Abstract
During a rotavirus surveillance conducted in Lulong County, Hebei Province, China, a total of 331 stool specimens collected in 2003 from children under 5 years old with diarrhea were screened. We identified a novel group A human rotavirus of genotype G5P[6]. Phylogenetic analysis confirmed that the VP7 protein of this newly identified strain, LL36755, was closely related to those of the G5 strains. As such, it has 95.4% homology with its counterparts in the porcine G5 strains C134 and CC117 at the amino acid sequence level. On the other hand, the VP4 protein of the LL36755 strain was 94.5% homologous to those of the porcine P[6] strains 134/04-10, 134/04-11, 221/04-7, and 221/04-13. Our findings indicate a dynamic interaction between human and porcine rotaviruses.
Group A rotaviruses are a major cause of acute gastroenteritis in infants and young children as well as a wide variety of domestic animals (2, 12, 36). In humans, rotavirus diarrhea results in significant morbidity and mortality, especially in developing countries. Globally, the World Health Organization has estimated that 611,000 (range, 454,000 to 705,000) children die each year due to rotavirus diarrhea (27).
Rotavirus, a member of the family Reoviridae, has a triple-layered capsid that contains 11 segments of double-stranded genomic RNA. Rotavirus genotypes are defined by genome segment 4 for the P (protease-sensitive protein) type and by genome segment 9 (or 7 or 8, depending on the strain) for the G (glycoprotein) type. Through extensive serological and genomic studies, 15 G and 26 P genotypes have been established for existing human and animal rotaviruses (11, 18, 20, 22, 23, 24, 29, 30). The most common rotavirus G genotypes found in humans are G1, G2, G3, and G4. P genotype P[8] is most common in humans, followed by P[4] and P[9]. Recently, P[6] rotavirus, which was first detected as an asymptomatic infection in neonates, has been increasingly found.
Human rotavirus strains of unusual G or P types and of rare combinations of G and P types have recently been identified. G6 has been detected in Italy, Australia, India, the United States, Belgium, and Hungary; G8 has been frequently isolated in Africa and sporadically in other countries; G10 has been reported in the United Kingdom, India, Thailand, Paraguay, and Brazil (5, 31); G11 has recently been detected in Dhaka, Bangladesh (29); and G12 has been identified in the Philippines (33), Thailand (28), the United States (16), India (9), Japan (32), Korea (7), Argentina (6), and Brazil (34). G5 strains, originally detected only in pigs, have been reported in Brazil, Argentina, Paraguay, Cameroon, and the United Kingdom (1, 5, 31). Although extensive surveys on the distribution of G genotypes in Asia have been conducted (4), the existence of G5 in humans has not been reported.
We collected a total of 331 fecal specimens from children under 5 years old with acute gastroenteritis in Lulong County, Hebei Province, China, in 2003. By using an enzyme-linked immunosorbent assay kit (DAKO), 130 out of 331 stool samples (39.3%) were found to be positive for group A rotavirus. Furthermore, viral RNA was extracted from these positive samples by using a QIAamp viral RNA minikit (QIAGEN) as per the manufacturer's instructions. G3P[8] was found to be the most common genotype in positive samples based on reverse transcription-PCR analysis. For specimens for which genotypes could not be determined by reverse transcription-PCR, DNA sequencing of the VP4 and VP7 genes was performed for genotyping. One rare G/P combination of G5P[6] was identified in specimen LL36755. This strain was isolated from an 18-month-old girl with high fever, vomiting (10 times a day), and watery diarrhea. To the best of our knowledge, this is the first identification of a human G5 rotavirus in Asia and of a G5P[6] genotype combination in the world. RNA polyacrylamide gel electrophoresis shows that the electrophoretic mobilities of genes 10 and 11 of this strain are similar to those of the Wa strain (data not shown).
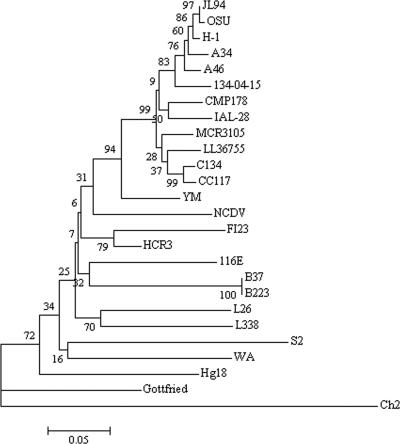
In comparison with representative rotavirus strains of the 15 known G genotypes, the VP7 sequence of strain LL36755 shared high homology of 88.6% to 89.9% at the nucleotide level and 92.6% to 95.4% at the amino acid level with those of the other G5 strains. For the remaining 14 G genotypes, the amino acid sequence homology ranged from 57.7% (Ch2 and G7) to 88% (YM and G11). Since strains sharing amino acid sequence identity of ≥90% belong to the same genotype (11, 13, 21), LL36755 qualifies as a G5 rotavirus. Phylogenetic analysis using the MEGA3.1 software further confirmed that the VP7 gene of the LL36755 strain was closely related to its counterparts in the G5 strains. In particular, it clustered with VP7 of the porcine G5 strains C134 and CC117 in the phylogenetic tree (Fig. 1). The VP7 protein of LL36755 shared 95.4% identical amino acid residues with the same protein in C134 and CC117. This degree of conservation is slightly higher than that compared to human strains MRC3105 and IAL-28 (94.0% and 93.3% identity at the amino acid level, respectively). Moreover, nine hypervariable regions (VR1 to VR9) in the linear amino acid sequence of VP7 were highly conserved within rotavirus strains of the G5 genotype but were highly polymorphic among strains of different G genotypes. Likewise, hydrophobic and hydrophilic regions in VP7 were also conserved within all G5 strains but not among strains of different G genotypes (data not shown).
FIG. 1.
Phylogenetic tree of the VP7 proteins of strain LL36755 and strains with other G genotypes. Evolutionary trees for deduced amino acid sequences were drawn by using the neighbor-joining method.
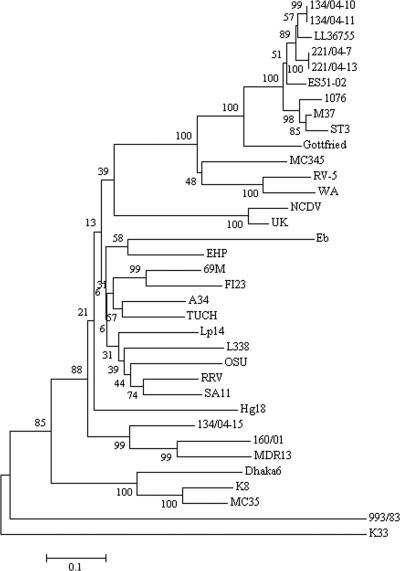
The deduced amino acid sequence of the VP4 gene of the LL36755 strain, encoding 290 residues representing the complete VP8 and the amino terminus of VP5, was compared with counterparts in rotavirus strains of all 26 P genotypes. The LL36755 VP4 had 83.2% to 94.5% amino acid sequence homology with those of the P[6] rotaviruses, whereas the homology with VP4 proteins from rotaviruses of other P genotypes was less than 74.2%. The VP4 sequence of the LL36755 strain was most closely related to those of the porcine strains 134/04-10, 134/04-11, 221/04-7, and 221/04-13 of genotype P[6] (Fig. 2), sharing 93.0% nucleotide identity and 94.5% amino acid identity with all four of these strains. In contrast, the amino acid sequence homology between the VP4 proteins of LL36755 and a rotavirus of a different P genotype ranged from 42.6% to 74.2%. Phylogenetic analysis demonstrated that the VP4 protein of strain LL36755 clustered with those of porcine viruses of the P[6] genotype (Fig. 2).
FIG. 2.
Phylogenetic tree of the VP4 proteins of strain LL36755 and strains with other P genotype. Evolutionary trees for deduced amino acid sequences were drawn by using the neighbor-joining method.
Globally, G1 to G4 as well as P[8] and P[4] are the most frequently distributed rotavirus G and P genotypes (10, 19). The four most common combinations of VP7 and VP4 are G1P[8], G2P[4], G3P[8], and G4P[8] (15). In addition, unusual G types, P types, and G-P combinations have also been reported. It is thought that immunity to rotavirus infection is predominantly homotypic initially and broadens after subsequent infections (3, 17). This eventually leads to protective immunity to all antigenic types. Reassortant rotavirus vaccine formulations have been targeted at the most commonly circulating human rotavirus strains (G1 to G4), with the ultimate goal of providing protection and minimizing severe rotavirus-associated disease. However, recent epidemiological studies in developing countries have shown increasing diversity of human rotaviruses. G5, G6, G8, G10, and G12 rotavirus strains are more frequently found in humans, but uncommon strains and high regional diversity among circulating rotaviruses have been increasingly documented over the past decade. G5 strains have been detected in Brazil, Argentina, Paraguay, Cameroon, and the United Kingdom. Particularly, in Brazil a surprising prevalence rate of 26% has been reported for G5 strains (14).
The LL36755 strain that we identified has a long RNA pattern and G5P[6] specificity. Epidemiological investigations and genotype analysis based on the VP7 and VP4 proteins are important for developing efficient rotavirus vaccines and elucidating rotavirus ecology and evolution. The amino acid sequence of the VP7 gene of the LL36755 strain showed the highest identity to those of porcine G5 strains C134 and CC117. In addition, the LL36755 VP4 is highly homologous to those of porcine strains 134/04-10, 134/04-11, 221/04-7, and 221/04-13 of genotype P[6]. P[6] specificity has been detected only in humans and pigs. The prevalence of human P[6] rotaviruses was low in Lulong County, since only 1 out of 331 cases of rotavirus infection during 2003 was identified to be caused by a P[6] strain. Lulong is a half-mountainous, half-rural county remote from cities. Direct transmission of the LL36755 strain from other areas previously recognized to have G5 rotaviruses seems unlikely. Thus, it will be of great interest to understand the origin of the LL36755 strain of genotype G5P[6]. The similarity of its VP7 and VP4 genes to those of porcine viruses (Fig. 1 and 2) suggested that the unusual human strains might have evolved gradually from other porcine and human strains. Cross-species transmission of rotaviruses has been documented (25) and represents one mechanism for genetic diversity of rotaviruses.
LL36755 is the first human G5P[6] rotavirus. The similarity of LL36755 to porcine strains suggests that the flux of genetic material between human and animal rotaviruses under natural conditions might be more common than expected (8, 26, 32, 35). Extensive surveillance of rotaviruses in humans as well as animals is therefore warranted, Although few G5 isolates have been found to date, further surveillance in different populations and geographical settings will likely reveal additional G5 rotaviruses. Thus, it remains to be determined whether this genotype should be included in future rotavirus vaccines. Rotaviruses are likely spread through different routes in developed and developing countries. Evidence for genetic reassortment between human and animal rotaviruses has been obtained for two particular animal species, cows and pigs, which are in close contact with humans. In Lulong County, people and domestic farm animals, especially domestic pigs, live in close proximity. The identification of a human G5P[6] rotavirus supports a dynamic interaction between human and animal rotaviruses. In this regard, further investigations are required to elucidate whether the LL36755 strain represents a natural reassortment between human and porcine viruses and is able to stabilize and spread successfully in humans. Simultaneous surveillance of animal and human rotavirus infections is therefore of paramount importance for studying the evolution of these viruses.
Nucleotide sequence accession numbers.
The nucleotide sequences of the VP7 and VP4 genes of human rotavirus strain LL36755 have been deposited in the GenBank database under accession numbers EF077484 and EF159569, respectively.
Acknowledgments
None of the authors has a conflict of interest.
Footnotes
Published ahead of print on 14 February 2007.
REFERENCES
- 1.Beards, G., and C. Graham. 1995. Temporal distribution of rotavirus G-serotypes in the West Midlands region of the United Kingdom, 1983-1994. J. Diarrhoeal Dis. Res. 13:235-237. [PubMed] [Google Scholar]
- 2.Bern, C., I. Martines, J. de Zoysa, and R. I. Class. 1992. The magnitude of theglobal problem of diarrhoeal disease: a ten-year update. Bull. W. H. O. 70:705-714. [PMC free article] [PubMed] [Google Scholar]
- 3.Bishop, R. F. 1996. Natural history of human rotavirus infection. Arch. Virol. Suppl. 12:119-128. [DOI] [PubMed] [Google Scholar]
- 4.Bresee, J. S., Z.-Y. Fang, B. Wang, et al. 2004. First report from the Asian Rotavirus Surveillance Network. Emerg. Infect. Dis. 10:988-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carmona, R. C., M. do C. Timenetsky, S. G. Morillo, and L. J. Richtzenhain. 2006. Human rotavirus serotype G9, Sao Paulo, Brazil, 1996-2003. Emerg. Infect. Dis. 12:963-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castello, A. A., B. Jiang, R. I. Glass, G. Glikmann, and J. R. Gentsch. 2004. Rotavirus G and P genotype prevalence in Argentina 1999-2003. Detection of P[9]G12 strains, abstr. P37-4. Abstr. 23rd Annu. Meet. Am. Soc. Virol., Montreal, Canada, 10 to 14 July 2004. American Society for Virology, Washington, DC.
- 7.Cheon, D. S., K. Lee, W. Kim, S. Lee, W. Choi, J. Ahn, et al. 2004. Genetic analysis of the VP7 gene of unusual genotypes of human group A rotavirus strains circulating in Korea, abstr. P37-3. Abstr. 23rd Annu. Meet. Am. Soc. Virol., Montreal, Canada, 10 to 14 July 2004. American Society for Virology, Washington, DC.
- 8.Cook, N., J. Bridger, K. Kendall, M. I. Gomara, L. El-Attar, and J. Gray. 2004. The zoonotic potential of rotavirus. J. Infect. 48:289-302. [DOI] [PubMed] [Google Scholar]
- 9.Das, S., V. Varghese, S. Chaudhury, P. Barman, S. Mahapatra, K. Kojima, et al. 2003. Emergence of novel human group A rotavirus G12 strain in India. J. Clin. Microbiol. 41:2760-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desselberger, U., M. Iturriza-Gómara, and J. Gray. 2001. Rotavirus epidemiology and surveillance. Novartis Found. Symp. 238:125-147. [DOI] [PubMed] [Google Scholar]
- 11.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott William & Wilikins, Philadelphia, PA. [Google Scholar]
- 12.Glass, R. I., J. S. Bressee, U. Parashar, M. Miller, and J. R. Gentsch. 1997. Rotavirus vaccines at the threshold. Nat. Med. 3:1324-1325. [DOI] [PubMed] [Google Scholar]
- 13.Gouvea, V., N. Santos, and M. C. Timenetsky. 1994. Identification of bovine and porcine rotavirus G types by PCR. J. Clin. Microbiol. 32:1338-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouvea, V., L. de Castro, M. C. Timenetsky, H. Greenberg, and N. Santos. 1994. Rotavirus serotype associated with diarrhea in Brazilian children. J. Clin. Microbiol. 32:1408-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin, D. D., C. D. Kirkwood, U. D. Parashar, P. A. Woods, J. S. Bresee, R. I. Glass, J. R. Gentsch, et al. 2000. Surveillance of rotavirus strains in the United States: identification of unusual strains. J. Clin. Microbiol. 38:2784-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin, D. D., T. Nakagomi, Y. Hoshino, O. Nakagomi, C. D. Kirkwood, U. D. Parashar, et al. 2002. Characterization of nontypeable rotavirus strains from the United Status: identification of a new rotavirus reassortant (P2A[6], G12) and rare P3[9] strains related to bovine rotavirus. Virology 294:256-269. [DOI] [PubMed] [Google Scholar]
- 17.Hoshino, Y., R. W. John, and A. Z. Kapikian. 1996. Classification of rotavirus VP4 and VP7 serotypes. Arch. Virol. Suppl. 12:99-111. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino, Y., R. W. Jone, and A. Z. Kapikian. 2002. Characterization of neutralization specificities of outer capsid spike protein VP4 of selected murine, lapine, and human rotavirus strains. Virology 299:64-71. [DOI] [PubMed] [Google Scholar]
- 19.Koopmans, M., and D. Brown. 1999. Seasonality and diversity of group A rotaviruses in Europe. Acta Paediatr. Suppl. 88:14-19. [DOI] [PubMed] [Google Scholar]
- 20.Liprandi, F., M. Gerder, Z. Bastidas, J. A. López, F. H. Pujol, J. E. Ludert, D. B. Joelsson, and M. Ciarlet. 2003. A novel type of VP4 carried by a porcine rotavirus strain. Virology 315:373-380. [DOI] [PubMed] [Google Scholar]
- 21.Martella, V., M. Ciarlet, R. Baselga, S. Arista, G. Elia, E. Lorusso, K. Banyai, V. Terio, A. Madio, F. M. Ruggeri, E. Falcone, M. Camero, N. Decaro, and C. Buonavoglia. 2005. Sequence analysis of theVP7 andVP4 genes identifies a novelVP7 gene allele of porcine rotaviruses, sharing a common evolutionary origin with human G2 rotaviruses. Virology 337:111-123. [DOI] [PubMed] [Google Scholar]
- 22.Martella, V., M. Ciarlet, A. Camarda, A. Pratelli, M. Tempesta, G. Greco, A. Cavalli, G. Elia, N. Decaro, V. Terio, G. Bozzo, M. Camero, and C. Buonavoglia. 2003. Molecular characterization of the VP4, VP6, VP7, and NSP4 genes of lapin rotaviruses identified in Italy: emergence of a novel VP4 genotype. Virology 314:358-370. [DOI] [PubMed] [Google Scholar]
- 23.Martella, V., M. Ciarlet, K. Banyai, E. Lorusso, A. Cavalli, M. Corrente, G. Elia, S. Aeista, M. Camero, C. Desario, M. Desario, A. Lavazza, and C. Buonavoglia. 2006. Identification of a novel VP4 genotype carried by a serotype G5 porcine rotavirus strain. Virology 346:301-311. [DOI] [PubMed] [Google Scholar]
- 24.McNeal, M. M., K. Sestak, A. H. Choi, M. Basu, M. J. Cole, P. P. Aye, R. P. Bohm, and R. L. Ward. 2005. Development of a rotavirus-shedding model in rhesus macaques, using a homologous wild-type rotavirus of a new P genotype. J. Virol. 79:944-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagomi, O., and T. Nakagomi. 1993. Interspecies transmission of rotaviruses studied from the perspective of genogroup. Microbiol. Immunol. 37:337-348. [DOI] [PubMed] [Google Scholar]
- 26.Palombo, E. A. 2003. Genetic reassortment and interspecies transmission of rotaviruses, p. 1-16. In N. Kobayashi (ed.), Genomic diversity and molecular epidemiology of rotaviruses. Research Signpost, Trivandrum, India.
- 27.Parashar, U. D., C. J. Gibson, J. S. Bresee, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pongsuwanna, Y., R. Guntapong, M. Chiwakul, R. Tacharoenmuang, N. Onvimala, M. Wakuda, et al. 2002. Detection of a human rotavirus with G12 and P[9] specificity in Thailand. J. Clin. Microbiol. 40:1390-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahman, M., J. Matthijnssens, S. Nahar, G. Podder, D. A. Sack, T. Azim, and M. Van Ranst. 2005. Characterization of a novel P[25], G11 human group A rotavirus. J. Clin. Microbiol. 43:3208-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao, C. D., K. Gowda, and B. S. Reddy. 2000. Sequence analysis of VP4 and VP7 genes of nontypeable strains identifies a new pair of outer capsid proteins representing novel P and G genotypes in bovine rotaviruses. Virology 276:104-113. [DOI] [PubMed] [Google Scholar]
- 31.Santos, N., and Y. Hoshino. 2005. Global distribution of rotavirus serotypes/genotypes and its implication for the development and implementation of an effective rotavirus vaccine. Rev. Med. Virol. 15:29-56. [DOI] [PubMed] [Google Scholar]
- 32.Shinozaki, K., M. Okada, S. Nagashima, I. Kaiho, and K. Taniguchi. 2004. Characterization of human rotavirus strains with G12 and P[9] detected in Japan. J. Med. Virol. 73:612-616. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi, K., T. Urasawa, N. Kobayashi, M. Gorziglia, and S. Urasawa. 1990. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new serotype specificity. J. Virol. 64:5640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Timenetsky, M. C. S. T., R. C. Carmona, S. G. Morillo, M. B. P. E. Eduardo, and L. J. Silva. 2005. Incidence of rotavirus G and P genotypes in children in southern Brazil. Emergence of genotype G9, abstr. V-330. Abstr. Int. Congr. Virol., Joint Meet. Int. Union Microbiol. Soc., San Francisco, CA, 23-28 July 2005. International Union of Microbiological Societies, Utrecht, The Netherlands.
- 35.Varghese, V., S. Das, N. B. Singh, K. Kojima, S. K. Bhattacharya, T. Krishnan, N. Kobayashi, and T. N. Naik. 2004. Molecular characterization of a human rotavirus reveals porcine characteristics in most of the genes including VP6 and NSP4. Arch. Virol. 149:155-172. [DOI] [PubMed] [Google Scholar]
- 36.Woode, G. N., and J. C. Bridger. 1975. Viral enteritis of calves. Vet. Rec. 96:85-88. [DOI] [PubMed] [Google Scholar]