Abstract
We developed a multiplex real-time PCR assay using 6-carboxyfluorescein, 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein, and carbocyanine 5-labeled probes to simultaneously quantify Epstein-Barr virus (EBV), cytomegalovirus (CMV), and human herpesvirus 6 (HHV-6) DNA. When previously tested and stored DNA samples were examined, results of the multiplex real-time PCR assay were as sensitive and specific as those of a single real-time PCR assay. The multiplex assay was used to quantify the EBV, CMV, and HHV-6 DNA in 46 transplant recipients. A total of 303 whole-blood and plasma specimens were collected and analyzed. According to the results of the multiplex assay, the detection rates for viral DNA in whole blood and plasma were 23.8% and 5.9% for EBV, 11.2% and 5.3% for CMV, and 12.5% and 2.0% for HHV-6, respectively. All forms of viral DNA were detected more frequently in whole blood than in plasma. During the symptomatic period, EBV DNA was detected in all whole-blood specimens but not in all plasma specimens. Furthermore, the EBV DNA load in whole blood was higher during the symptomatic period than during the asymptomatic period, whereas the EBV DNA load in plasma was similar for both periods. These results demonstrate that whole blood is more suitable for the quantification of EBV DNA in transplant patients. However, a cutoff value with clinical relevance still needs to be determined.
Herpesviruses are ubiquitous in the human population and often become reactivated in latently infected immunocompromised patients (4, 22). Herpesvirus reactivation frequently occurs after hematopoietic stem cell or solid-organ transplantation and occasionally results in symptomatic diseases (2, 4, 28). Among human herpesviruses, Epstein-Barr virus (EBV), cytomegalovirus (CMV), and human herpesvirus 6 (HHV-6) may cause life-threatening complications, such as lymphoproliferative disorders (3, 13, 27), interstitial pneumonia (7, 15), and encephalitis (2, 12, 23, 25). Therefore, to ensure the success of transplantation, it is essential to monitor for these viruses and diagnose any virus-related diseases as early as possible.
With the advent of real-time PCR technology, quantitative PCR assays are becoming widespread methodologies for diagnostic purposes (9, 11). We initially developed quantitative real-time PCR systems to detect EBV (5), CMV (18), and HHV-6 DNA (19). However, because each system was able to detect only one form of viral DNA, simultaneous virus monitoring was cost, time, and labor intensive. It was recently shown that quantification of more than one target per well is possible with the use of different fluorochromes (1, 14, 21, 24). One aim of the present study was to establish a system for the simultaneous quantification of EBV, CMV, and HHV-6 DNA by using a multiplex real-time PCR assay. Another goal was to identify specimens that would optimize virus monitoring. While peripheral blood cells, whole blood, and plasma are commonly used in quantitative real-time PCR, whole blood and plasma are more convenient to use and are suitable for clinical analysis. Since whole blood contains peripheral blood cells, we considered that whole blood could be used instead of peripheral blood cells. In the present study, we developed a multiplex real-time PCR assay to quantify EBV, CMV, and HHV-6 DNA. In addition, we sought to determine whether whole blood or plasma was more suitable for simultaneous virus monitoring in samples from transplant recipients.
MATERIALS AND METHODS
Patients and clinical specimens.
In total, 27 hematopoietic stem cell transplantation recipients (16 men, 11 women) and 19 liver transplantation recipients (9 men, 10 women) were enrolled in the study. Beginning 1 week after transplantation, EDTA blood samples were prospectively obtained from subjects weekly until they were discharged. From 46 patients, 1 to 13 samples per patient (total 303, mean 6.6) were collected from 1 to 15 weeks after transplantation (mean 7.8 weeks). From some patients, only a few samples could be obtained because of their early death or follow-up loss. Informed consent was obtained from all patients or guardians. The institutional review board of Nagoya University Hospital approved the use of specimens included in this study.
Among the hematopoietic stem cell transplantation recipients, 10 had acute leukemia, 6 had severe aplastic anemia, 4 had solid tumors, 2 had chronic leukemia, and 5 had other diseases. The patients underwent transplantation between November 2003 and October 2005 at Nagoya University Hospital or at Japanese Red Cross Nagoya First Hospital. The median age of the patients was 8.0 years (range, 1 to 22 years). Eight of 27 patients received antithymocyte globulin, which is known to be a risk factor for EBV-related lymphoproliferative disorders (3, 13).
The liver transplantation recipients received their organs from living donors at Nagoya University Hospital between February 2004 and December 2005. Of 19 patients, 8 had hepatic cirrhosis (hepatitis B virus, 3; hepatitis C virus, 2; other viruses, 3), 3 had biliary atresia, and 8 had other diseases. The median age of the patients was 48.0 years (range, 6 months to 62 years).
Symptomatic EBV infections were diagnosed from clinical findings (fever, enlarged lymph nodes, and hematochezia) and serological examinations. A lymph node biopsy provided histological confirmation that one patient with symptomatic EBV infection had posttransplant lymphoproliferative disorder. CMV hepatitis was diagnosed from the clinical findings and serological examination and confirmed by liver biopsy.
A total of 303 consecutive blood specimens were obtained from transplant recipients and divided into whole-blood or plasma samples, and the 303 paired samples were tested with the multiplex real-time PCR assay. Viral DNA was extracted from 200 μl of whole blood or 200 μl of plasma, using QIAamp DNA blood kits (QIAGEN, Hilden, Germany) and eluted in 100 μl of water.
Specificity and sensitivity studies were performed with 111 DNA samples that had been obtained from other transplantation recipients. These DNA samples had been extracted from either whole-blood or plasma samples and used to monitor viruses by either qualitative PCR or real-time PCR, and stored at −30°C.
The specificity was also confirmed with viral DNA from standard strains (KOS for herpes simplex virus type 1, 186 for herpes simplex virus type 2, Kawaguchi strain for varicella-zoster virus, B95-8 for EBV, AD169 for CMV, Sato strain for HHV-7, and a clinical isolate from a patient with exanthem subitum for HHV-6). Viral DNA was extracted from the supernatant of each virus culture and was used for the cross-reactivity study.
EDTA blood was taken from a patient who was seronegative for EBV, CMV, and HHV-6, and plasma was separated. A DNA extraction solution from either the whole blood or the plasma fraction was used for reconstruction studies.
Primers and probes.
The sequences of the primers and probes used for the multiplex real-time PCR assay are listed in Table 1. The primer and probe sets for the viruses have been described previously (5, 18, 19). Each probe was labeled with different fluorochromes, as follows: the EBV probe was labeled with 6-carboxyfluorescein and quenched with Black-Hole-Quencher 1a (BHQ1a); the CMV probe was labeled with 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein and quenched with BHQ1a; the HHV-6 probe was labeled with carbocyanine 5 and quenched with BHQ3a. All primers (Fasmac, Kanagawa, Japan) and probes (Operon Biotechnologies, Huntsville, AL) were synthesized commercially.
TABLE 1.
Sequences of primers and probes used in the real-time PCR assay
| Target gene (GenBank accession no.) | Primer or probe | Sequence (5′→3′)a | Genome coordinate |
|---|---|---|---|
| EBV BALF5 (AJ507799) | Forward primer | CGGAAGCCCTCTGGACTTC | 156007-156025 |
| Reverse primer | CCCTGTTTATCCGATGGAATG | 155936-55956 | |
| Probe | FAM-TGTACACGCACGAGAAATGCGCC-BHQ1a | 155959-155981 | |
| CMV IE (NC001347) | Forward primer | GACTAGTGTGATGCTGGCCAAG | 172435-172456 |
| Reverse primer | GCTACAATAGCCTCTTCCTCATCTG | 172256-172270 | |
| Probe | JOE-AGCCTGAGGTTATCAGTGTAATGAAGCGCC-BHQ1a | 172389-172418 | |
| HHV-6 U31 (AF157706) | Forward primer | TTTGCAGTCATCACGATCGG | 46661-46680 |
| Reverse primer | AGAGCGACAAATTGGAGGTTTC | 46862-46883 | |
| Probe | Cy5-AGCCACAGCAGCCATCTACATCTGTCAA-BHQ3a | 46753-46780 |
FAM, 6-carboxyfluorescein; JOE, 6-carboxy-4′,5′-dichloro-2′,7′-dimethoxyfluorescein; Cy5, carbocyanine 5.
Quantification of viral DNA by multiplex real-time PCR.
Multiplex and independent real-time PCR were performed with a QuantiTect multiplex PCR kit (QIAGEN). The multiplex real-time PCR assay was performed in a total reaction mixture (25 μl) containing 5 μl of DNA extracts, 12.5 μl of 2× QuantiTect multiplex PCR master mix, each forward and reverse primer, and each probe. To determine the optimal concentrations of the primers and probes, we evaluated various concentrations of primer and probe sets using the multiplex real-time PCR assay. The optimal concentration of forward primer was 200 nM for EBV and 100 nM for CMV and HHV-6. The optimal concentration of reverse primer was 200 nM for EBV, 100 nM for CMV, and 200 nM for HHV-6. The optimal concentration of each probe was 100 nM. The independent real-time PCR was performed in the same way as the multiplex real-time PCR assay except that only one primer/probe set was included. Amplification and real-time fluorescence detection were performed with a model Mx3000P real-time PCR system (Stratagene, La Jolla, CA) using the following protocol: an initial denaturation and polymerase activation step for 15 min at 95°C, followed by 50 cycles of denaturation at 95°C for 15 s and 60°C for 1 min. Real-time fluorescence measurements were taken, and a threshold cycle value for each sample was calculated by determining the point at which the fluorescence exceeded a threshold limit. Each real-time PCR assay contained the dilution series of a standard for the calibration curve, and all samples and the standard were run in duplicate. The standards were plasmid controls that contained the PCR products amplified by each primer set, as described previously (5, 18, 19). For the multiplex real-time PCR, each plasmid control was mixed, diluted, and used to make calibration curves. The number of viral DNA copies was calculated from the standard curves and expressed as copies per 1 ml of whole blood or plasma.
CMV antigenemia assay.
The CMV antigenemia assay was performed as previously described (18). The approximate antigenemia threshold was 1 positive cell per 5 × 104 leukocytes, as determined by guidelines from the Japanese Society for Hematopoietic Cell Transplantation. When the antigenemia assay was positive, patients were administered ganciclovir preemptively.
Statistical analysis.
StatView J 4.02 (Abacus Concepts Inc., Berkeley, CA) was used to perform the data analysis. A regression analysis compared the multiplex assay with the single assay. Fisher's exact test was used to compare the viral DNA detection rates, and Student's t test was used to compare the mean viral DNA log10 copy numbers. To determine the minimum detection level of each assay, 35 replicates each of one, two, and five copies of the plasmid standard were quantified. The 95% confidence interval (CI) was calculated from the t distribution using the following formula: 95% CI = the mean of the estimated copy number ± t × standard error, where t was estimated to be 2.042 from the Student's t table.
RESULTS
Specificity and sensitivity of the multiplex quantification using an Mx3000P real-time PCR system.
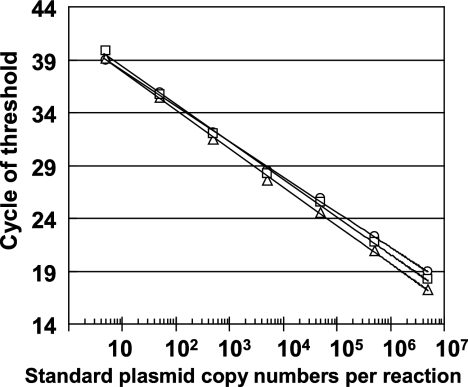
Serial dilutions of mixed viral standard plasmids were tested with the multiplex assay, and three standard curves were constructed from the cycle of threshold values. The assay was able to detect each viral DNA form over a linear span between 5 and 5 × 106 copies per reaction mixture without interference (Fig. 1). The standard curves generated from the multiplex real-time PCR were almost equal to those generated from the single assay. Because amplification efficiency may be influenced by the background DNA of the clinical specimen, we evaluated the performance of the multiplex assay using standard plasmids diluted in water or in water containing EBV-, CMV-, and HHV-6-negative human genomic DNA. The standard curves generated from the standard plasmids in background DNA were almost equal to those generated from the standard plasmids in water, indicating that amplification efficiency was not influenced by the background DNA (data not shown). We also performed reconstruction studies to confirm the absence of the inhibitor from whole-blood or plasma samples. DNA-extraction solution from either whole blood or plasma was added to serially diluted plasmid controls. The DNA solutions from both whole blood and plasma did not inhibit the amplification efficiency, indicating the absence of inhibitors in these samples. To confirm the specificity of the multiplex assay, viral DNA from standard strains was tested. None of the primer/probe sets reacted with the other viral DNA, indicating that no cross-reactivity occurred. Furthermore, the quantitative linearity, which was made using a standard plasmid, was not influenced by the presence of two other kinds of viral DNA.
FIG. 1.
Standard curves generated by multiplex real-time PCR. Serial dilutions of each viral standard ranging from 5 to 5 × 106 copies per reaction were used to generate the standard curves. The cycle of threshold values that corresponded to the PCR cycle number was plotted against the copy number of each viral standard. Circles, EBV DNA standard plasmid; triangles, CMV DNA standard plasmid; squares, HHV-6 standard plasmid.
The minimum detection level established with this multiplex assay was 2 copies per reaction for EBV (95% CI, 1.16 to 3.89), 2 copies for CMV (95% CI, 1.01 to 2.95), and 2 copies for HHV-6 (95% CI, 0.49 to 2.64). By contrast, the minimum detection level with the single assay was 2 copies for EBV (95% CI, 1.39 to 3.96), 2 copies for CMV (95% CI, 1.01 to 2.71), and 5 copies for HHV-6 (95% CI, 4.35 to 7.58). The multiplex assay had an overall dynamic range of 200 to 5 × 108 copies/ml of specimen.
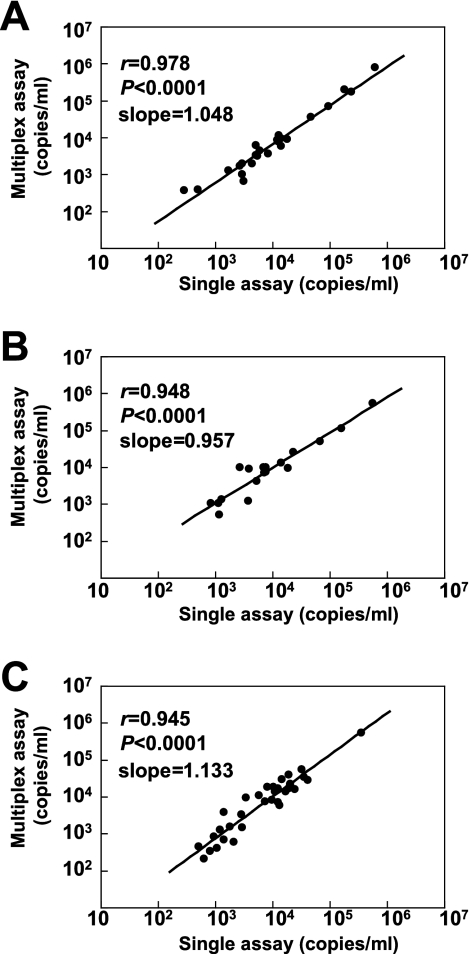
DNA samples (111), which had been tested and stored previously, were evaluated with the multiplex and the single assays. Compared to the results of the single assay, the sensitivity and specificity values of the multiplex assay were 96.0 and 100% for EBV, 94.7 and 95.7% for CMV, and 89.2 and 94.6% for HHV-6, respectively (Table 2). Some discordant results, however, were obtained between the multiplex and single assays. The viral loads of all of these discordant samples were low and around the detection limits. The viral DNA copy numbers were compared using all samples determined to be positive according to both assays. Strong correlations were detected between the viral DNA copy numbers determined by the multiplex assays and those by single assays (Fig. 2). The slopes of the correlation curves ranged from 0.957 to 1.133, indicating that no significant shifts occurred in the actual quantitative values, using the multiplex assay.
TABLE 2.
Comparison of multiplex PCR assay and single PCR assay of previously tested DNA samples
| Multiplex PCR assay | Single PCR assay (copies/ml)
|
Sensitivity (%) | Specificity (%) | |
|---|---|---|---|---|
| Positive | Negative | |||
| EBV | ||||
| No. positive | 24 | 0 | 96.0 | 100 |
| No. negative | 1 | 86 | ||
| CMV | ||||
| No. positive | 18 | 4 | 94.7 | 95.7 |
| No. negative | 1 | 88 | ||
| HHV-6 | ||||
| No. positive | 33 | 4 | 89.2 | 94.6 |
| No. negative | 4 | 70 | ||
FIG. 2.
Correlation of each viral DNA load determined by the multiplex and single real-time PCR assays. (A) Correlation of EBV DNA copy number (n = 24). (B) Correlation of CMV DNA copy number (n = 18). (C) Correlation of HHV-6 DNA copy number (n = 33).
Detection of EBV, CMV, and HHV-6 DNA in whole-blood and plasma specimens from transplant recipients.
Using the multiplex real-time PCR assay, we serially measured the EBV, CMV, and HHV-6 DNA levels in whole-blood and plasma specimens from 46 transplant recipients. In total, 303 paired samples (6.6 paired samples per patient) were tested. Positives were defined as any positive samples for a given patient. Because we had different numbers of samples from each patient, it was possible that the variety of sample numbers per patient would introduce some bias in the results. Using the whole-blood specimens, at least one form of viral DNA was detected in 36 of 46 recipients (78.3%), plural viral DNA forms were detected in 13 recipients (28.3%), and all three viral DNA forms were detected in 3 recipients (6.5%). Using the plasma specimens, at least one viral DNA form was detected in 18 of 46 recipients (39.1%), and plural viral DNA forms were detected in 5 recipients (10.9%).
Four patients who underwent hematopoietic stem cell transplantation developed symptomatic EBV infections and were preconditioned with antithymocyte globulin. The patients had prolonged fever, lymphadenopathy, diarrhea, or hematochezia, which could not be explained by other causes. One patient who underwent liver transplantation developed CMV hepatitis despite preemptive ganciclovir therapy. None of the patients developed HHV-6-related diseases. Each form of viral DNA was detected in both the whole-blood and plasma specimens of these five symptomatic patients, and the peaks of the viral DNA loads were concordant with the symptoms observed.
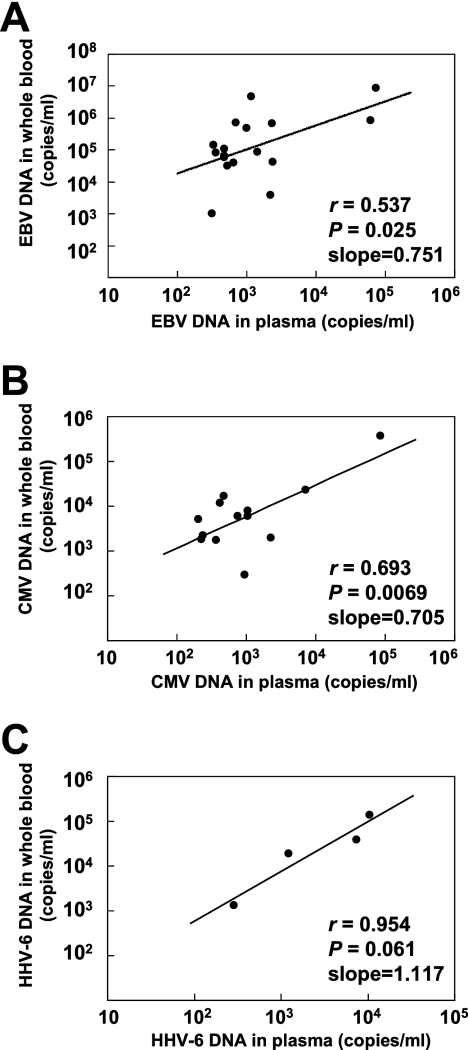
Of the 303 whole-blood samples tested, 112 were positive for at least one viral DNA form, and 29 were positive for plural viral DNA forms. Conversely, 37 of 303 plasma samples were positive for at least one viral DNA form, and only 3 samples were positive for plural viral DNA forms (Table 3). The detection rates of viral DNA in whole blood and plasma were 23.8% and 5.9% for EBV, 11.2% and 5.3% for CMV, and 12.5% and 2.0% for HHV-6, respectively. The three viral DNA forms of interest were detected more frequently in whole blood than in plasma. EBV DNA loads ranged from 220 to 8.6 × 106 copies/ml in whole blood and from 250 to 7.5 × 104 copies/ml in plasma. CMV DNA loads ranged from 220 to 3.8 × 105 copies/ml in whole blood and from 200 to 8.5 × 104 copies/ml in plasma. HHV-6 DNA loads ranged from 240 to 1.3 × 105 copies/ml in whole blood and from 290 to 1.1 × 104 copies/ml in plasma. The viral DNA copy numbers were compared using samples determined to be positive in both whole blood and plasma (Fig. 3). Weak correlations were seen between the viral loads in whole blood and those in plasma.
TABLE 3.
Detection of EBV, CMV, and HHV-6 DNA in whole blood and plasma using multiplex real-time PCR assay
| Detected viral DNA | No. of samples detected from:
|
P valuea | |||
|---|---|---|---|---|---|
| Whole blood (n = 303)
|
Plasma (n = 303)
|
||||
| n | % | n | % | ||
| EBV or CMV or HHV-6 | 112 | 37.0 | 37 | 12.2 | <0.0001 |
| EBV | 72 | 23.8 | 18 | 5.9 | <0.0001 |
| CMV | 34 | 11.2 | 16 | 5.3 | 0.0114 |
| HHV-6 | 38 | 12.5 | 6 | 2.0 | <0.0001 |
| EBV only | 48 | 15.8 | 15 | 5.0 | |
| CMV only | 16 | 5.3 | 13 | 4.3 | |
| HHV-6 only | 19 | 6.3 | 6 | 2.0 | |
| EBV and CMV | 10 | 3.3 | 3 | 1.0 | |
| CMV and HHV-6 | 5 | 1.7 | 0 | 0 | |
| EBV and HHV-6 | 11 | 3.6 | 0 | 0 | |
| EBV, CMV, and HHV-6 | 3 | 1.0 | 0 | 0 | |
| Below detection limits in all viruses | 191 | 63.0 | 266 | 87.8 | |
Fisher's exact test.
FIG. 3.
Correlation of each viral DNA load between whole blood and plasma. (A) Correlation of EBV DNA copy number (n = 17). (B) Correlation of CMV DNA copy number (n = 13). (C) Correlation of HHV-6 DNA copy number (n = 4). The viral DNA copy numbers were compared using samples determined to be positive in both whole blood and plasma.
Comparison of EBV DNA loads between whole blood and plasma.
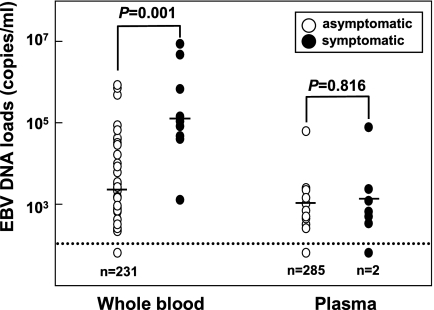
We divided the 303 samples into two groups based on EBV infection-related symptoms, symptomatic and asymptomatic. In the symptomatic group, EBV DNA was detected in all nine whole-blood samples with high viral loads but not in two plasma samples (Fig. 4). Among the whole-blood samples, the viral loads of the symptomatic group were significantly higher than those of the asymptomatic group. However, among the plasma samples, the viral loads were similar between the two groups (Fig. 4).
FIG. 4.
Comparison of EBV DNA loads between whole-blood and plasma specimens. The dotted line indicates the detection limit in this assay. Bars show the mean copy numbers of EBV DNA. n, number of undetectable samples.
Comparison of antigenemia and multiplex real-time PCR assays.
The antigenemia assay was applied to 235 of 303 blood specimens that had been evaluated with the multiplex assay. CMV antigen was detected in 13 of 235 blood samples (5.5%). With the multiplex assay, CMV DNA was detected in 32 of 235 whole-blood specimens (13.6%; P = 0.002 versus antigenemia) and 14 of 235 plasma specimens (6.0%; P = 0.5 versus antigenemia). When the antigenemia assay was defined as the standard, the specificity of CMV DNA detection was higher for the plasma specimens than for the whole-blood specimens (Table 4). On the other hand, the sensitivity seemed to be higher for the whole-blood specimens, although statistical significance was not achieved, probably due to the small sample size.
TABLE 4.
Comparison of multiplex real-time PCR assay and antigenemia assay of 235 blood specimens
| Multiplex real-time PCR | Antigenemia assay
|
% of sensitivity (95% CI) | % of specificity (95% CI) | |
|---|---|---|---|---|
| No. of positive samples | No. of negative samples | |||
| Whole-blood specimens | ||||
| No. positive | 12 | 20 | 92.3%a | 91.0%b |
| No. negative | 1 | 202 | (77.8-100) | (97.2-94.8) |
| Plasma specimens | ||||
| No. positive | 9 | 5 | 69.2%a | 97.7%b |
| No. negative | 4 | 217 | (44.1-94.3) | (95.8-99.7) |
P = 0.32 by Fisher's exact test.
P = 0.003 by Fisher's exact test.
The 235 specimens were classified into four categories by the antigenemia values (negative, 0; low, 1 to 10; intermediate, 11 to 100; high, >100), and CMV DNA loads were compared (Table 5). Although the sample numbers for each group were small, it appeared that CMV DNA loads increased in proportion to increases in the antigenemia values. Correlations between antigenemia values and CMV DNA loads were then analyzed using the positive samples. Antigenemia values were significantly correlated with CMV DNA loads, both in whole blood (n = 12; r = 0.776; P = 0.002) and in plasma (n = 9; r = 0.861; P = 0.002).
TABLE 5.
CMV DNA loads determined by multiplex real-time PCR assay in comparison with the antigenemia assay
| Categories of antigenemia values (positive cells/5 × 104 cells) | Median antigenemia values (range) | No. of tested specimens | CMV DNA loads (copies/ml)
|
|||
|---|---|---|---|---|---|---|
| Whole blood
|
Plasma
|
|||||
| No. of positive samples | Median (range) | No. of positive samples | Median (range) | |||
| 0 | 222 | 20 | 1,420 (250-11,810) | 5 | 280 (230-950) | |
| 1-10 | 2.0 (1-4) | 10 | 9 | 5,040 (430-17,000) | 6 | 620 (200-1,060) |
| 11-100 | 17.5 (16-19) | 2 | 2 | 12,450 (1,980-22,920) | 2 | 4,740 (2,250-7,230) |
| >100 | 103 | 1 | 1 | 375,500 | 1 | 85,480 |
Monitoring of EBV, CMV, and HHV-6 DNA loads in a transplant recipient.
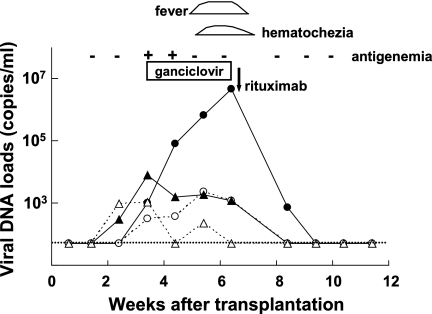
Figure 5 shows changes in the viral DNA loads in a representative case. The patient was a 4-year-old girl with severe aplastic anemia who underwent bone marrow transplantation. Two weeks after transplantation, CMV DNA was detected in her whole-blood and plasma samples (Fig. 5). Shortly after the detection of CMV DNA, the patient's blood tested positive for the CMV antigen. Although the patient had no symptoms associated with CMV infection, ganciclovir was administered preemptively. Her CMV DNA load decreased in accordance with the ganciclovir therapy. During treatment, a high level of EBV DNA was detected in her blood. In the fifth week after transplantation, she developed a fever and hematochezia. Since EBV DNA had been detected at a high level in her whole blood and plasma and other pathogens and causes that may have explained her symptoms were excluded, an EBV-related lymphoproliferative disorder was suspected. Immunosuppression was tapered, but that was not effective. Therefore, the monoclonal anti-CD20 antibody rituximab was administered. Immediately after rituximab administration, her EBV DNA load decreased in conjunction with her clinical symptoms. During the period of viral monitoring, HHV-6 DNA was undetectable in her whole blood and plasma.
FIG. 5.
Monitoring of viral DNA loads by multiplex real-time PCR in a 4-year-old girl who underwent bone marrow transplantation. Closed circles, EBV DNA copy number in whole blood; open circles, EBV DNA in plasma; closed triangles, CMV DNA in whole blood; open triangles, CMV DNA in plasma. The dotted line indicates the detection limit in this assay.
DISCUSSION
Real-time PCR is a powerful tool for quantifying gene targets using fluorogenic probes and real-time laser scanning. Although multiplex real-time PCR is theoretically possible using probes with spectrally different fluorophores, the overlap of fluorophores prevents accurate quantification and limits amplification to two genes per tube (21). Only two targets could be quantified reliably in a single tube. However, recent advances in the development of real-time PCR platforms and master mix have made it possible to quantify more than three different genes in a single tube (21).
To our knowledge, this is the first study using the multiplex real-time PCR assay in the quantification of EBV, CMV, and HHV-6 DNA. Our results showed that the multiplex assay was as sensitive and specific as the single real-time PCR assay. The viral DNA loads determined by the multiplex real-time PCR assay were consistent with those described previously using other assays (6-8, 12, 26). Monospecific PCR assays require separate amplification of each target and are therefore more cost, time, and labor intensive than multiplex assays. The multiplex real-time PCR assay offers a major advantage in the field of clinical virology as it permits simultaneous amplification of several viruses in a single reaction mixture (1, 14, 21). The multiplex real-time PCR assay is particularly useful in the management of posttransplant patients, in whom frequent viral monitoring is required. The multiplex assay facilitates cost-effective diagnosis and may contribute to a decrease in the use of antiviral agents and in viral complications and hospitalizations.
One aim of the present study was to determine whether whole blood or plasma was more suitable for simultaneous virus monitoring in transplant recipients. EBV latently infects B lymphocytes. In EBV-associated lymphoproliferative disorders, EBV-infected B lymphocytes are usually found in the blood (27). Therefore, peripheral blood mononuclear cells are the best specimens to use in quantifying the EBV DNA load in transplant patients. However, whole blood, which contains mononuclear cells and is convenient to use, has been suggested as an acceptable alternative (17, 26). CMV latently infects a variety of leukocytes but predominantly cells in the monocyte/macrophage lineage. CMV quantification can be performed with acellular fractions of the blood, such as plasma and serum (7, 20, 29); however, in transplant recipients, the quantity of viral DNA is greater in leukocytes than in plasma (6, 10). HHV-6, which is closely related to CMV, infects mainly CD4+ T lymphocytes and is predominantly found in latently infected monocytes/macrophages (28). Since considerable amounts of the HHV-6 genome persist in monocytes/macrophages, detection of HHV-6 DNA in whole blood may reflect both latent and active viral infection. Therefore, it has been suggested that the HHV-6 viral load in plasma is an effective indicator of active infection (16, 28).
In the present study, we compared whole-blood and plasma specimens in the simultaneous detection of EBV, CMV, and HHV-6 DNA. The detection rate for each viral DNA form was higher in the whole-blood specimens than in the plasma specimens. During the symptomatic periods, EBV DNA was found in all whole-blood specimens obtained but not in all plasma specimens. EBV DNA loads in whole blood were higher during the symptomatic period than during the asymptomatic period, whereas EBV DNA loads in plasma were similar during both periods. These results support the use of whole-blood specimens for multiplex real-time PCR assays in transplant patients, although we have insufficient data to conclude that whole blood is preferable for assaying CMV and HHV-6 loads. On the other hand, EBV, CMV, and HHV-6 latently infect blood corpuscles, and asymptomatic reactivation may occur in transplant patients. Therefore, it is necessary to determine a cutoff value that reflects clinical relevance. In this study, we could not determine the cutoff value because of the sample size and heterogeneity of the transplantations. Since the number of peripheral blood cells varies, especially after stem cell transplantation, this may influence the viral load in whole blood. A large prospective study to determine the clinical cutoff value for each virus and each transplantation type is currently under way.
In summary, we developed a multiplex real-time PCR assay for the simultaneous detection of EBV, CMV, and HHV-6 DNA. The results of the multiplex assay were as sensitive and specific as those of the single real-time PCR assay. Compared to plasma, whole blood was more suitable for quantifying EBV DNA in transplant patients. The savings in cost, time, and labor associated with multiplex real-time PCR validate its use in the management of transplant recipients.
Acknowledgments
We thank the following individuals for their contributions to this study: Yukiko Watanebe, Kenichiro Kaneko, Youhei Yamauchi (Nagoya University Graduate School of Medicine), and Hideki Muramatsu (Japanese Red Cross Nagoya First Hospital).
This study was supported by grants from the Ministry of Education, Culture, Sport, Science and Technology of Japan (17209037).
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Adelson, M. E., M. Feola, J. Trama, R. C. Tilton, and E. Mordechai. 2005. Simultaneous detection of herpes simplex virus 1 and 2 by real-time PCR and pyrosequencing. J. Clin. Virol. 33:25-34. [DOI] [PubMed] [Google Scholar]
- 2.Hentrich, M., D. Oruzio, G. Jäger, M. Schlemmer, M. Schleuning, X. Schiel, W. Hiddemann, and H.-J. Kolb. 2005. Impact of human herpesvirus-6 after haematopoietic stem cell transplantation. Br. J. Haematol. 128:66-72. [DOI] [PubMed] [Google Scholar]
- 3.Hoshino, Y., H. Kimura, N. Tanaka, I. Tsuge, K. Kudo, K. Horibe, K. Kato, T. Matsuyama, A. Kikuta, S. Kojima, and T. Morishima. 2001. Prospective monitoring of the Epstein-Barr virus DNA by a real-time quantitative polymerase chain reaction after allogenic stem cell transplantation. Br. J. Haematol. 115:105-111. [DOI] [PubMed] [Google Scholar]
- 4.Jenkins, F. J., D. T. Rowe, and C. R. Rinaldo, Jr. 2003. Herpesvirus infections in organ transplant recipients. Clin. Diagn. Lab. Immunol. 10:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura, H., M. Morita, Y. Yabuta, K. Kuzushima, K. Kato, S. Kojima, T. Matsuyama, and T. Morishima. 1999. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 37:132-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li, H., J. S. Dummer, W. R. Estes, S. Meng, P. F. Wright, and Y.-W. Tang. 2003. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J. Clin. Microbiol. 41:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Limaye, A. P., M.-L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 183:377-382. [DOI] [PubMed] [Google Scholar]
- 8.Macé, M., C. Manichanh, P. Bonnafous, S. Précigout, D. Boutolleau, A. Gautheret-Dejean, and H. Agut. 2003. Real-time PCR as a versatile tool for investigating the susceptibility of human herpesvirus 6 to antiviral agents. Antimicrob. Agents Chemother. 47:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mackay, I. M., K. E. Arden, and A. Nitsche. 2002. Real-time PCR in virology. Nucleic Acids Res. 30:1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mengelle, C., K. Sandres-Sauné, C. Pasquier, L. Rostaing, J.-M. Mansuy, M. Marty, I. Da Silva, M. Attal, P. Massip, and J. Izopet. 2003. Automated extraction and quantification of human cytomegalovirus DNA in whole blood by real-time PCR assay. J. Clin. Microbiol. 41:3840-3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niesters, H. G. M. 2004. Molecular and diagnostic clinical virology in real time. Clin. Microbiol. Infect. 10:5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogata, M., H. Kikuchi, T. Satou, R. Kawano, J. Ikewaki, K. Kohno, K. Kashima, E. Ohtsuka, and J. Kadota. 2006. Human herpesvirus 6 DNA in plasma after allogeneic stem cell transplantation: incidence and clinical significance. J. Infect. Dis. 193:68-79. [DOI] [PubMed] [Google Scholar]
- 13.Peres, E., S. Savasan, J. Klein, M. Abidi, R. Dansey, and E. Abella. 2005. High fatality rate of Epstein-Barr virus-associated lymphoproliferative disorder occurring after bone marrow transplantation with rabbit anti-thymocyte globulin conditioning regimens. J. Clin. Microbiol. 43:3540-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pradeau, K., L. Couty, J.-C. Szelag, P. Turlure, F. Rolle, P. Ferrat, D. Borsessoule, Y. L. Meur, F. Denis, and S. Ranger-Rogez. 2006. Multiplex real-time PCR assay for simultaneous quantitation of human cytomegalovirus and herpesvurus-6 in polymorphonuclear and mononuclear cells of transplant recipients. J. Virol. Methods 132:77-88. [DOI] [PubMed] [Google Scholar]
- 15.Razonable, R. R., R. A. Brown, A. Humar, E. Covington, E. Alecock, C. V. Paya, and the PV16000 Study Group. 2005. Herpesvirus infections in solid organ transplant patients at high risk of primary cytomegalovirus disease. J. Infect. Dis. 192:1331-1339. [DOI] [PubMed] [Google Scholar]
- 16.Secchiero, P., D. R. Carrigan, Y. Asano, L. Benedetti, R. W. Crowley, A. L. Komaroff, R. C. Gallo, and P. Lusso. 1995. Detection of human herpesvirus 6 in plasma of children with primary infection and immunosuppressed patients by polymerase chain reaction. J. Infect. Dis. 171:273-280. [DOI] [PubMed] [Google Scholar]
- 17.Stevens, S. J. C., S. A. W. M. Verkuijilen, A. J. C. van den Brule, and J. M. Middeldorp. 2002. Comparison of quantitative competitive PCR with LightCycler-based PCR for measuring Epstein-Barr virus DNA load in clinical specimens. J. Clin. Microbiol. 40:3986-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuga, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60:445-462. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka, N., H. Kimura, Y. Hoshino, K. Kato, T. Yashikawa, Y. Asano, K. Horibe, S. Kojima, and T. Motishima. 2000. Monitoring four herpesviruses in unrelated cord blood transplantation. Bone Marrow Transplant. 26:1193-1197. [DOI] [PubMed] [Google Scholar]
- 20.Tedder, R. S., U. Ayliffe, W. Preiser, N. S. Brink, P. R. Grant, K. S. Peggs, S. Mackinnon, F. Kreig-Schneider, S. Kirk, and J. A. Garson. 2002. Development and evaluation of an internally controlled semiautomated PCR assay for quantification of cell-free cytomegalovirus. J. Med. Virol. 66:518-523. [DOI] [PubMed] [Google Scholar]
- 21.Templeton, K. E., S. A. Scheltinga, N. F. C. Beersma, A. C. M. Kroes, and E. C. J. Claas. 2004. Rapid and sensitive method using multiplex real-time PCR for diagnosis of infections by influenza A and influenza B viruses, respiratory syncytial virus, and parainfluenza viruses 1, 2, 3, and 4. J. Clin. Microbiol. 42:1564-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timbury, M. C., and E. Edmond. 1979. Herpesviruses. J. Clin. Pathol. 32:859-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torre, D., R. Mancuso, and P. Ferrante. 2005. Pathogenic mechanisms of meningitis/encephalitis caused by human herpesvirus 6 in immunocompetent adult patients. Clin. Infect. Dis. 41:422-423. [DOI] [PubMed] [Google Scholar]
- 24.Traverso, M., M. Malnati, C. Minetti, S. Regis, S. Tedeschi, M. Pedemonte, C. Bruno, R. Biassoni, and F. Zara. 2006. Multiplex real-time PCR for detection of deletions and duplications in dystrophin gene. Biochem. Biophyse Res. Commun. 339:145-150. [DOI] [PubMed] [Google Scholar]
- 25.Visser, A. M., G. J. J. van Doornum, J. J. Cornelissen, and M. J. van den Bent. 2005. Severe amnesia due to HHV-6 encephalitis after allogenic stem cell transplantation. Eur. Neurol. 54:233-234. [DOI] [PubMed] [Google Scholar]
- 26.Wadowsky, R. M., S. Laus, M. Green, S. A. Webber, and D. Rowe. 2003. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J. Clin. Microbiol. 41:5245-5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang, J., Q. Tao, I. W. Flinn, P. G. Murray, L. E. Post, H. Ma, S. Piantadosi, M. A. Caligiuri, and R. F. Ambinder. 2000. Characterization of Epstein-Barr virus-infected B cells in patients with posttransplantation lymphoproliferative disease: disappearance after rituximab therapy does not predict clinical response. Blood 96:4055-4063. [PubMed] [Google Scholar]
- 28.Yoshikawa, T. 2004. Human herpesvirus 6 infection in hematopoietic stem cell transplant patients. Br. J. Haematol. 124:421-432. [DOI] [PubMed] [Google Scholar]
- 29.Zipeto, D., S. Morris, C. Hong, A. Dowling, R. Wolitz, T. C. Merigan, and L. Rasmussen. 1995. Human cytomegalovirus (CMV) DNA in plasma reflects quantity of CMV DNA present in leukocytes. J. Clin. Microbiol. 33:2607-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]