Abstract
Thermoanaerobacter tengcongensis is a rod-shaped, gram-negative, anaerobic eubacterium that was isolated from a freshwater hot spring in Tengchong, China. Using a whole-genome-shotgun method, we sequenced its 2,689,445-bp genome from an isolate, MB4T (Genbank accession no. AE008691). The genome encodes 2588 predicted coding sequences (CDS). Among them, 1764 (68.2%) are classified according to homology to other documented proteins, and the rest, 824 CDS (31.8%), are functionally unknown. One of the interesting features of the T. tengcongensis genome is that 86.7% of its genes are encoded on the leading strand of DNA replication. Based on protein sequence similarity, the T. tengcongensis genome is most similar to that of Bacillus halodurans, a mesophilic eubacterium, among all fully sequenced prokaryotic genomes up to date. Computational analysis on genes involved in basic metabolic pathways supports the experimental discovery that T. tengcongensis metabolizes sugars as principal energy and carbon source and utilizes thiosulfate and element sulfur, but not sulfate, as electron acceptors. T. tengcongensis, as a gram-negative rod by empirical definitions (such as staining), shares many genes that are characteristics of gram-positive bacteria whereas it is missing molecular components unique to gram-negative bacteria. A strong correlation between the G + C content of tDNA and rDNA genes and the optimal growth temperature is found among the sequenced thermophiles. It is concluded that thermophiles are a biologically and phylogenetically divergent group of prokaryotes that have converged to sustain extreme environmental conditions over evolutionary timescale.
[Supplemental material is available online at http://www.genome.org.]
Thermoanaerobacter tengcongensis, isolated from a hot spring in Tengchong, Yunnan, China, is a rod-shaped, gram-negative (by empirical definitions) bacterium that grows anaerobically under extreme environment. It propagates at temperatures ranging from 50° to 80°C (optimally at 75°) and at pH values ranging between 5.5 and 9 (optimally from 7 to 7.5). It shares several key genomic and physiological features common to the genus Thermoanaerobacter, such as a relatively low genomic G + C content (<40%), reduction of thiosulfate/sulfur to hydrogen sulfide, and fermentation of glucose to acetate/ethanol (Xue et al. 2001).
T. tengcongensis, however, has several important phenotypic properties that contradict its membership to the genus. Some of the examples include the absence of spore production, negative gram-staining result, lack of motility under cultural conditions, and exclusive metabolic pathways (such as deficiencies in lactate production and xylan utilization; Cayol et al. 1995; Xue et al. 2001). To obtain a global view of genes possessed by the organism and to resolve some of the controversies at molecular levels, as well as to understand the biology of thermophilic prokaryotes in general through comparative genomics, we set out to sequence the T. tengcongensis genome.
Using a whole-genome-shotgun method, we acquired sequence data at high-genome coverage (9.87×) and assembled the complete sequence of the T. tengcongensis genome of a laboratory strain, MB4T (Genbank accession no. AE008691; see also http://btn.genomics.org.cn/tten/). Computational analyses of the high-quality genomic sequence not only confirmed many of the early experimental observations, but also uncovered the heterogeneous nature of thermophilic prokaryotic genomes. The T. tengcongensis genome sequence should provide vital information for understanding cellular and molecular mechanisms that are employed by microorganisms under extreme environments.
RESULTS AND DISCUSSION
General Features
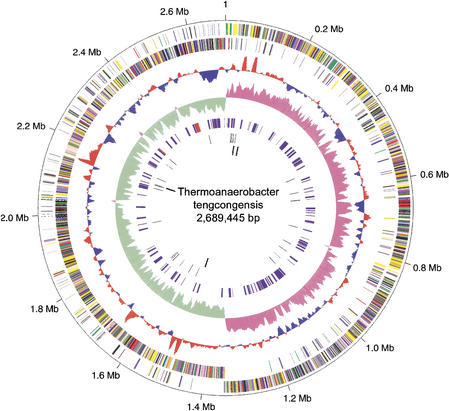
T. tengcongensis has a single, circular chromosome of 2,689,445 bp (base pairs) in length (Fig. 1a,b; Table 1). Second only to the Sulfolobus solfataricus genome, it is one of the largest genomes of thermophiles sequenced to date (Bult et al. 1996; Klenk et al. 1997; Smith et al. 1997; Deckert et al. 1998; Kawarabayasi et al. 1998, 1999; Nelson et al. 1999; Kawashima et al. 2000; Ruepp et al. 2000; She et al. 2001; Heilig, unpubl.). The genomic sequence has an average G + C content of 37.6%, similar to those of other members of the genus Thermoanaerobacter (Table 1; Supplemental Table A [available at http://www.genome.org]). The genome has 4 rRNA gene clusters (12 rRNA genes) and each cluster encompasses a single copy of 5S, 16S, and 23S RNA genes. The G + C content of the rRNA genes or rDNAs varies from 58.2% to 60.3%. There are 55 tRNA genes scattered over the genome in 28 loci (1–8 tRNAs in each locus). The G + C content of tDNAs has a broader distribution than that of rDNAs, from 52.6% to 69.3%. The characteristically high G + C content of rDNA and tDNA genes found in T. tengcongensis appears common to all thermophiles (discussed in detail later; also see Supplemental Table A). The elevated G + C content of rDNAs and tDNAs as a function of genomic G + C content increase is also evident in most of the mesophiles, albeit less pronounced (Supplemental Table B).
Figure 1.
(a) Circular representation of the Thermoanaerobacter tengcongensis genome. Circles display (from the outside): (1) Physical map scaled in megabases from base 1, the start of the putative replication origin. (2) Coding sequences transcribed in the clockwise direction. (3) Coding sequences transcribed in the counterclockwise direction. (4) G + C percent content (in a 10-kb window and 1-kb incremental shift); values >37.6% (average) are in red and smaller in blue. (5) GC skew (G-C/G + C, in a 10-kb window and 1-kb incremental shift); values greater than zero are in magenta and smaller in green. (6) Repeated sequences; short 30-bp repeats are in red and other types in blue. (7) tRNA genes. (8) rRNA genes. Genes displayed in 2 and 3 are color-coded according to different functional categories: translation/ribosome structure/biogenesis, pink; transcription, olive drab; DNA replication/recombination/repair, forest green; cell division/chromosome partitioning, light blue; posttranslational modification/protein turnover/chaperones, purple; cell envelope biogenesis/outer membrane, red; cell motility/secretion, plum; inorganic ion transport/metabolism, dark sea green; signal transduction mechanisms, medium purple; energy production/conversion, dark olive green; carbohydrate transport/metabolism, gold; amino acid transport/metabolism, yellow; nucleotide transport/metabolism, orange; coenzyme metabolism, tan; lipid metabolism, salmon; secondary metabolites biosynthesis/transport/catabolism, light green; general function prediction only, dark blue; conserved hypothetical, medium blue; hypothetical, black; unclassified, light blue; pseudogenes, gray. (b) Linear representation of the T. tengcongensis genome. Genes are color-coded according to different functional categories as described above for a , with above character-string representing gene names or IDs. Arrows indicate the direction of transcription. Genes with authentic frameshift and point mutations are indicated with X. Paralogous gene families are indicated by family ID in a box above the predicted genes. Numbers next to GES (Goldman-Engleman-Steitz) represent the number of membrane-spanning domains predicted by Goldman-Engleman-Steitz scale calculated by TMHMM. Proteins with five or more GES are indicated. The 305 copies of the 30-bp short repeat, clustered in two regions, are indicated with the greater-than symbol. RNA genes, including those of rRNA, tRNA, and other RNA genes, signal peptides and long repeats are also indicated. Numbers on the tRNA symbols represent the number of tRNAs in the cluster.
Table 1.
General Features of the Thermoanaerobacter tengcongensis Genome
| Genome size (bp) | 2,689,445 | |
| G+C content | 37.6% | |
| Protein coding | 87.1% | |
| Average CDS length (bp) | 905 | |
| Predicted CDS | 2,588 | |
| Known proteins | 1494 (57.8%) | |
| Homologous to | Protein domains/motifs | 270 (10.4%) |
| Hypothetical proteins | 301 (11.6%) | |
| No homology | 523 (20.2%) | |
| Stable RNAs | 0.9% | |
| rRNA operons | 4 | |
| rRNAs | 55 | |
| Major repetitive elements | ||
| Short noncoding repeats | 0.5% | |
| Long coding repeats | 8.6% |
CDS, coding sequences.
Repetitive Sequences
The T. tengcongensis genome has a significant fraction (9.1%) of repetitive sequences that include simple repeats of a few dozen base pairs in length as a limited number of clusters to complex ones, such as transposase coding (Tables 1, 2). In this study, all repeats were categorized by the means of a suffix tree algorithm (Rocha et al. 1999; Kurtz et al. 2001), coupled with intensive manual alignment and visual inspection.
Table 2.
Selected List of Repetitive Elements in the Thermoanaerobacter tengcongensis Genome
| Repeat ID | Length (bp) | Number of copies | Identity (%) | Short noncoding repeats | |
|---|---|---|---|---|---|
| TSR001 | 30 | 305 (67/238) | 100 | TSR001a(GTTTTTAGCCTACCTAAAAGGGATTGAAAC) | |
| TSR001b(GTTTTTAGCCTACCTAAGAGGGATTGAAAC) | |||||
| TSR027 | −250 | 18 | <87 | ||
| Copies | |||||
| Complete | Partial | Long coding repeats | |||
| TLR028b | 3565 | 4 | 5 | >99 | Transposase + hypothetical protein + transposase |
| TLR393c | 3045 | 2 | 1 | >98 | ABC transporters + hypothetical protein |
| TLR315 | 2603 | 2 | >94 | ABC transporters + permease component + conserved hypothetical protein | |
| TLR408 | 2490 | 2 | >98 | Ferredoxin oxidoreductases α subunit + β subunit + γ subunit | |
| TLR076 | 2021 | 2 | >91 | Hypothetical protein | |
| TLR271 | 2020 | 2 | >92 | ABC transporters | |
| TLR264 | 1986 | 5 | 1 | >98 | Transposase |
| TLR294 | 1851 | 2 | >98 | ABC transporters + permease component | |
| TLR004 | 1819 | 14 | >98 | Transposase | |
| TLR005 | 1800 | 7 | >98 | Transposase | |
| TLR158 | 1774 | 1 | 2 | >89 | TPR-repeat-contaning proteins |
| TLR048 | 1711 | 2 | >99 | Transposase | |
| TLR223 | 1629 | 2 | >97 | Transposase | |
| TLR008 | 1596 | 21 | >92 | Hypothetical protein | |
| TLR014 | 1592 | 14 | 3 | >87 | Hypothetical protein |
| TLR073 | 1571 | 6 | >93 | Transposase | |
| TLR488 | 1549 | 2 | >98 | ABC transporters | |
| TLR533 | 1506 | 2 | >99 | Pseudotransposase | |
| TLR354 | 1400 | 2 | >91 | Arylsulfatase regulator | |
| TLR152 | 1347 | 2 | 100 | Transposase | |
| TLR478 | 1199 | 2 | >98 | GTPases | |
| TLR107 | 1141 | 2 | >99 | Methyl-accepting chemotaxis protein | |
| TLR070 | 1037 | 3 | 1 | >88 | Hypothetical protein |
| TLR177d | 978 | 2 | 1 | >97 | Hypothetical protein + permeases |
| TLR500 | 885 | 2 | >94 | Hypothetical protein | |
| TLR403 | 848 | 2 | >98 | Pyruvate carboxylase | |
| TLR211 | 663 | 2 | >94 | CheY-like receiver domains | |
| TLR115 | 623 | 8 | 9 | >87 | Predicted site-specific integrase-resolvase |
| TLR250 | 527 | 2 | 100 | Hypothetical protein | |
| TLR429 | 502 | 2 | >95 | Methylmalonyl-CoA mutase | |
| TLR384 | 496 | 3 | >90 | Hypothetical protein | |
| TLR509 | 479 | 2 | >93 | Hypothetical protein | |
| TLR098e | 428 | 2 | 1 | >98 | Hypothetical protein |
| TLR434 | 403 | 2 | >97 | Lactoylglutathione lyase | |
| TLR311f | 369 | 2 | 2 | >95 | Partial transposase |
| TLR537 | 361 | 2 | >98 | ABC transporters | |
| TLR349 | 352 | 2 | 1 | >95 | Hypothetical protein |
A copy is complete if the length of the repeat is ε ≥90% of the consensus, otherwise, the copy is partial.
Two partial copies with 96% identity.
A 1300 bp deletion in the partial copy.
One partial copy with 89% identity.
One partial copy with 94% identity.
Two partial copies with identities of 92% and 94%, respectively.
The most characteristic repeat family of the T. tengcongensis genome consists of 305 copies of a unique 30-bp AT-rich repeat, TSR001 (Fig. 1b). They are further grouped into two subfamilies, TSR001a and TSR001b. The two subfamilies differ from each other only by a single substitution at position 18, an adenosine (67 copies) in TSR001a and a guanine (238 copies) in TSR001b, respectively. Sixty-five copies of TSR001a are clustered between 2,326,770 bp and 2,331,141 bp and all units are oriented in the same direction. The two remaining copies are arrayed together with a single cluster of TSR001b (238 copies) from 2,537,291 bp to 2,555,096 bp. The repeat units are not attenuated directly but interrupted by nonrepetitive sequence spacers, most of which are 34 to 41 bp in length. However, three of the spacers are longer than 100 bp (2,329,533–2,329,637 bp, 2,538,340–2,538,450 bp, and 2,550,689–2,550,793 bp) and another one is 1632 bp (2,540,469–2,538,790 bp) in length, which encodes a transposase (TTE2646). Repeats of similar types are found in other thermophiles, from both archaea and eubacteria. Most of them are distinct, short (20 to ∼60 bp), relatively abundant, and organized in a single cluster or multiple clusters (Bult et al. 1996; Klenk et al. 1997; Smith et al. 1997; Kawarabayasi et al. 1998, 1999; Nelson et al. 1999). The function of such repeats is yet to be defined and they might play important roles in chromosome anchorage and segregation in these thermophilic organisms.
Thirty-seven families of protein-coding repetitive sequences longer than 300 bp were also categorized. Most of them are related to transposases (10 families; 54 copies) and ABC transporters (6 families; 13 copies). Others are unknown or hypothetical (11 families; 62 copies). The largest repeated sequence, TLR028 (3565-bp in length), is composed of two different transposases flanking a hypothetical protein. The most abundant one, a 1596-bp repeat (TLR008), consisting of a single hypothetical gene and a 200-bp noncoding region, occurs 21 times over the entire genome.
Origin of Replication
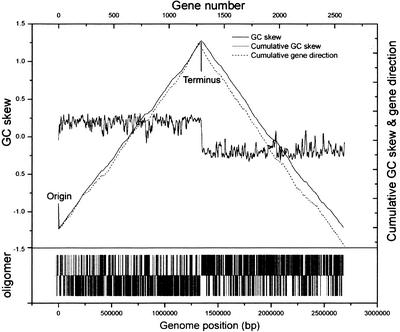
Of a half dozen methods for determining origins and termini of DNA replication, including asymmetric distribution of oligomers (Salzberg et al. 1998), GC-skew (G-C/G + C; Lobry 1996), accumulated GC-skew (Grigoriev 1998), and orientation of coding sequences (CDS), all worked satisfactorily in determining the origin of replication for T. tengcongensis. Figure 2 depicts results from some of the analyses. The predicted origin is defined between ribosomal protein L34 (TTE2802) and dnaA (TTE0001) genes, which is dictated by the asymmetry of the nucleotide composition between the leading and the lagging strands. The first base of an octamer repeat (TTTTTCTT)1423, 307-bp upstream of dnaA, is assigned as base-pair number one, whereas the terminus is about halfway into the genome, ∼1345-kbp from the origin (Fig. 1a).
Figure 2.
The replication origin of the Thermoanaerobacter tengcongensis. GC skew [(G-C)/(G + C)] was calculated with a nonoverlapping sliding window of 10 kb for a single strand over the length (upper horizontal line). Cumulative GC skew was plotted from position 1 of the genome (upper solid line). Cumulative gene direction (upper dotted line) was plotted from position 1 of the genome sequence, showing that the majority of genes transcribe along the same direction following the replication forks. In the skewed oligomer (TTTTTCTT)1423 part (lower), vertical lines above the center represent the location of this octamer on one DNA strand, and lines below the center indicate the positions on the complementary strand. The transition in GC and oligomer skews, maxima of the curves at the middle of the genome sequence, is identified as the putative terminus of replication.
T. tengcongensis has the most biased gene distribution on the leading strand, in the same direction as genome replication, among all sequenced prokaryotic genomes known to date (Fig. 1a). Of the genes, 86.7% (41.9% and 44.8% from the two replication forks) are transcribed along the leading strand from the two halves of the genome divided by the replication origin. The lagging strand encodes only 13.3% (7% and 6.3% from the two replication forks). The biases in gene orientation have been observed in many other bacteria (Karlin 1999), but only three of them exceed 80% of the total encoded genes. The extreme case is not seen in prokaryotes but in a eukaryotic organism, Leishmania major, in which the leading strands of all chromosomes encode all the genes (Myler et al. 1999). Further analysis and experimentation are of essence to address what is the driving force that instigates such extreme gene distributions.
Coding Sequences
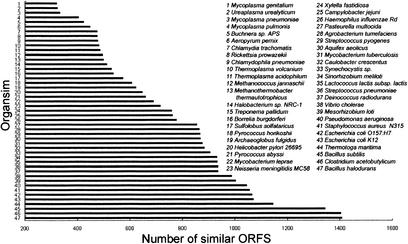
Identified were 2588 predicted CDS, covering 87.1% of the genome (Table 1; for functional classifications, see Table 3 and Supplemental Table C). Genes for stable RNAs populates 0.9% of the genome. The average length of the CDS is 905 bp, slightly longer than that of a mesophile, Bacillus halodurans (880 bp; Takami et al. 2000). Of the CDS, 72.9% start with ATG, 13.2% with TTG, and 13.9% with GTG. Such a distribution is similar to that of the B. halodurans genome, of which 78% of the CDS begin with ATG, 10% with TTG, and 12% with GTG. There are 1764 CDS (68.2%) that are homologous to known proteins or protein domains/motifs in public databases; thus, their biological functions are putatively assigned. Identified were 301 CDS (11.6%) in other sequenced prokaryotic genomes as conserved protein sequences of unknown function; 523 CDS (20.2%) have no homologous counterparts in all public databases. When protein similarity was scored in a genome-wide fashion, 54.4 % of T. tengcongensis genes have extensive similarity (BLASTP; 1e-10) to those of B. halodurans. Their overall genome similarity ranks the highest among all the sequenced genomes, regardless if they are thermophiles or mesophiles (Fig. 3).
Figure 3.
Relative distance of the Thermoanaerobacter tengcongensis genome with those of other 47 completely sequenced genomes, measured by a collective similarity score of the 2588 predicted coding sequences (CDS). All the sequences were retrieved from NCBI databases. A tally was kept of which genome produces the significant similarity with the BLASTP program above an expected value of 1e-10. The number of T. tengcongensis CDS matched to those of each genome is tabulated. Bacillus halodurans has the highest value of 54.4%, indicating its highest similarity to T. tengcongensis.
Replication, Recombination, and DNA Repair
Genes for the primary replication machinery, the DNA polymerase III complex in T. tengcongensis, are similar to those of well-characterized components in Escherichia coli, which is composed of α-subunit (dnaE, TTE1818), β-subunit (dnaN, TTE0002), γ-τ-subunit (dnaX, TTE0039), and δ-subunit (holA, TTE0942). In addition, a polC-like gene encoding an alternative DNA polymerase III α-subunit was also identified in T. tengcongensis (TTE1398). The presence of two α-subunits is not exceptional for T. tengcongensis: this function of polC gene has been reported in Bacillus subtilis (Dervyn et al. 2001). Both dnaE and polC genes are found in several fully sequenced bacterial genomes of the Bacillus/Clostridium group. The thermophilic Thermotoga maritima also harbors these two genes. Although the essential DNA polymerase I homolog is present in T. tengcongensis (TTE0874), DNA polymerase II—recently being shown to be involved in replication-related DNA damage repair in E. coli (Bonner et al. 1988; Napolitano et al. 2000) but not being essential—is absent. Many other essential DNA replication-related genes are readily determined by sequence homology. For instance, topoisomerases I/II (topA, TTE1449; gyrA, TTE0011; and gyrB, TTE0010), single-stranded DNA- binding protein (Ssb), DNA helicase (dnaB, TTE2774), and primase (dnaG, TTE1757) are all readily defined by sequence homology.
Homologs of recombination and DNA repair-related genes, such as recA/B/D/F/G/N/O/R (TTE1374, TTE0264, TTE0489, TTE0004, TTE1492, TTE1302, TTE0976, and TTE0041, respectively), and >20 genes that are involved in postreplicational mismatch/excision, ultraviolet-induced damage and transcription-coupled DNA repairs, including the mutT/mutS gene families, uvrA/B/C (TTE1970, TTE1971, and TTE1966, respectively) gene cluster, and the uvrD (TTE0604) gene, were found in T.tengcongensis. Although none of the methylation-related dam/dcm homologs was found, suggesting that the genome DNA has no dam/dcm methyl modification, the T. tengcongensis genome possesses seven putative endonuclease genes and a type-I restriction-modification system that is composed of four genes in a single operon. The functions of these putative genes are currently being evaluated.
Transcription and Translation
Three RNA polymerase core-enzyme genes (rpoA, TTE2263; rpoB, TTE2301; and rpoC, TTE2300), which encode subunits α, β and β', and another gene that encodes polymerase subunit Ω (rpoZ, TTE1510) are all documented. Seventeen σ factors belonging to four groups that constitute the holoenzyme of the RNA polymerase complex are found. The first group contains four of the rpoD (σ70)-like genes, believed to have housekeeping functions. rpoN (σ54)-like gene (Lonetto et al. 1992) stands alone. The third group, the largest of all, is composed of seven rpoE (σ24) homologs of the Extracytoplasmic function (ECF) subfamily, whose function is postulated as stress-related, and they perhaps are responsive to the high-temperature environment (Hiratsu et al. 1995; Schurr et al. 1995; Petersohn et al. 2001). The last five fliA-like genes as a group (σfliA) are alternative σ factors. Additional transcription-related factors, such as the elongation factor (greA), the rho factor, the termination factors (nugA, nugB), and three antitermination factors (nugG-like genes) are all unambiguously recognized. Among these documented genes, greA, nugB, and rho have homologs only in Eubacteria.
T. tengcongensis has >50 transcriptional regulators acting as activators or repressors involved in many physiological and metabolic pathways. There are ∼15 response regulators also related to transcriptional regulation. Twelve of them are two-component response regulators (Kunst et al. 1997), characterized by a CheY-like receiver domain and an HTH (helix-turn-helix) DNA-binding domain. Two of them are serine phosphatases (encoded by rsbU) with orthologs found in B. subtilis and T. maritime. The last one is a ppGpp synthetase/hydrolase (TTE1195) whose product is believed to be the effector involved in bacterial stringent response (Sarubbi et al. 1988; Metzger et al. 1989).
All translation-related genes are highly conserved as seen in other prokaryotes, and shared by both Eubacteria and Archea. Twenty-three genes that encode 20 essential tRNA synthetases are predicted. Two copies (TTE1394 and TTE2299) of an archaeal gene that encodes the ribosomal subunit RpL8A protein are identified in the T. tengcongensis genome. This gene has been found in two other related eubacterial genomes, a thermophile, Thermotoga maritina, and a mesophile, B. halodurans. Many gene products involved in posttranslational processes are also inevitable, including those heat-shock proteins (such as GroES, GroEL, DnaJ/K, and HslU) and chaperones (such as Hsp33 and Hsp20, ATPases associated with various cellular acts and peptidase). A homolog of cold-shock protein, CspC, (Schroder et al. 1993) and a protein that has a regulatory function in transcription and stationary phase survival, SurE, (Nelson et al. 1999) is also present in the genome.
Respiratory Pathways
T. tengcongensis gains energy anaerobically by sulfur respiration and uses thiosulfate or element sulfur as electron receptors because its growth increases in the presence of thiosulfate or sulfur but not in the presence of sulfate (Xue et al. 2001). Such an observation seems to contradict a common feature observed in most sulfur-respiratory prokaryotes, a heterogeneous group of microorganisms that have the ability to use sulfate as a terminal electron acceptor (Hansen 1994), including both eubacteria and archaea.
What has happened to the sulfate pathway in the T. tengcongensis genome? First, neither the genes related to sulfate transport systems, nor the key genes involved in the sulfate reduction (such as sulfate adenylate transferase, 3′-phosphoadenosine 5′-phosphosulfate sulfotransferase and adenylylsulfate kinase) are present. Secondly, in the reduction process, thiosulfate is generally reduced to sulfite and further to sulfide. Thiosulfate reductase and sulfite reductase, which play crucial roles in these steps, are not found in the T. tengcongensis genome. Instead, a rhodanese-related sulfurtransferase (TTE1148), which employs thiosulfate as electron acceptor in the presence of cyanide ion (Alexander and Volini 1987), is identified. Because sulfite is not an end product of sulfur metabolism and cannot be reduced to sulfide, it might be recycled back to thiosulfate through a thiosulfate-synthesis pathway in T. tengcongensis as it has been described in Desulfovibrio vulgaris (Kim and Akagi 1985; Hansen 1994). In D. vulgaris, a trithionate reductase system consisting of two proteins was identified. One is bisulfite reductase, which reduces bisulfite to trithionate, and the other putative protein is designated as TR-1. Both enzymes are required to reduce trithionate to thiosulfate. If this is also the case in T. tengcongensis, it is expected to find flavodoxin (TTE0566, TTE0694, TTE1329, and TTE1531) and cytochrome c3 (TTE1025), which are essential to this pathway. Indeed, the two genes are present in T. tengcongensis. Moreover, two putative ancient conserved regions (ACR) (TTE0085 and TTE0087, stress proteins believed to be involved in the bacillary response to adverse conditions and in non-replicating persistence) related to intracellular sulfur reduction and oxidation also exist in the genome. Although most of the sequenced bacterial genomes have rhodanese-related sulfurtransferases, the two ACR genes are detectable only in a few other bacterial genomes, including Methanobacterium thermoautotrophicum (Smith et al. 1997), T. maritime (Nelson et al. 1999), E. coli (Blattner et al. 1997), Pseudomonas aeruginosa (Stover et al. 2000), and Vibrio cholerae (Heidelberg et al. 2000). M. thermoautotrophicum is a methanogen that utilizes CO2 as the electron acceptor (Kral et al. 1998), and T. maritima is a thermophile that has an ability to gain energy through a fermentation pathway in the presence of Fe (III) (Vargas et al. 1998) and utilizes sulfur as electron acceptor but does not consequently produce any ATP (Janssen and Morgan 1992). No rhodanese-related sulfurtransferase has been recognized in the T. maritima genome either. P. aeruginosa and V. cholerae are oxygenic-respiration bacteria. E. coli has both aerobic and anaerobic respiratory pathways, and the pathway involving formate oxidation and nitrate reduction constitutes a major anaerobic respiratory pathway in E. coli (Berg and Stewart 1990), which is completely absent in T. tengcongensis.
Metabolisms
As an anaerobic and heterotrophic eubacterium, T. tengcongensis utilizes both monosaccharides and polysaccharides as carbon sources and yields H2, CO2, and acetate as its major metabolic end products (Xue et al. 2001). Among the complex sugars, it is capable of metabolizing starch but not cellulose or xylan. It is known that thiosulfate reducers, such as T. brockii and T. thermohydrosul furicus, as well as several other thermoanaerobacteria, consume a variety of sugars, including polymeric sugars (Cayol et al. 1995; Xue et al. 2001). However, only a few sulfate-reducers are known to grow on sugars, including Archaeoglobus fulgidus, D. nigrificans, D. geothermicum, D. simplex, D. termitidis, and D. fructosovorans (Qatibi et al. 1998; Labes and Schonheit 2001). A. fulgidus is the only one among the group capable of utilizing polymeric sugars.
T. tengcongensis has a complete set of genes constituting the glycolysis and the pentose phosphate pathways. It, however, has a few key metabolic enzymes yet to be found for other related pathways. One of the examples is fructose-1,6-biphosphatase, a key enzyme in the gluconeogenesis pathway. Such a depletion is not extraordinary, as similar cases are encountered in all other sequenced thermophiles and certain nonthermophilic bacteria, such as B. subtilis (Kunst et al. 1997), Deinococcus radiodurans (White et al. 1999), and Xylella fastidiosa (Simpson et al. 2000). Another example is the absence of 2-keto-3-deoxy-6-phosphogluconate aldolase in the Entner-Doudoroff pathway.
The metabolism of pyruvate reflects the microaerophilic nature of T. tengcongensis. Neither the aerobic pyruvate dehydrogenase (COG0567; Tatusov et al. 2001) nor the strictly anaerobic pyruvate formate lyase (COG1882) is present in T. tengcongensis. Similar to the cases of Helicobacter pylori (Tomb et al. 1997) and Campylobacter jejuni (Parkhill et al. 2000), T. tengcongensis has 12 genes (TTE0445, TTE0960, TTE0961, TTE1209, TTE1210, TTE1211, TTE1340, TTE1341, TTE1342, TTE2193, TTE2194, and TTE2198) related to the pyruvate:ferredoxin oxidoreductases and 2-oxoacid:ferredoxin oxidoreductases. The conversion of pyruvate to acetyl coenzyme A (acetyl CoA) is performed by the pyruvate ferrodoxin oxidoreductase (POR; Cayol et al. 1995; Menon and Ragsdale 1997), a four-subunit enzyme described in H. pylori and other hyperthermophilic organisms (Hughes et al. 1995). Acetyl CoA is converted to acetate and this process is catalyzed by four enzymes, phosphate acetyltransferase (TTE1482, TTE2195, and TTE2204), acetate kinase (TTE1481), NADH:flavin oxidoreductase (TTE0012, TTE0988, TTE2131, and TTE2625), and Acyl-CoA dehydrogenase (TTE0545; Bock et al. 1999). These four enzymes are identified in T. tengcongensis.
Anaerobic acetogenic bacteria with acetate as their primary reduced end product are capable of utilizing H2 and CO2 to produce acetyl CoA in an autotrophic biosynthetic scheme known as the Wood-Ljungdahl pathway (or the acetyl-CoA pathway). This pathway, catalyzed by enzymes of carbon monoxide dehydrogenase (CODH), formyltetrahydrofolate synthetase, and acetyl-CoA synthetase, synthesizes acetyl CoA from two molecules of CO2 (Ragsdale 1991; Kuhner et al. 1997). The key enzymes for the acetyl-CoA pathway, such as a CODH subunit CooS (TTE1708) and a formyltetrahydrofolate synthetase (TTE2391), are identified in T. tengcongensis. The existence of this pathway might reflect the acetogenic aspect of T. tengcongensis. The same pathway was described in A. fulgidus, a thermophilic, anaerobic sulfate-reducing archaeon that grows chemolithoautotrophically on H2 and CO2 with sulfate or thiosulfate as electron acceptor and grows chemoorganoheterotrophically with sulfate and lactate, as well as other carbohydrates (Labes and Schonheit 2001). Many chemolithoautotrophic sulfate-reducing prokaryotes, such as those of the genus Desulfobacterium, are acetogenic bacteria (Janssen and Schink 1995), whereas no acetogenic features have been clearly reported so far about the thermophilic anaerobic thiosulfate-reducing Thermoanaerobacter bacteria, including T. tengcongensis (Cayol et al. 1995; Xue et al. 2001).
The tricarboxylic acid cycle (TCA) is also incomplete in T. tengcongensis and only half of the relevant clusters of Orthologous groups (COG), 8 out of 16, are present. The absence of the TCA-cycle enzymatic components have only been seen in other anaerobic bacteria, such as Pyrococcus horikoshii (Kawarabayasi et al. 1998), Methanococcus jannaschii (Bult et al. 1996), and A. fulgidus (Klenk et al. 1997). These three bacteria have only 3, 9, and 7 of the COGs, respectively.
T. tengcongensis has a complete collection of genes involved in most of the amino acid biosynthetic pathways for threonine, valine, leucine, histidine, phenylalanine/tyrosine, tryptophan, arginine, and methionine. However, it lacks a few key genes such as threonine dehydratase for isoleucine biosynthesis and ornithine cyclodeaminase for proline biosynthesis. For nucleotide metabolism, it also has a complete set of genes for purine biosynthesis, purine salvage, and pyrimidine biosynthesis pathways, but an enzyme, ribonucleotide reductase β-subunit for either pyrimidine salvage or thymidylate biosynthesis, appears absent. Similarly, the genes involving in coenzyme metabolism, such as ubiquinone and thiamine biosynthesis, are also incomplete. It is in fact quite common in other sequenced bacteria genomes that one or more genes in certain metabolic pathways are unidentifiable as gene identification and classification are based solely on sequence homology.
Transporters
Coping with a heated aquatic environment, T. tengcongensis evolves to have a complex ion transport system and a large number of functionally defined transporter genes, crucial for acquiring essential substrates. It encodes ion transporters, not only for monovalent cations, such as K+/Na+, but also for divalent cations, such as Mn2+, Zn2+, and Ca2+. It also encodes transporters for both Fe2+ and Fe3+, as well as for other heavy-metal cations, such as cobalt and nickel, often serving as components of coenzymes. In addition, four undefined cation-transporting ATPases and three anion ion transporter genes for formate/nitrite, phosphate, and nitrate/sulfonate/taurine/bicarbonate are identified. Most of these genes are clustered in the genome, and the majority is composed of ABC-type transporters that require ATP as energy source, such as seven nickel-chelating ABC-type transporters that are involved in the uptake of di- or oligopeptide. Furthermore, 15 genes encoding permeases, members of the major facilitator superfamily, are found scattered over the genome. Finally, as the growth of T. tengcongensis takes place on many carbohydrate substrates (Xue et al. 2001), the operons for related substrate transport, including maltose, lactose, galactose, and spermidine/putrescine, are all readily identifiable.
Cell Structure
Genes contributing to the cellular structure of T. tengcongensis are quite complex, especially those related to flagellar formation and gram staining. Despite the fact that flagella were not found in the cultured cells (Xue et al. 2001), T. tengcongensis does appear to be well equipped with all essential genes for flagellar biogenesis and with nearly all the genes for the chemotaxis signaling pathways. However, it remains puzzling why T. tengcongensis does not assemble functional flagellar under the culture conditions.
Bacteria sense a wide range of environmental cues, including nutrients, toxins, and compounds that alter electron transport, pH, temperature, and even Earth's magnetic field (Armitage 1999). Histidine protein kinase (CheA, TTE1039 and TTE1417) plays a central role in bacterial chemotaxis signaling. Autophosphorylated CheA passes its phosphoryl group onto CheY (TTE0136, TTE0288, TTE1038, TTE1063, TTE1101, TTE1203, TTE1302, and TTE1428), and phosphoryl CheY (CheY-P) then acts on the flagellar motor/switch complex, FliG/FliM/FliN (TTE1441 and TTE1430). Consequently, the complex switches on and controls the flagellar movement. Two auxiliary proteins, CheW (TTE0700, TTE1034, TTE1136, and TTE1416) and CheZ, and two receptor modification enzymes, methylesterase (CheB, TTE1035 and TTE1418) and methyltransferase (CheR, TTE1037 and TTE1135), manipulate the fluctuation of phosphoryl groups within this central pathway (Djordjevic and Stock 1998). All genes in the chemotaxis signaling pathways except CheZ are unambiguously found in the T. tengcongensis genome. CheZ, a protein known to accelerate dephosphorylation of the responsive regulator phosphoryl CheY, has only been found in a few nonthermophilic eubacteria, such as E. coli (Blattner et al. 1997), P. aeruginosa (Stover et al. 2000), and V. cholerae (Heidelberg et al. 2000), and it neither affects the flagellar motors directly nor sequesters the CheY (Scharf et al. 1998). The presence of these “silent” components involved in flagellar structure and movement in T. tengcongensis suggests a possibility that they might be activated only under certain environmental conditions or they used to be active not long before the present day.
Another controversy is that T. tengcongensis, as a gram-negative rod by staining, shares many genes that are characteristic of gram-positive bacteria but lacks some characteristics of gram-negative bacteria. First, sporulation is generally one of the important features for certain gram-positive and rod-shaped bacteria (Kim et al. 2001; Sokolova et al. 2001). There are, surprisingly, 23 CDS, which are related to sporulation, in the T. tengcongensis genome. Even with such a remarkable number, only next to the genus Bacillus, which has an additional CDS of polysaccharide biosynthesis protein F (COG 1861) involved in spore-coat formation (Takami et al. 2000), no spore formation has been observed in T. tengcongensis culture. None of the other prokaryotes sequenced to date have more than 15 CDS implicated in sporulation. Secondly, gram-negative organisms have lipopolysaccharides (LPS), which gram-positive lacks. In the gram-negative organisms, lipopolysaccharides not only offer structural rigidity, but also affect surface permeability, charges, and hydrophobicity. Consequently, they alter the way bacteria interact with the environment. Biosynthesis of O-antigen polysaccharides takes place in multiple steps involved in synthesis of sugar precursors in the cytoplasm, formation and polymerization of the repeating units, and export to the cell surface (Xu et al. 1998). The T. tengcongensis genome, though having a few CDS related to lipopolysaccharide biosynthesis (TTE0652 and TTE0199), does not possess three of the key genes: the one related to lipopolysaccharide biosynthesis (LPS:glycosyltransferase, COG1442), and the two related to lipopolysaccharide transport (i.e., a periplasmic protein involved in polysaccharide export, COG1596) and an ATPase component of ABC-type polysaccharide/polyol phosphate transport system, COG1134. At least one of these three CDS is present in most of the gram-negative prokaryotes, such as P. aeruginosa, V. choleraeserotype, Neisseria meningitidis, X. fastidiosa, and E. coli. Thermophiles of archaea and eubacteria are not exceptional, such as A. fulgidus, Aquifex aeolicus, and T. maritima. Of the sequenced gram-positive bacteria, only the genus Bacillus contains two of the key proteins. Thirdly, none of the four CDS involved in lipid A synthesis are found in the T. tengcongensis genome, although they are well documented in most of the gram-negative prokaryotes, including a thermophilic eubacterium, A. aeolicus. Finally, CDS for porins unique to gram-negative bacteria also appear absent in T. tengcongensis.
Less complicated but relevant examples, in which a decision was made for gram staining, do exist. For instance, T. wiegelii, a thermophilic, spore-forming and rod-shaped bacterium in the same genus of T. tengcongensis, is in fact gram-negative by the gram-staining protocol (Cook et al. 1996). Members of the genus Mycobacteria, believed to be phylogenetically closer to T. tengcongensis, are also recalcitrant to gram staining under standard conditions. Similar cases are encountered when staining other sulfur/sulfate-reducing species, such as the bacteria of the genus Desulfotomaculum. Although stained as gram-negative, they have many features related to the gram-positive organisms, such as that they form endospores and can be grouped according to their 16S rDNA sequences with the genus Clostridium. Some of them are indeed thermophilic acetogens (Janssen and Schink 1995). Sporomusa sphaeroides represents another similar case (Kamlage and Blaut 1993). It is clear that sensitivity to gram staining is a delicate feature of the bacterial world and the staining results are not readily explained at molecular levels.
Features Associated with Thermophily
Only 15 CDS predicted in T. tengcongensis appear unique to thermophiles, which are found in various thermophlic genomes but not shared by all of them. Only a single copy of reverse gyrase (TTE1745) seems common to most, if not all, thermophiles. Other genes include CODH maturation factor (TTE1709), MinD superfamily P-loop ATPases (TTE1891 and TTE1892), metal-dependent hydrolase of the β-lactamase superfamily II (TTE1889), predicted methyltransferase (TTE1898), uncharacterized Fe-S center proteins (TTE0177), uncharacterized Fe-S protein PflX (TTE1779), and conserved hypothetical proteins (TTE0285, TTE1224, TTE1505, TTE2664, TTE2667, TTE2636, and TTE2662). It is unlikely that thermophiles would have unique cellular machinery to make themselves capable of living in the extreme environment; rather it could be a result of an evolutionary process leading to the changes at many levels of their biochemical makeup (i.e., proteins and RNAs) and physiology (Lindsay 1995; Jaenicke and Bohm 1998).
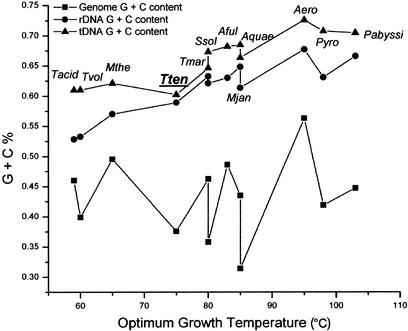
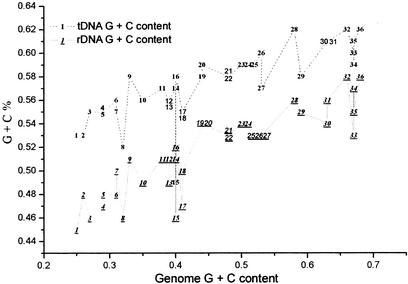
A strong correlation is observed between G + C contents of tDNA/rDNA clusters and the optimal growth temperatures (OGT) in all 12 sequenced thermophiles (Fig. 4). Similar finding has been reported recently in thermophilic archaea (Kawashima et al. 2000). No correlation has been observed between G + C contents of the overall genomic average and OGTs in these thermophiles. In hyperthermophilic archaea, the chromosomes exist as relaxed to positively supercoiled in vivo due to the action of the enzyme, reverse gyrase, and this peculiarity is believed relevant to the stabilization of DNA double-helix against heat-denaturation (Napoli et al. 2001). In mesophiles, a correlation between G + C contents of rDNA/tDNA and the genome average becomes noticeable (Fig. 5). When G + C contents of all the sequenced mesophiles are analyzed, the linear regression coefficients are R = 0.88 for rDNA and R = 0.8 for tDNA, respectively. Nevertheless, especially in the case of mesophiles, G + C content changes not only affect the stability of functional RNAs but also have potential effects on amino acid composition of proteins. However, the interpretation of the underlying mechanism is expected to be statistical and multifaceted (Jaenicke and Bohm 1998).
Figure 4.
Correlation of G + C contents and optimum growth temperatures (OGT) of thermophilic bacteria. G + C contents of genomes (solid squares), rDNAs (solid circles), and tDNAs (solid triangles) of 12 thermophilic archaea and eubacteria are plotted against the corresponding OGT. G + C contents of tDNAs and rDNAs show significant correlation with OGTs (linear regression coefficients R = 0.9 and R = 0.92, respectively), but no significant correlation is observed between genomic G + C contents and OGT (R = 0.09).
Figure 5.
Correlation of G + C contents between the genome average and rDNA/tDNA clusters from 36 mesophiles. G + C contents of tDNA and rDNA (underlined) show significant correlation with genome G + C contents (linear regression coefficients R = 0.88 and R = 0.8, respectively). Numbers in the figure stand for the sequenced prokaryotes: 1, Uure; 2, Buch; 3, Mpul; 4, Bbur; 5, Rpxx; 6, Cjej; 7, Cace; 8, Mgen; 9, SaurN; 10, Llact; 11, Hinf; 12, Spyo; 13, Hpyl; 14, Spneu; 15, Mpneu; 16, Pmul; 17, Cpneu; 18, Ctra; 19, Bsub; 20, Bhal; 21, Vcho; 22, Synecho; 23, Ecoli_O157; 24, Ecoli; 25, Nmen; 26, Xfas; 27, Tpal; 28, Mlep; 29, Atum; 30, Smel; 31, Mlot; 32, Mtub; 33, Paer; 34, Drad; 35, Ccre; and 36, Hbsp.
The addition of the T. tengcongensis genome sequence to the growing list of sequenced microbes provides a pivotal view on the genome biology of thermophilic prokaryotes. However, to understand how thermophiles adapt themselves to the ever-changing environment over evolutionary timescale is still an ongoing effort. Systematic computational analysis and experimental verification of complex cellular and molecular mechanisms are essential for understanding the conservation and diversification of bacterial genomes regarding to their many specialized lifestyles. Valuable hypotheses and insights from such endeavors will be applied to medical research and the developing biotech industry.
METHODS
Sequence Assembly and Quality Control
Genomic DNA libraries were made in pUC18 carrying insert sizes from 1.5 to 10 kb. The genomic DNA was isolated from a laboratory strain of T. tengcongensis, MB4T. To avoid cloning bias and to achieve optimal genome coverage, DNA inserts were prepared in two different ways, physical shearing (sonication) and enzyme digestion (Sau3AI). There were 75,971 successful sequence reads (>50 bp at Phred value Q20; Ewing and Green 1998; Ewing et al. 1998) generated, which gave rise to an overall genome coverage 9.87×, of which 2084 were from large insert libraries (∼10 kb) and sequenced from both ends. Phred/Phrap/Consed software package (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998) was used for quality assessment and sequence assembly. The initial assembly yielded 273 contigs. The number of gaps was effectively reduced to 46 by two basic steps. One is to resequence the low-quality reads flanking the contig ends. The other is to carry out intensive primer walking, based on the sequence information from the initial contig assembly and by using plasmid clones that extend outwards from the contigs as PCR templates. The remaining gaps were closed by a random primer-walking strategy against each contig ends. Some of the larger gaps were closed by long-range PCRs (Advantage Genomic PCR Kit, CLONTECH). In the latter cases, genomic DNA was used as template for PCR amplifications. All gap-closing clones and PCR products were sequenced from both directions to ensure high-sequence quality. The low-quality regions, often a few dozen base pairs, were improved by PCR-based methods. The overall sequence quality of the genome was further improved by insisting the following: (1) three independent, high-quality reads as minimal coverage, (2) sequence coverage accountable from both strands, and (3) Phred quality value >Q40 for each given base. Collectively, an additional 4089 finishing reactions were added to the final assembly at the finishing stage. Based on the final consensus quality scores generated by Phrap, we estimated an overall error rate of 0.86 in 10,000 bases for the final gap-free genome assembly.
Physical Map Verification
The complete sequence assembly was verified based on restriction digests of genomic DNA with a panel of three restriction enzymes. DNA fragments were resolved on 1% agarose gels in a pulse-field electrophoresis system (Bio-Rad) at 4 volts/cm in 0.5× TBE buffer for 23 h at 14°C. Lambda DNA concatemer was used as molecular-weight markers. All major fragments resolved by the electrophoresis system were unambiguously identified, including fragments for Sfi I (790, 760, 530, 279, 270, and 58 kb), Asc I (1398, 594, 498, 104, 40, 30, and 23 kb), and SgrA I (504, 447, 354, 327, 291, 158, 153, 145, 109, 56, 40, 37, 28, 17, 12, 4, 2, and 1 kb). The result was in complete agreement with the predicted physical map based on the fully assembled genome sequence to the extent that the restriction fragments were resolvable within the dynamic range of the electrophoretic system.
Sequence Annotation
The first set of potential CDS were established with GLIMMER 2.0 (Delcher et al. 1999) trained with a set of CDS larger than 500 bp from the genomic sequence and with ORPHEUS (Frishman et al. 1998) at their default settings. Both predicted CDS and putative intergenic sequences were subjected to further manual inspections. Exhaustive BLAST (Altschul et al. 1997) searches with an incremental stringency against NCBI nonredundant protein database were performed to determine homology. Translational start codons were identified based on protein homology, proximity to ribosome-binding site, relative positions to predicted signal peptide, and putative promoter sequences. Rho-independent transcription terminators were identified based on TransTerm (Ermolaeva et al. 2000) in nonprotein coding regions. A few methodological criteria were followed to resolve problematic cases. For instance, when two translation starts were identified, the first was always chosen to yield a larger predicted protein. When frameshifts and point mutations were discovered from two adjacent CDS, they were classified as inactive or pseudogene after careful inspections of the raw sequence data. When significant overlaps of two predicted CDS were encountered, those showing similarity to known genes or protein motifs/domains were preferentially taken, and the longer one was always the choice unless a biological argument favored the shorter. CDS <150 bp, which lack detectable similarity to known protein motifs/domains and distinguishable promoter/termination regions, were also excluded from the annotated CDS. The results were assembled together with manual refinements with Artemis sequence viewer (Rutherford et al. 2000). Each gene or CDS was then assigned with a unique numeric identifier prefixed with “TTE”. The first CDS from the origin of replication, the putative dnaA gene, was assigned as TTE0001, and each subsequent CDS was numbered consecutively in a clockwise direction.
To find putative orthologs in other completed genome sequences, CDS from the genomes were identified based on the COG database and classified accordingly (Tatusov et al. 2001). Protein motifs and domains of all CDS were documented based on intensive searches against publicly available databases and by using their application tools, including Pfam, PRINTS, PROSITE, ProDom, and SMART. The results were summarized with InterPro (Apweiler et al. 2001). Transfer RNAs, together with tRNA-like and mRNA-like sequences (such as 10Sa RNA or SsrA; see also www.indiana.edu/∼tmrna/), and RNase P genes were predicted with tRNAscan-SE (Lowe and Eddy 1997). The program was trained with a prokaryotic dataset and by using suggested procedures at tmRDB (Knudsen et al. 2001) and RNase P databases (Brown 1999). Signal peptides, transmembrane domains, putative membrane proteins, and ABC transporters were defined with TMHMM (Krogh et al. 2001) and SIGNALP-2.0 (Nielsen et al. 1999) after intensive trainings with a dataset of gram-negative bacteria.
Sequence data for comparative analyses were obtained from NCBI databases (ftp://ncbi.nlm.nih.gov/genbank/genomes/Bacteria). When there was more than one strain sequenced for a given species, only one was chosen arbitrarily for the comparative study. Forty-seven fully sequenced genomes were used in the analyses. Their full names and abbreviations (in parentheses) are as follows: Agrobacterium tumefaciens (Atum), Aeropyrum pernix (Aero), Aquifex aeolicus (Aquae), Archaeoglobus fulgidus (Aful), Bacillus halodurans (Bhal), Bacillus subtilis (Bsub), Borrelia burgdorferi (Bbur), Buchnera sp APS (Buch), Campylobacter jejuni (Cjej), Caulobacter crescentus (Ccre), Chlamydia trachomatis (Ctra), Chlamydophila pneumoniae CWL029 (Cpneu), Clostridium acetobutylicum (Cace), Deinococcus radiodurans (Drad), Escherichia coli K12 (Ecoli), Escherichia coli O157:H7 EDL933 (Ecoli_O157), Haemophilus influenzae (Hinf), Halobacterium sp. NRC-1 (Hbsp), Helicobacter pylori 26695 (Hpyl), Helicobacter pylori J99 (Hpyl99), Lactococcus lactis (Llact), Mesorhizobium loti (Mlot), Methanobacterium thermoautotrophicum (Mthe), Methanococcus jannaschii (Mjan), Mycobacterium leprae (Mlep), Mycobacterium tuberculosis H37Rv (Mtub), Mycoplasma genitalium (Mgen), Mycoplasma pneumoniae (Mpneu), Mycoplasma pulmonis (Mpul), Neisseria meningitidis MC58 (Nmen), Neisseria meningitidis Z2491 (NmenA), Pasteurella multocida (Pmul), Pseudomonas aeruginosa (Paer), Pyrococcus abyssi (Pabyssi), Pyrococcus horikoshii (Pyro), Rickettsia prowazekii (Rpxx), Sinorhizobium meliloti (Smel), Staphylococcus aureus N315 (SaurN), Streptococcus pneumoniae (Spneu), Streptococcus pyogenes (Spyo), Sulfolobus solfataricus (Ssol), Synechocystis PCC6803 (Synecho), Thermoplasma acidophilum (Tacid), Thermoplasma volcanium (Tvol), Thermotoga maritima (Tmar), Treponema pallidum (Tpal), Ureaplasma urealyticum (Uure), Vibrio cholerae (Vcho), and Xylella fastidiosa (Xfas).
To handle recursive-input sequences with efficiency, several custom-designed, perl-based scripts were also developed. The raw data were imported into an Oracle relational database. The user interface for this database was a series of web pages that allow frequent access to the databases.
WEB SITE REFERENCE
http://btn.genomics.org.cn/tten/; Beijing Genomics Institute's T. tengcongensis genome project web site.
http://www.indiana.edu/∼tmrna/; Tmrna information web site.
Acknowledgments
We thank Min Sun, Wei Tian, Jinsong Liao, Tingting Wu, Huiqiang Lou, Wenli Li, Liping Nie, Yanwei Huang, Hongnian Guo, Yong Shi, Wenzhong Wei, Zheng Sun, Xianhua Cao, and Junyong Jia for their contribution to DNA sequencing and early stages of library construction. We also thank other faculty and staff for their help in many aspects during the course of this project at Beijing Genomics Institute/Genomics and Bioinformatics Center, Institute of Genetics and Developmental Biology, Institute of Microbiology and Institute of Biophysics, Chinese Academy of Sciences. We are grateful to the two reviewers for their critical reading of the manuscript and many instructive comments. We thank Dr. Shouguang Jin for expert comments on the manuscript. This work is supported by a special grant from the Chinese Academy of Sciences.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hmyang@genetics.ac.cn; FAX 86-10-6488 9329.
E-MAIL tanhr@sun.im.ac.cn; FAX 86-10-6265 4083.
E-MAIL crs@sun5.ibp.ac.cn; FAX 86-10-6487 1293.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.219302.
REFERENCES
- Alexander K, Volini M. Properties of an Escherichia coli rhodanese. J Biol Chem. 1987;262:6595–6604. [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 2001;29:37–40. doi: 10.1093/nar/29.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage JP. Bacterial tactic responses. Adv Microb Physiol. 1999;41:229–289. doi: 10.1016/s0065-2911(08)60168-x. [DOI] [PubMed] [Google Scholar]
- Berg BL, Stewart V. Structural genes for nitrate-inducible formate dehydrogenase in Escherichia coli K-12. Genetics. 1990;125:691–702. doi: 10.1093/genetics/125.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bock AK, Glasemacher J, Schmidt R, Schonheit P. Purification and characterization of two extremely thermostable enzymes, phosphate acetyltransferase and acetate kinase, from the hyperthermophilic eubacterium Thermotoga maritima. J Bacteriol. 1999;181:1861–1867. doi: 10.1128/jb.181.6.1861-1867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner CA, Randall SK, Rayssiguier C, Radman M, Eritja R, Kaplan BE, McEntee K, Goodman MF. Purification and characterization of an inducible Escherichia coli DNA polymerase capable of insertion and bypass at abasic lesions in DNA. J Biol Chem. 1988;263:18946–18952. [PubMed] [Google Scholar]
- Brown JW. The ribonuclease P database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Cayol JL, Ollivier B, Patel BK, Ravot G, Magot M, Ageron E, Grimont PA, Garcia JL. Description of Thermoanaerobacter brockii subsp. lactiethylicus subsp. nov., isolated from a deep subsurface French oil well, a proposal to reclassify Thermoanaerobacter finnii as Thermoanaerobacter brockii subsp. finnii comb. nov., and an emended description of Thermoanaerobacter brockii. Int J Syst Bacteriol. 1995;45:783–789. doi: 10.1099/00207713-45-4-783. [DOI] [PubMed] [Google Scholar]
- Cook GM, Rainey FA, Patel BK, Morgan HW. Characterization of a new obligately anaerobic thermophile, Thermoanaerobacter wiegelii sp. nov. Int J Syst Bacteriol. 1996;46:123–127. doi: 10.1099/00207713-46-1-123. [DOI] [PubMed] [Google Scholar]
- Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, et al. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dervyn E, Suski C, Daniel R, Bruand C, Chapuis J, Errington J, Janniere L, Ehrlich SD. Two essential DNA polymerases at the bacterial replication fork. Science. 2001;294:1716–1719. doi: 10.1126/science.1066351. [DOI] [PubMed] [Google Scholar]
- Djordjevic S, Stock AM. Structural analysis of bacterial chemotaxis proteins: Components of a dynamic signaling system. J Struct Biol. 1998;124:189–200. doi: 10.1006/jsbi.1998.4034. [DOI] [PubMed] [Google Scholar]
- Ermolaeva MD, Khalak HG, White O, Smith HO, Salzberg SL. Prediction of transcription terminators in bacterial genomes. J Mol Biol. 2000;301:27–33. doi: 10.1006/jmbi.2000.3836. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- Frishman D, Mironov A, Mewes HW, Gelfand M. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 1998;26:2941–2947. doi: 10.1093/nar/26.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Abajian C, Green P. Consed: A graphical tool for sequence finishing. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- Grigoriev A. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 1998;26:2286–2290. doi: 10.1093/nar/26.10.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TA. Metabolism of sulfate-reducing prokaryotes. Antonie Van Leeuwenhoek. 1994;66:165–185. doi: 10.1007/BF00871638. [DOI] [PubMed] [Google Scholar]
- Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Amemura M, Nashimoto H, Shinagawa H, Makino K. The rpoE gene of Escherichia coli, which encodes sigma E, is essential for bacterial growth at high temperature. J Bacteriol. 1995;177:2918–2922. doi: 10.1128/jb.177.10.2918-2922.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes NJ, Chalk PA, Clayton CL, Kelly DJ. Identification of carboxylation enzymes and characterization of a novel four-subunit pyruvate:flavodoxin oxidoreductase from Helicobacter pylori. J Bacteriol. 1995;177:3953–3959. doi: 10.1128/jb.177.14.3953-3959.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenicke R, Bohm G. The stability of proteins in extreme environments. Curr Opin Struct Biol. 1998;8:738–748. doi: 10.1016/s0959-440x(98)80094-8. [DOI] [PubMed] [Google Scholar]
- Janssen PH, Morgan HW. Heterotrophic sulfur reduction by Thermotoga sp. strain FjSS3.B1. FEMS Microbiol Lett. 1992;75:213–217. doi: 10.1016/0378-1097(92)90406-e. [DOI] [PubMed] [Google Scholar]
- Janssen PH, Schink B. Metabolic pathways and energetics of the acetone-oxidizing, sulfate- reducing bacterium, Desulfobacterium cetonicum. Arch Microbiol. 1995;163:188–194. doi: 10.1007/BF00305352. [DOI] [PubMed] [Google Scholar]
- Kamlage B, Blaut M. Isolation of a cytochrome-deficient mutant strain of Sporomusa sphaeroides not capable of oxidizing methyl groups. J Bacteriol. 1993;175:3043–3050. doi: 10.1128/jb.175.10.3043-3050.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin S. Bacterial DNA strand compositional asymmetry. Trends Microbiol. 1999;7:305–308. doi: 10.1016/s0966-842x(99)01541-3. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y, Sawada M, Horikawa H, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, et al. Complete sequence and gene organization of the genome of a hyper- thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi Y, Hino Y, Horikawa H, Yamazaki S, Haikawa Y, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, et al. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 1999;6:83–101. doi: 10.1093/dnares/6.2.83. , 145–152. [DOI] [PubMed] [Google Scholar]
- Kawashima T, Amano N, Koike H, Makino S, Higuchi S, Kawashima-Ohya Y, Watanabe K, Yamazaki M, Kanehori K, Kawamoto T, et al. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium. Proc Natl Acad Sci. 2000;97:14257–14262. doi: 10.1073/pnas.97.26.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BC, Grote R, Lee DW, Antranikian G, Pyun YR. Thermoanaerobacter yonseiensis sp. nov., a novel extremely thermophilic, xylose-utilizing bacterium that grows at up to 85 degrees C. Int J Syst Evol Microbiol. 2001;51:1539–1548. doi: 10.1099/00207713-51-4-1539. [DOI] [PubMed] [Google Scholar]
- Kim JH, Akagi JM. Characterization of a trithionate reductase system from Desulfovibrio vulgaris. J Bacteriol. 1985;163:472–475. doi: 10.1128/jb.163.2.472-475.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate- reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Knudsen B, Wower J, Zwieb C, Gorodkin J. tmRDB (tmRNA database) Nucleic Acids Res. 2001;29:171–172. doi: 10.1093/nar/29.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kral TA, Brink KM, Miller SL, McKay CP. Hydrogen consumption by methanogens on the early Earth. Orig Life Evol Biosph. 1998;28:311–319. doi: 10.1023/a:1006552412928. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuhner CH, Frank C, Griesshammer A, Schmittroth M, Acker G, Gossner A, Drake HL. Sporomusa silvacetica sp, nov., an acetogenic bacterium isolated from aggregated forest soil. Int J Syst Bacteriol. 1997;47:352–358. doi: 10.1099/00207713-47-2-352. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Choudhuri JV, Ohlebusch E, Schleiermacher C, Stoye J, Giegerich R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labes A, Schonheit P. Sugar utilization in the hyperthermophilic, sulfate-reducing archaeon Archaeoglobus fulgidus strain 7324: Starch degradation to acetate and CO2 via a modified Embden-Meyerhof pathway and acetyl-CoA synthetase (ADP-forming) Arch Microbiol. 2001;176:329–338. doi: 10.1007/s002030100330. [DOI] [PubMed] [Google Scholar]
- Lindsay JA. Is thermophily a transferrable property in bacteria? Crit Rev Microbiol. 1995;21:165–174. doi: 10.3109/10408419509113539. [DOI] [PubMed] [Google Scholar]
- Lobry JR. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:660–665. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- Lonetto M, Gribskov M, Gross CA. The σ 70 family: Sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Ragsdale SW. Mechanism of the Clostridium thermoaceticum pyruvate:ferredoxin oxidoreductase: Evidence for the common catalytic intermediacy of the hydroxyethylthiamine pyropyrosphate radical. Biochemistry. 1997;36:8484–8494. doi: 10.1021/bi970403k. [DOI] [PubMed] [Google Scholar]
- Metzger S, Sarubbi E, Glaser G, Cashel M. Protein sequences encoded by the relA and the spoT genes of Escherichia coli are interrelated. J Biol Chem. 1989;264:9122–9125. [PubMed] [Google Scholar]
- Myler PJ, Audleman L, deVos T, Hixson G, Kiser P, Lemley C, Magness C, Rickel E, Sisk E, Sunkin S, et al. Leishmania major Friedlin chromosome 1 has an unusual distribution of protein-coding genes. Proc Natl Acad Sci. 1999;96:2902–2906. doi: 10.1073/pnas.96.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli A, Kvaratskelia M, White MF, Rossi M, Ciaramella M. A novel member of the bacterial-archaeal regulator family is a nonspecific dna-binding protein and induces positive supercoiling. J Biol Chem. 2001;276:10745–10752. doi: 10.1074/jbc.M010611200. [DOI] [PubMed] [Google Scholar]
- Napolitano R, Janel-Bintz R, Wagner J, Fuchs RP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Clayton RA, Gill SR, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Nelson WC, Ketchum KA, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Brunak S, von Heijne G. Machine learning approaches for the prediction of signal peptides and other protein sorting signals. Protein Eng. 1999;12:3–9. doi: 10.1093/protein/12.1.3. [DOI] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Petersohn A, Brigulla M, Haas S, Hoheisel JD, Volker U, Hecker M. Global analysis of the general stress response of Bacillus subtilis. J Bacteriol. 2001;183:5617–5631. doi: 10.1128/JB.183.19.5617-5631.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qatibi AI, Bennisse R, Jana M, Garcia JL. Anaerobic degradation of glycerol by desulfovibrio fructosovorans and D. carbinolicus and evidence for glycerol-dependent utilization of 1,2– propanediol. Curr Microbiol. 1998;36:283–290. doi: 10.1007/s002849900311. [DOI] [PubMed] [Google Scholar]
- Ragsdale SW. Enzymology of the acetyl-CoA pathway of CO2 fixation. Crit Rev Biochem Mol Biol. 1991;26:261–300. doi: 10.3109/10409239109114070. [DOI] [PubMed] [Google Scholar]
- Rocha EP, Danchin A, Viari A. Analysis of long repeats in bacterial genomes reveals alternative evolutionary mechanisms in Bacillus subtilis and other competent prokaryotes. Mol Biol Evol. 1999;16:1219–1230. doi: 10.1093/oxfordjournals.molbev.a026212. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Salzberg SL, Salzberg AJ, Kerlavage AR, Tomb JF. Skewed oligomers and origins of replication. Gene. 1998;217:57–67. doi: 10.1016/s0378-1119(98)00374-6. [DOI] [PubMed] [Google Scholar]
- Sarubbi E, Rudd KE, Cashel M. Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol Gen Genet. 1988;213:214–222. doi: 10.1007/BF00339584. [DOI] [PubMed] [Google Scholar]
- Scharf BE, Fahrner KA, Berg HC. CheZ has no effect on flagellar motors activated by CheY13DK106YW. J Bacteriol. 1998;180:5123–5128. doi: 10.1128/jb.180.19.5123-5128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Zuber P, Willimsky G, Wagner B, Marahiel MA. Mapping of the Bacillus subtilis cspB gene and cloning of its homologs in thermophilic, mesophilic and psychrotrophic bacilli. Gene. 1993;136:277–280. doi: 10.1016/0378-1119(93)90479-m. [DOI] [PubMed] [Google Scholar]
- Schurr MJ, Yu H, Boucher JC, Hibler NS, Deretic V. Multiple promoters and induction by heat shock of the gene encoding the alternative σ factor AlgU (σ E) which controls mucoidy in cystic fibrosis isolates of Pseudomonas aeruginosa. J Bacteriol. 1995;177:5670–5679. doi: 10.1128/jb.177.19.5670-5679.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc Natl Acad Sci. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson AJ, Reinach FC, Arruda P, Abreu FA, Acencio M, Alvarenga R, Alves LM, Araya JE, Baia GS, Baptista CS, et al. The genome sequence of the plant pathogen Xylella fastidiosa. The Xylella fastidiosa Consortium of the Organization for Nucleotide Sequencing and Analysis. Nature. 2000;406:151–157. doi: 10.1038/35018003. [DOI] [PubMed] [Google Scholar]
- Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova TG, Gonzalez JM, Kostrikina NA, Chernyh NA, Tourova TP, Kato C, Bonch-Osmolovskaya EA, Robb FT. Carboxydobrachium pacificum gen. nov., sp. nov., a new anaerobic, thermophilic, CO-utilizing marine bacterium from Okinawa Trough. Int J Syst Evol Microbiol. 2001;51:141–149. doi: 10.1099/00207713-51-1-141. [DOI] [PubMed] [Google Scholar]
- Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, Brinkman FS, Hufnagle WO, Kowalik DJ, Lagrou M, et al. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Takami H, Nakasone K, Takaki Y, Maeno G, Sasaki R, Masui N, Fuji F, Hirama C, Nakamura Y, Ogasawara N, et al. Complete genome sequence of the alkaliphilic bacterium Bacillus halodurans and genomic sequence comparison with Bacillus subtilis. Nucleic Acids Res. 2000;28:4317–4331. doi: 10.1093/nar/28.21.4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- Vargas M, Kashefi K, Blunt-Harris EL, Lovley DR. Microbiological evidence for Fe(III) reduction on early Earth. Nature. 1998;395:65–67. doi: 10.1038/25720. [DOI] [PubMed] [Google Scholar]
- White O, Eisen JA, Heidelberg JF, Hickey EK, Peterson JD, Dodson RJ, Haft DH, Gwinn ML, Nelson WC, Richardson DL, et al. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Murray BE, Weinstock GM. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. Infect Immun. 1998;66:4313–4323. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Xu Y, Liu Y, Ma Y, Zhou P. Thermoanaerobacter tengcongensis sp. nov., a novel anaerobic, saccharolytic, thermophilic bacterium isolated from a hot spring in Tengcong, China. Int J Syst Evol Microbiol. 2001;51:1335–1341. doi: 10.1099/00207713-51-4-1335. [DOI] [PubMed] [Google Scholar]