin, and J. P. Latgé. 1994. Molecular epidemiology of nosocomial invasive aspergillosis. J. Clin. Microbiol. 32:684-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leenders, A., A. van Belkum, S. Janssen, S. de Marie, J. Kluytmans, J. Wielenga, B. Lowenberg, and H. Verbrugh. 1996. Molecular epidemiology of apparent outbreak of invasive aspergillosis in a hematology ward. J. Clin. Microbiol. 34:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loudon, K. W., J. P. Burnie, A. P. Coke, and R. C. Matthews. 1993. Application of polymerase chain reaction to fingerprinting Aspergillus fumigatus by random amplification of polymorphic DNA. J. Clin. Microbiol. 31:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menotti, J., J. Waller, O. Meunier, V. Letscher-Bru, R. Herbrecht, and E. Candolfi. 2005. Epidemiological study of invasive pulmonary aspergillosis in a haematology unit by molecular typing of environmental and patient isolates of Aspergillus fumigatus. J. Hosp. Infect. 60:61-68. [DOI] [PubMed] [Google Scholar]
- 16.Perrone, G., G. Mulè, A. Susca, P. Battilani, A. Pietri, and A. Logrieco. 2006. Ochratoxin A production and amplified fragment length polymorphism analysis of Aspergillus carbonation, Aspergillus tubingensis, and Aspergillus niger strains isolated from grapes in Italy. Appl. Environ. Microbiol. 72:680-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schmidt, H., M. Ehrmann, R. F. Vogel, M. H. Taniwaki, and L. Niessen. 2003. Molecular typing of Aspergillus ochraceus and construction of species specific SCAR-primers based on AFLP. Syst. Appl. Microbiol. 26:138-146. [DOI] [PubMed] [Google Scholar]
- 18.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. A. Murray, D. H. Persin, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verweij, P. E., J. F. Meis, J. Sarfati, J. A. Hoogkamp-Korstanje, J. P. Latgé, and W. J. Melchers. 1996. Genotypic characterization of sequential Aspergillus fumigatus isolates from patients with cystic fibrosis. J. Clin. Microbiol. 34:2595-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warris, A., C. H. W. Klaassen, J. F. G. M. Meis, M. T. de Ruiter, H. A. de Valk, T. G. Abrahamsen, P. Gaustad, and P. E. Verweij. 2003. Molecular epidemiology of Aspergillus fumigatus isolates recovered from water, air, and patients shows two clusters of genetically distinct strains. J. Clin. Microbiol. 41:4101-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]