Abstract
Mycobacterium abscessus has been isolated increasingly often from the respiratory tracts of cystic fibrosis (CF) patients. It is not known whether these organisms are transmitted from person to person or acquired from environmental sources. Here, colony morphology and pulsed-field gel electrophoresis (PFGE) pattern were examined for 71 isolates of M. abscessus derived from 14 CF patients, three non-CF patients with chronic respiratory M. abscessus infection or colonization, one patient with mastoiditis, and four patients with infected wounds, as well as for six isolates identified as environmental contaminants in various clinical specimens. Contaminants and wound isolates mainly exhibited smooth colony morphology, while a rough colony phenotype was significantly associated with chronic airway colonization (P = 0.014). Rough strains may exhibit increased airway-colonizing capacity, the cause of which remains to be determined. Examination by PFGE of consecutive isolates from the same patient showed that they all represented a single strain, even in cases where both smooth and rough isolates were present. When PFGE patterns were compared, it was shown that 24 patients had unique strains, while four patients harbored strains indistinguishable by PFGE. Two of these were siblings with CF. The other two patients, one of whom had CF, had not had contact with each other or with the siblings. Our results show that most patients colonized by M. abscessus in the airways have unique strains, indicating that these strains derive from the environment and that patient-to-patient transmission rarely occurs.
Mycobacterium abscessus is an environmental mycobacterial species ubiquitous in soil and water (4, 43). It can grow in distilled water and has been isolated from water treatment plants, domestic water supply systems, and aquaria, where it may participate in biofilm formation (34).
M. abscessus may cause cutaneous infections after contact with contaminated water in public baths. Such infections usually resolve without complications, but medical or surgical treatment is sometimes needed (7, 20). M. abscessus is also capable of colonizing the respiratory tract and causes disease in certain groups of individuals. One such group is Caucasian females above 60 years of age. While most of them have no apparent underlying abnormality, 40% have some predisposing condition such as lung disease, lung transplantation, achalasia, or recurrent vomiting (5). Another group at risk for M. abscessus colonization is patients with cystic fibrosis (CF). In some CF patients, acquisition of M. abscessus is associated with poor clinical outcome, while in others colonization may persist for several years without evident disease or deterioration of pulmonary function (9, 27, 28, 30). M abscessus is inherently multiresistant and difficult to treat, and consequently its isolation in CF is of great concern.
M. abscessus has been reported to be isolated increasingly often from CF patients in several countries (8, 14, 30, 35). This appears to be true also in western Sweden, where the first case in the CF population was discovered in 1997. After that year, an increase in M. abscessus-colonized patients gave rise to the suspicion that person-to-person transmission had occurred between individuals at the West Swedish CF Centre at the Queen Silvia Children's Hospital in Göteborg. In the present study, pulsed-field gel electrophoresis (PFGE) was used to identify respiratory M. abscessus isolates from CF patients treated in western Sweden and at other Swedish CF centers, as well as respiratory isolates from other patient groups. We also examined wound isolates acquired from environmental sources during the tsunami catastrophe in Thailand in 2004, as well as M. abscessus isolates from various patient samples, where the bacterium was identified as an environmental contaminant. The aim was to determine whether CF patients cared for at a certain center shared strains, suggesting patient-to-patient spread. A second aim was to determine whether M. abscessus strains colonizing the respiratory tract were more related to each other than to wound isolates or contaminants.
MATERIALS AND METHODS
Patient groups and M. abscessus isolates.
In all, 71 isolates of M. abscessus, isolated from 28 patients, were included in the study (Table 1). Fourteen patients had CF; nine of them were seen at the West Swedish CF Centre at the Queen Silvia Children's Hospital in Göteborg. Three were cared for at the Stockholm CF Centre. Two were cared for at the Uppsala CF Centre and referred to the Sahlgrenska University Hospital in Göteborg for lung transplantation.
TABLE 1.
Description of Mycobacterium abscessus isolates used in this study
| Clinical condition | No. of patients | No. of isolates
|
|||
|---|---|---|---|---|---|
| Total | Respiratory | Wound/abscess | Blood | ||
| CF | 14 | 51 | 37a | 9 | 5b |
| Non-CF lung infection/chronic colonization | 3 | 9 | 9c | 0 | 0 |
| Mastoiditis | 1 | 1 | 0 | 1 | 0 |
| Wounds (tsunami victims) | 4 | 4 | 0 | 4 | 0 |
| Transient colonization/ contaminatione | 6 | 6 | 6d | 0 | 0 |
| Total | 28 | 71 | 52 | 14 | 5 |
Includes 33 sputum, 3 bronchioalveolar lavage, and 1 pleural fluid sample.
Includes four blood cultures and one sample from a central venous catheter.
Includes eight sputum and one bronchial lavage sample.
Includes two sputum, two bronchial lavage, and two pleural fluid samples.
Defined as an M. abscessus isolate appearing on a single culture occasion in a sample from a non-CF patient, without associated clinical symptoms, and followed by negative sample.
Three non-CF patients with M. abscessus lung infection/colonization (two women, 56 and 84 years of age, and one 54-year-old male) contributed a total of nine isolates (Table 1). An isolate from a patient with M. abscessus mastoiditis was also included. Four victims of the tsunami catastrophe in Asia on 26 December 2004 contributed one wound isolate each.
Six additional patients contributed one isolate each, derived from various clinical samples. Follow-up samples from these patients were negative for M. abscessus, whose presence was regarded as the result of transient colonization without clinical significance.
The type strain of M. abscessus ATCC 19977 was obtained from the Culture Collection University of Göteborg (CCUG 20993).
Isolation and identification of M. abscessus from patient samples.
Direct microscopy for mycobacteria and culture were done according to routine procedures at the laboratory (18). M. abscessus was isolated on Lowenstein-Jensen solid medium. Mycobacteria were identified by the Hallberg staining technique (16). Briefly, bacterial slides were heated to 80°C in a solution containing Nachtblau, cooled to room temperature, rinsed in distilled water, decolorized in 8% HNO3 and 70% ethanol, and counterstained with fuchsine.
Species were identified by using a biochemical test panel (tolerance to 5% NaCl, iron uptake, Tween hydrolysis, urease, nitrate reduction) (24), by examination of the mycolic acid pattern by high-pressure liquid chromatographic analysis (1, 2), and by sequencing of the 16S RNA gene (19). For the most recent isolates (2005 onward), these methods were complemented with the GenoType Mycobacterium CM test (HAIN Lifescience, Nehren, Germany) (21).
All isolates were stored at room temperature in vials with Lowenstein-Jensen medium until analyzed.
Special procedures for processing of sputum samples from CF patients.
All patients with CF were screened yearly for mycobacteria in sputum. Respiratory specimens were prepared according to routine procedures employed at the Bacteriological Laboratory, Sahlgrenska University Hospital (18), and the Karolinska University Hospital in Stockholm. To kill unwanted bacteria in the samples, aliquots were decontaminated by the sodium dodecyl sulfate-NaOH method (16) and treated with a cocktail of amphotericin B (250 μg/ml), carbenicillin (1.25 mg/ml), polymyxin B-sulfate (5 mg/ml), and trimethoprim-lactate (500 μg/ml) to eliminate overgrowth of fungi and unwanted bacteria. After 1993, sputum samples were also treated with 5% oxalic acid to minimize the risk of overgrowth with Pseudomonas aeruginosa (39, 40).
PFGE for strain typing of M. abscessus. (i) Preparation of bacterial DNA.
M. abscessus isolates were grown in Middlebrook 7H10 agar vials until rich growth was achieved, usually after 1 week. The colony morphology (rough or smooth) was noted. DNA was prepared according to a published method (12), but with minor modifications. A loopful of bacteria was transferred to 5 ml Middlebrook 7H9 broth (Difco) modified by the addition of 0.5 M sucrose, 0.05% (wt/vol) Tween 80, 0.2% (wt/vol) d-glucose, and 10% oleic acid-albumin complex (Becton-Dickinson), and incubated on a shaker for 72 h at 37°C. This broth permitted adequate growth of isolates forming rough colonies on solid medium. For disruption of the bacteria, 400 μl of a solution containing 0.2 M glycine, 60 μg d-cycloserine/ml, 20 mM LiCl, 200 mg lysozyme/ml, and 5 mM EDTA was added to the cultures, which were incubated for 16 h and then centrifuged at 2,300 × g for 15 min at room temperature. The bacteria were resuspended in 1.2 ml TS buffer (50 mM Tris, 0.5 M sucrose; pH 7.6), and 125 μl of the suspension was transferred to Eppendorf tubes and frozen at −20°C. The suspensions were thawed in room temperature, heated to 75°C for 20 min, suspended in 125 μl of 2% low-melting-point agarose (SeaPlaque; Cambrex Bio Science Inc., Baltimore, MD) mixed with EC buffer (6 mM Tris-HCl, 1 M NaCl, 100 mM EDTA, 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% sodium lauroyl sarcosine; pH 7.5) at 55°C, and cast in plugs. Bacteria in plugs were lysed in TE buffer (10 mM Tris, 1 mM EDTA; pH 7.6) with 1 mg lysozyme/ml overnight at 37°C, followed by incubation in 250 μl ESP buffer (200 μl 0.5 M EDTA [pH 9.0], 25 μl sodium lauroyl sarcosine 10%, 25 μl proteinase K [5 mg/ml]) for 24 h at 55°C. The ESP buffer was decanted and replaced with HE buffer (10 mM HEPES-NaOH [pH 8.0], 1 mM EDTA) containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF). After incubation at 4°C for 1 h, the PMSF solution was decanted, and plugs were washed five times for 30 min each time at 4°C in HE buffer without PMSF. Using HE buffer instead of Tris buffer in the washing step gave better results and was described previously for PFGE analysis of Streptomyces lividans strains (10). Plugs were stored in 0.2 M EDTA (HE and TE buffers were also tested) at 4°C until used.
(ii) Restriction endonuclease digestion.
Before digestion, plugs containing embedded DNA were washed in HE buffer six times for 15 min each time, at 4°C. Genomic DNA in the plugs was digested overnight at 37°C with 20 U of restriction endonuclease AseI or XbaI in buffer as recommended by the manufacturer (New England BioLabs, Inc.).
(iii) Gel electrophoresis.
Digested plugs were loaded into a 1% agarose gel prepared and run in HEPES buffer (16 mM HEPES-NaOH [pH 7.5], 16 mM sodium acetate, 0.8 mM EDTA) (32). PFGE was performed with the Genepath system (Bio-Rad Laboratories, Sundbyberg, Sweden) at 14°C and with a reduced voltage gradient (4 V/cm) to compensate for the higher ionic strength of HEPES than Tris buffer. Due to problems with DNA degradation during electrophoresis, HEPES was used as the electrophoresis buffer instead of Tris buffer (31, 32). The programs used were modifications of the ones described by Wallace et al. (38). The program used for AseI was a combination of two programs, the first consisting of an initial switch time of 1 s, a final switch time of 23 s, a run time of 23 h, an angle of 120°, and a linear ramping factor. This program was immediately followed by the second program, consisting of an initial switch time of 10 s, a final switch time of 17 s, a run time 4 h, an angle of 120°, and a linear ramping factor. The program following XbaI digestion had an initial switch time of 3 s, a final switch time of 12 s, a run time of 23 h, an angle of 120°, and a linear ramping factor. In every run, two plugs containing genomic DNA of Staphylococcus aureus (NCTC 8325), digested with SmaI, were used as a reference. A lambda ladder PFG marker (New England BioLabs, Inc.) was used as a molecular weight standard.
(iv) Interpretation of PFGE patterns.
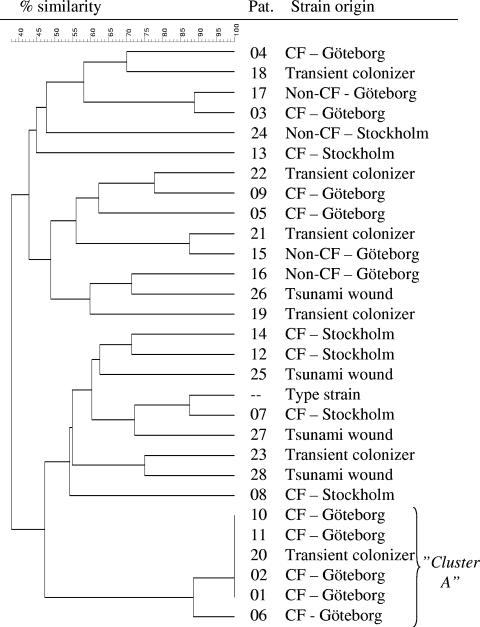
Gel electrophoresis of the restriction endonuclease-treated DNA yielded a band pattern specific for each bacterial clone. The PFGE types were interpreted as described by Tenover et al. (37). Isolates with patterns differing by three bands or less were considered related or probably related. The gels were also digitized for computer-aided analysis using Fingerprinting II software (Bio-Rad Laboratories, Hercules, CA). Calculation of the similarity matrix was done with the Jaccard algorithm after defining each band between 674 and 117 kb, and clustering was achieved with the unweighted pair group method (UPGMA). Two isolates were considered to belong to the same bacterial clone if the similarity was ≥90% according to the cluster analysis (see Fig. 2).
FIG. 2.
PFGE analysis of AseI-digested DNA from 29 M. abscessus isolates. Relationships among isolates from 28 Swedish patients and type strain ATCC 19977 are shown in a similarity dendrogram constructed after calculation of the similarity matrix with the Jaccard algorithm (optimization, 1.20%; position tolerance, 1.2% to 1.2%; (minimum height > 0.0%; minimum > 0.0%) and clustering according to UPGMA. CF-Göteborg, isolates obtained from CF patients in the Västra Götaland region; CF-Stockholm, isolates obtained from CF patients in the Stockholm/Uppsala area; non-CF-Göteborg, isolates from other patients with lung infection/colonization from the Västra Götaland region; non-CF-Stockholm, isolates from a patient with mastoiditis in the Stockholm area; tsunami wound, isolate from Swedish tsunami victim (strain obtained in Thailand); transient colonizer, isolate found on a single occasion in a clinical specimen followed by a negative sample and/or lack of associated clinical conditions, suggesting that the isolate was an environmental contaminant.
Statistics.
Comparisons between groups were made using Fisher's exact test.
Ethics.
This study was approved by the Ethical Committee at the Göteborg University, Göteborg, Sweden.
RESULTS
Occurrence of M. abscessus in CF patients and clinical correlates.
Of 140 patients with CF included in the regular sputum screening programs at the Queen Silvia Children's Hospital, nine (6%) had been positive for M. abscessus on at least one occasion by the end of 2005. Details for these patients are shown in Table 2. The first case appeared in 1997. Among the nine CF patients positive for M. abscessus, five (patients 1, 2, 3, 4, and 5) (Table 2) were considered to have clinical disease caused by this bacterium and were treated for this for at least some period. For patient 6, the contribution of M. abscessus to the clinical disease was difficult to assess, but he received antimycobacterial treatment for at least 3 months. Since 2005, this patient has had five negative cultures. One CF patient (no. 9) was persistently colonized by M. abscessus without clear clinical impact. This patient was treated for Mycobacterium avium complex (MAC) infection in 1996. This organism was eradicated, but in 1997 the patient was infected with M. abscessus and has been colonized since. Two CF patients (no. 10 and 11) were thought to have M. abscessus as a transient colonizer and did not receive any antimycobacterial treatment.
TABLE 2.
M. abscessus colonization in patients with CF at the western Swedish CF center in Göteborg, Sweden
b SA, Staphylococcus aureus; SM, Stenotrophomonas maltophilia; BC, Burkholderia cepacia; AF, Aspergillus fumigatus; PA, Pseudomonas aeruginosa; HI, Haemophilus influenzae.
c ◊, M. abscessus found in sputum not analyzed by PFGE (nonviable or not stored); ⧫, M. abscessus found in sputum analyzed by PFGE.
d Eradicated.
Two CF patients (a female born in 1976 and a male born in 1992) from the Stockholm/Uppsala region were referred to Göteborg for lung transplantation. They had been colonized by M. abscessus since 2000 and 2002, respectively. Both developed disseminated M. abscessus disease posttransplantation, for which they received treatment. Three CF patients, cared for at the Stockholm CF Centre, were interpreted as having persistent airway colonization with M. abscessus, probably of clinical significance. One of these patients (no. 14; see the Fig. 2 dendrogram) was treated for a MAC infection 10 years previously with rifabutin, ethambutol, and amikacin. Ethambutol was switched to clarithromycin and clofazimine because of side effects. The MAC strain was eradicated, but M. abscessus was isolated from sputum from this patient during MAC treatment. The M. abscessus strain developed in vitro resistance to all tolerated drugs, but the patient was given gamma interferon with clinical effect. The M. abscessus strain was never eradicated, but this patient has been without mycobacterial treatment for the past 5 years.
Colony morphology of M. abscessus in relation to clinical picture.
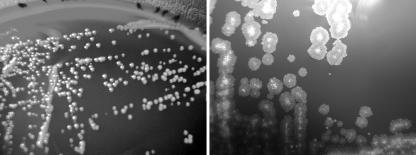
Two types of colony appearances were noted after culture of M. abscessus on either horse blood or Middlebrook 7H10 agar (Fig. 1). One phenotype was smooth and shiny and resembled that of the type strain, ATCC 19977. The other form had larger, rough, and markedly waxier colonies. The two forms were stable during subculture.
FIG. 1.
Colony morphology of M. abscessus after 5 days of incubation on horse blood agar at 37°C. The same magnification was used for both images. (Left) Type strain CCUG 20993 with smooth colonies. (Right) Strain with rough colonies from a CF patient. Images were obtained with a Nikon Coolpix 4300 and Adobe Photoshop Elements 2.0.
The clinical origin of smooth and rough isolates is shown in Table 3. Sixteen patients, most of whom were CF and non-CF patients with chronic colonization of the airways, yielded only rough isolates. A few isolates were isolates causing transient colonization of the respiratory tract.
TABLE 3.
Colony morphologya among M. abscessus strains
| Sample type or patient group | No. of patients yielding:
|
||
|---|---|---|---|
| Rough isolates | Smooth isolates | Rough and smooth isolates | |
| Respiratory samples | |||
| Clinical disease, CF | 9 | 1 | 1 |
| Clinical disease, non-CF | 1 | 1 | |
| Chronic colonization, CF | 1 | ||
| Chronic colonization, non-CF | 1 | ||
| Mastoiditis, non-CF | 1 | ||
| Transient colonization without symptoms, CF | 2 | ||
| Transient colonization without symptoms, non-CF | 1 | 5 | |
| Wound infectionb samples | 4 | ||
| Total (n = 28) | 16 | 10 | 2 |
Colony appearance (rough or smooth) when cultured on Middlebrook 7H10 agar or horse blood agar.
Wound infections acquired during the tsunami in Thailand in 2004.
Two patients yielded a mixture of rough and smooth isolates. Both patients had clinical respiratory disease due to M. abscessus (Table 3). Ten patients consistently yielded isolates of a smooth phenotype (Table 3). One of these was a CF patient with respiratory disease, while four were patients who sustained wounds that were contaminated by environmental sources during the tsunami catastrophe in Thailand in 2004. Five isolates were found in airway samples from patients without related symptoms and were categorized as transient. Follow-up samples were negative.
Among the rough isolates, 81% (13/16) were isolated from patients with chronic colonization/disease in the airways, while only 10% (1/10) of the smooth ones were from this patient group. This difference was significant (P = 0.0014, Fisher's exact test).
PFGE analysis of M. abscessus isolates.
Seventy-one M. abscessus isolates from 28 patients and the M. abscessus type strain were examined by PFGE after digestion of bacterial DNA by AseI to determine their strain identity (Fig. 2).
From each of 10 patients, several isolates of M. abscessus were obtained from successive specimens. The longest interval between the first and last isolates analyzed was almost 4 years (CF patient 2) (Table 2). All isolates analyzed from a single patient yielded the same PFGE pattern. Smooth and rough isolates from the same patient also had identical PFGE patterns.
Figure 2 shows the relatedness of the 28 patient strains after digestion with AseI. Twenty-two of the patients had isolates with unique PFGE patterns. Six patients harbored strains that appeared to be identical or nearly identical. We designated these strains cluster A. In five patients (CF patients 1, 2 10, and 11 and non-CF patient 20), the strain had identical PFGE patterns within cluster A (named A1), while the strain colonizing the sixth patient (CF patient 6) differed by a single band and was termed A2. Except for the cluster, the isolates from CF patients living in the Göteborg area were no more related to one another than to isolates from CF patients living in the Stockholm/Uppsala area or to isolates acquired in Thailand (Fig. 2). Furthermore, isolates causing a certain clinical condition (e.g., lung infection in CF patients) were no more related to one another than to isolates causing other clinical conditions (e.g., wound isolates).
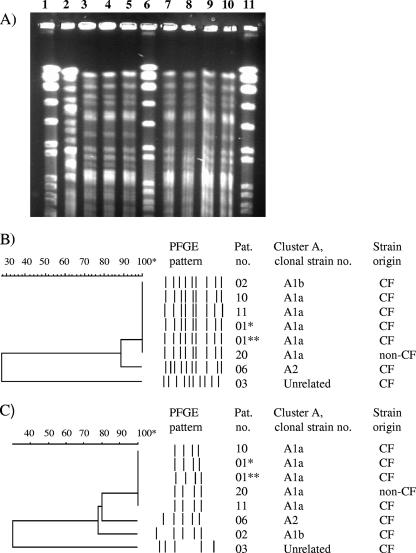
To further characterize cluster A, we used another enzyme, XbaI, to digest DNA in the PFGE analysis. Using this enzyme, four strains belonging to cluster A1 (termed A1a) were still identical, while A2 and one A1 strain (termed A1b) differed by one additional band from the A1 cluster and by two bands from one another (Fig. 3). All isolates belonging to cluster A exhibited a rough phenotype.
FIG. 3.
PFGE analyses of cluster A after AseI and XbaI digestion of DNA. (A) AseI PFGE gel corresponding to the dendrogram in panel B. Lanes 1, 6, and 11, reference strain S. aureus NCTC 8325 (not shown in dendrogram). Lane 2, patient 3, isolate 05*1230; lane 3, patient 6, isolate 05*2299; lane 4, patient 10, isolate 05*2316; lane 5, patient 1, isolate 01*5153; lane 7, patient 1, isolate 03*4569; lane 8, patient 2, isolate 04*2226; lane 9, patient 20, isolate 02*1284; lane 10, patient 11, isolate 01* 1526. (B and C) AseI (B) and XbaI (C) PFGE patterns presented as similarity dendrograms and schematic views of bands analyzed. *, isolate 01*5153; **, isolate 03*4569.
Two of the four patients (no. 1 and 11) harboring cluster A1 were siblings, both suffering from CF. The older sibling (no. 1) has been permanently colonized by M. abscessus since 2000 and has clinical disease caused by this bacterium (Table 2). The younger sibling (no. 11) was positive for M. abscessus in sputum on one occasion only, in 2001. Thereafter, screening cultures have been negative. She never received antimycobacterial treatment. The third patient with a strain belonging to cluster A1 (no.10) is a CF patient, living in western Sweden but no longer cared for at the CF center in Göteborg. She has had only one M. abscessus-positive culture. To our knowledge, she has had no social contacts with the CF siblings. The fourth patient (no. 20) having an A1 cluster strain is an elderly woman from another town. She was seen at the local hospital for unspecified pleuritis, and M. abscessus was isolated from the pleural fluid on a single occasion in 2002 but was not interpreted as the etiology of her pleuritis. We received no other patient samples harboring the cluster strain from this hospital during this year. Patient no. 2, whose strain belonged to cluster A and was termed A1b, is a CF patient also colonized by Burkholderia cepacia. Due to this colonization, she has been kept segregated from other CF patients.
DISCUSSION
During the last decade we have noted an increasing prevalence of M. abscessus in sputum samples from CF patients cared for at the West Swedish CF Centre. Since 1997, when the first case of M. abscessus colonization was discovered in the CF population of 140 patients, one new case per year has occurred on average. The staff has changed little in our laboratory over the past 15 years, and samples have been prepared and cultured almost in the same manner since 1993. Furthermore, because M. abscessus grows more rapidly than most other environmental mycobacteria, it is less easily overlooked than other mycobacterial species in sputum samples. Taking these observations together, we consider it likely that there has been a true increase in the prevalence of respiratory colonization by M. abscessus in the last decade. A similar increase has also been reported from several CF centers in Europe and the United States (9, 15, 26, 28, 30, 35).
In total, 6% of the individuals in the western Swedish CF population had been positive for M. abscessus in sputum on at least one occasion by the end of 2005. Other mycobacteria isolated from CF patients were the MAC (3%) and Mycobacterium lentiflavum (1%). This makes M. abscessus the predominant mycobacterial species recovered from CF patients in western Sweden today.
In patients without CF from whose airway samples environmental mycobacteria have been isolated (n = 62 during 2005), the MAC is far more common than M. abscessus (33 versus 4 patients). Thus, M. abscessus appears to display a predilection for colonization of the airways in patients with CF.
We hypothesized that colonization by M. abscessus in CF patients could have occurred because one or a few clones had spread via social contacts or through contaminated equipment used for clinical examination at the CF clinic. PFGE has previously been used to retrospectively analyze M. abscessus outbreaks, all of which were found to involve a single strain usually originating from the water supply system, with no spread between patients (38, 41, 42). However, in our study most CF patients carried unique strains of M. abscessus. The only exception was a cluster of two siblings and two other patients who harbored a strain indistinguishable by PFGE (cluster A1a). The samples from the patients harboring the cluster strain were never processed in the same month in the laboratory, and samples from the siblings were not obtained on the same occasion, making contamination via the laboratory highly unlikely. The siblings both had CF. One of them had been permanently colonized since 2000, while the other was positive on a single occasion only, in 2001. It is reasonable to believe that the colonized sibling may have passed the strain to her sister. Transfer of P. aeruginosa strains between siblings with CF has been observed (36), which supports our statement.
The third patient with a cluster A1a strain, a CF patient not cared for in Göteborg, had no known contacts with the siblings. The fourth patient with a cluster A1a strain was an elderly woman cared for at the local hospital in another town for other causes. She was positive for M. abscessus on a single occasion only, without associated symptoms. Thus, except for the possible transmission of clone A1a between the two siblings, we found no evidence of spread of M. abscessus between CF patients cared for at the West Swedish CF Centre. Furthermore, isolates from western Sweden were no more similar to one another than isolates from other geographical locations.
As M. abscessus is ubiquitous in nature, our results suggest that this bacterium is acquired from the environment by individual patients and that spread between patients is uncommon.
Such “spontaneous” colonization by M. abscessus might have increased in CF patients. One reason may be that better control of “classical” respiratory pathogens such as S. aureus and P. aeruginosa might have paved the way for opportunists with lower inherent pathogenicity, including M. abscessus. One example of this was the two CF siblings; one of them was permanently colonized by M. abscessus, harboring no other pathogen besides Aspergillus fumigatus, whereas the other sibling, positive only once for M. abscessus, had all classical CF pathogens in sputum, i.e., S. aureus, Haemophilus influenzae, P. aeruginosa, and Stenotrophomonas maltophilia. The patients colonized with M. abscessus might also have subtle deficiencies in their defense systems. Two CF patients who were both colonized/infected with M. abscessus underwent lung transplantation. Both developed disseminated disease postoperatively but survived. Both fatal and nonfatal post-lung-transplant infections with M. abscessus have been described (3, 11, 22, 33). Clearly, immunosuppression is involved in aggravation of the disease, but whether any abnormalities underlie respiratory colonization per se remains to be shown.
The three non-CF patients with chronic pulmonary M. abscessus colonization/infection studied had no known immunological deficiencies, besides old age in one case. One patient had previously suffered from MAC disease and had developed bronchiectatic changes and A. fumigatus colonization. Allergic bronchopulmonary aspergillosis and systemic steroids have been identified as risk factors for mycobacterial disease (26). The second patient was a smoker with chronic obstructive pulmonary disease (COPD), while the third was a healthy nonsmoking middle-aged woman. This fits with the previously identified risk groups for pulmonary colonization with M. abscessus (middle-aged female nonsmokers, COPD patients, and patients previously treated for mycobacterial disease) (5).
We observed no apparent relatedness between strains causing a certain type of disease manifestation. Four isolates were from patients who sustained injuries which became contaminated with dirty water in the tsunami catastrophe in Thailand in 2004. These infections manifested as skin abscesses. These four strains were no more related to one another than to Swedish strains from CF or non-CF patients. When M. abscessus was cultured on solid media, colonies displayed either of two phenotypes: smooth and shiny, or rough and waxy. The M. abscessus ATCC type strain was smooth, as were all but one of the isolates interpreted as contaminants or transient colonizers of the airways and all wound isolates. In contrast, all isolates colonizing the airways of CF patients were rough. The only exception was an isolate from a newly infected patient, who presented abruptly with clinical disease and heavy mycobacterial growth in sputum. Indeed, we found a significant association between a rough phenotype and chronic colonization of the airways. In a case report of a fatal pulmonary infection due to M. abscessus in a patient with CF, it was noted that the bacterial strain exhibited a rough phenotype (33). Two patients in our study had a mixture of smooth and rough isolates. In both cases, all isolates from the same patient belonged to a single strain, regardless of morphology, which shows that strains may switch between rough and smooth phenotype in vivo. Both phenotypes were stable upon subculture.
A rough phenotype of M. abscessus was shown to display increased virulence in both a human monocyte model and a murine pulmonary assay (17). Conversion from the smooth to the rough phenotype was associated with alterations in the glycopeptidolipid composition of the M. abscessus cell wall (17). In Mycobacterium chelonae, a species closely related to M. abscessus, rough-colony mutants were more hydrophobic than the smooth wild type and had reduced susceptibility to glutaraldehyde and ethambutol (23). In our study, we saw that a rough phenotype was associated with persistent airway colonization. The rough phenotype might increase the ability of M. abscessus to form biofilms, a growth form thought to be advantageous in airway colonization of CF patients (29). Rough-phenotype strains of P. aeruginosa formed biofilms faster than the smooth wild type when cultivated in flow chambers (6), and rough Vibrio cholerae variants were shown to have increased abilities to form biofilms in a broth model compared to the normal smooth strains (25).
Although PFGE is time-consuming and expensive, it has high reproducibility, and band patterns may be stored in a database for future comparisons. The finding that M. abscessus is part of the microbial flora in CF patients increases the need for improved surveillance for this species to enable prompt diagnosis and treatment. There are indications that eradication is possible when treatment is started early (13).
Acknowledgments
We acknowledge the kind assistance of Erja Chryssanthou of the Clinical Microbiological Laboratory, Karolinska University Hospital, Stockholm, who submitted the isolates from the Stockholm area, and Anne Geborek at the Stockholm CF Centre, Karolinska University Hospital, Huddinge, Pia Appelgren at the Infection Clinic, Karolinska University Hospital, Stockholm, and Marie Johannesson at the Uppsala CF Centre, Uppsala University Hospital, for valuable discussions concerning the CF patients and tsunami victims in the Stockholm region. We also acknowledge Leif Dotevall at the infection clinic, Sahlgrenska University Hospital, Göteborg, for clinical information regarding tsunami victims cared for in western Sweden.
This work was supported by grants from the Västra Götaland region, Sweden, the Sahlgrenska University Hospital, Göteborg, Göteborg Medical Society and The Oscar & Hanna Björkbom Foundation, Sweden.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Butler, W., M. Floyd, V. Silcow, G. Cage, E. Desmond, P. Duffey, L. Guthertz, W. Gross, K. C. Jost, Jr., L. Ramos, L. Thibert, and N. Warren. 1996. Standardized method for HPLC identification of mycobacteria. Centers for Disease Control and Prevention, Atlanta, GA. http://cdc.gov/ncidod/publications/hplc.pdf.
- 2.Butler, W. R., and J. O. Kilburn. 1990. High-performance liquid chromatography patterns of mycolic acids as criteria for identification of Mycobacterium chelonae, Mycobacterium fortuitum, and Mycobacterium smegmatis. J. Clin. Microbiol. 28:2094-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang, L., K. Hohneker, M. Knowles, P. Gilligan, and N. Peader. 2005. Mycobacterium abscessus infection in cystic fibrosis. Pediatr. Pulmonol. Suppl. 28:291-292. [Google Scholar]
- 4.Covert, T. C., M. R. Rodgers, A. L. Reyes, and G. N. Stelma, Jr. 1999. Occurrence of nontuberculous mycobacteria in environmental samples. Appl. Environ. Microbiol. 65:2492-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daley, C. L., and D. E. Griffith. 2002. Pulmonary disease caused by rapidly growing mycobacteria. Clin. Chest Med. 23:623-632. [DOI] [PubMed] [Google Scholar]
- 6.Drenkard, E., and F. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 7.Dytoc, M., L. Honish, C. Shandro, P. Ting, L. Chui, L. Fiorillo, J. Robinson, A. Fanning, G. Predy, and R. Rennie. 2005. Clinical, microbiological, and epidemiological findings of an outbreak of Mycobacterium abscessus hand-and-foot disease. Diagn. Microbiol. Infect. Dis. 53:39-45. [DOI] [PubMed] [Google Scholar]
- 8.Ebert, D. L., and K. N. Olivier. 2002. Nontuberculous mycobacteria in the setting of cystic fibrosis. Clin. Chest Med. 23:655-663. [DOI] [PubMed] [Google Scholar]
- 9.Esther, C., Jr., M. Henry, P. Molina, and M. Leigh. 2005. Nontuberculous mycobacterial infection in young children with cystic fibrosis. Pediatr. Pulmonol. 40:39-44. [DOI] [PubMed] [Google Scholar]
- 10.Evans, M., and P. Dyson. 1993. Pulsed-field gel electrophoresis of Streptomyces lividans DNA. Trends Genet. 9:72. [DOI] [PubMed] [Google Scholar]
- 11.Fairhurst, R., B. Kubak, R. Shpiner, M. Levine, D. Pegues, and A. Ardehali. 2002. Mycobacterium abscessus empyema in a lung transplant recipient. J. Heart Lung Transplant 21:391-394. [DOI] [PubMed] [Google Scholar]
- 12.Galamba, A., K. Soetaert, X. M. Wang, J. De Bruyn, P. Jacobs, and J. Content. 2001. Disruption of adhC reveals a large duplication in the Mycobacterium smegmatis mc2155 genome. Microbiology 147:3281-3294. [DOI] [PubMed] [Google Scholar]
- 13.Gilljam, M., S. E. Berning, C. A. Peloquin, B. Strandvik, and L. O. Larsson. 1999. Therapeutic drug monitoring in patients with cystic fibrosis and mycobacterial disease. Eur. Respir. J. 14:347-351. [DOI] [PubMed] [Google Scholar]
- 14.Griffith, D. E. 2003. Emergence of nontuberculous mycobacteria as pathogens in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167:810-812. [DOI] [PubMed] [Google Scholar]
- 15.Hjelte, L., B. Petrini, G. Kallenius, and B. Strandvik. 1990. Prospective study of mycobacterial infections in patients with cystic fibrosis. Thorax 45:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffner, S. 1993. Referensmetodik för laboratoriediagnostik vid kliniskt bakteriologiska laboratorier, p. 34-36, 39-40. Swedish Institute of Infectious Disease Control, Stockholm, Sweden.
- 17.Howard, S. T., E. Rhoades, J. Recht, X. Pang, A. Alsup, R. Kolter, C. R. Lyons, and T. F. Byrd. 2006. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 152:1581-1590. [DOI] [PubMed] [Google Scholar]
- 18.Jonsson, B., and M. Ridell. 2003. The Cobas Amplicor MTB test for detection of Mycobacterium tuberculosis complex from respiratory and non-respiratory clinical specimens. Scand. J. Infect. Dis. 35:372-377. [DOI] [PubMed] [Google Scholar]
- 19.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Bottger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, W. J., T. W. Kim, K. B. Shur, B. J. Kim, Y. H. Kook, J. H. Lee, and J. K. Park. 2000. Sporotrichoid dermatosis caused by Mycobacterium abscessus from a public bath. J. Dermatol. 27:264-268. [DOI] [PubMed] [Google Scholar]
- 21.Makinen, J., A. Sarkola, M. Marjamaki, M. K. Viljanen, and H. Soini. 2002. Evaluation of genotype and LiPA MYCOBACTERIA assays for identification of Finnish mycobacterial isolates. J. Clin. Microbiol. 40:3478-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malouf, M., and A. Glanville. 1999. The spectrum of mycobacterial infection after lung transplantation. Am. J. Respir. Crit. Care Med. 160:1611-1616. [DOI] [PubMed] [Google Scholar]
- 23.Manzoor, S. E., P. A. Lambert, P. A. Griffiths, M. J. Gill, and A. P. Fraise. 1999. Reduced glutaraldehyde susceptibility in Mycobacterium chelonae associated with altered cell wall polysaccharides. J. Antimicrob. Chemother. 43:759-765. [DOI] [PubMed] [Google Scholar]
- 24.Metchock, B. G., F. S. Nolte, and R. J. Wallace, Jr. 1999. Mycobacterium, p. 399-437. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 25.Mizunoe, Y., S. Wai, A. Takade, and S. Yoshida. 1999. Isolation and characterization of rugose form of Vibrio cholerae O139. Infect. Immun. 67:958-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mussaffi, H., J. Rivlin, I. Shalit, M. Ephros, and H. Blau. 2005. Nontuberculous mycobacteria in cystic fibrosis associated with allergic bronchopulmonary aspergillosis and steroid therapy. Eur. Respir. J. 25:324-328. [DOI] [PubMed] [Google Scholar]
- 27.Olivier, K. N., D. J. Weber, J. H. Lee, A. Handler, G. Tudor, P. L. Molina, J. Tomashefski, and M. R. Knowles. 2003. Nontuberculous mycobacteria. II. Nested-cohort study of impact on cystic fibrosis lung disease. Am. J. Respir. Crit. Care Med. 167:835-840. [DOI] [PubMed] [Google Scholar]
- 28.Olivier, K. N., D. J. Weber, R. J. Wallace, Jr., A. R. Faiz, J. H. Lee, Y. Zhang, B. A. Brown-Elliot, A. Handler, R. W. Wilson, M. S. Schechter, L. J. Edwards, S. Chakraborti, and M. R. Knowles. 2003. Nontuberculous mycobacteria. I. Multicenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 167:828-834. [DOI] [PubMed] [Google Scholar]
- 29.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 30.Pierre-Audigier, C., A. Ferroni, I. Sermet-Gaudelus, M. Le Bourgeois, C. Offredo, H. Vu-Thien, B. Fauroux, P. Mariani, A. Munck, E. Bingen, D. Guillemot, G. Quesne, V. Vincent, P. Berche, and J. Gaillard. 2005. Age-related prevalence and distribution of nontuberculous mycobacterial. J. Clin. Microbiol. 43:3467-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ray, T., A. Mills, and P. Dyson. 1995. Tris-dependent oxidative DNA strand scission during electrophoresis. Electrophoresis. 16:888-894. [DOI] [PubMed] [Google Scholar]
- 32.Ray, T., J. Weaden, and P. Dyson. 1992. Tris-dependent site-specific cleavage of Streptomyces lividans DNA. FEMS Microbiol. Lett. 75:247-252. [DOI] [PubMed] [Google Scholar]
- 33.Sanguinetti, M., F. Ardito, E. Fiscarelli, M. La Sorda, P. D'Argenio, G. Ricciotti, and G. Fadda. 2001. Fatal pulmonary infection due to multidrug-resistant Mycobacterium abscessus in a patient with cystic fibrosis. J. Clin. Microbiol. 39:816-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schulze-Robbecke, R., B. Janning, and R. Fischeder. 1992. Occurrence of mycobacteria in biofilm samples. Tuber. Lung Dis. 73:141-144. [DOI] [PubMed] [Google Scholar]
- 35.Sermet-Gaudelus, I., L. B. M., C. Pierre-Audigier, C. Offredo, D. Guillemot, S. Halley, C. Akoua-Koffi, V. Vincent, V. Sivadon-Tardy, A. Ferroni, P. Berche, P. Scheinmann, G. Lenoir, and J. L. Gaillard. 2003. Mycobacterium abscessus and children with cystic fibrosis. Emerg. Infect. Dis. 9:1587-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speert, D. P., M. E. Campbell, D. A. Henry, R. Milner, F. Taha, A. Gravelle, A. G. Davidson, L. T. Wong, and E. Mahenthiralingam. 2002. Epidemiology of Pseudomonas aeruginosa in cystic fibrosis in British Columbia, Canada. Am. J. Respir. Crit. Care Med. 166:988-993. [DOI] [PubMed] [Google Scholar]
- 37.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wallace, R. J., Jr., Y. Zhang, B. A. Brown, V. Fraser, G. H. Mazurek, and S. Maloney. 1993. DNA large restriction fragment patterns of sporadic and epidemic nosocomial strains of Mycobacterium chelonae and Mycobacterium abscessus. J. Clin. Microbiol. 31:2697-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whittier, S., R. L. Hopfer, M. R. Knowles, and P. H. Gilligan. 1993. Improved recovery of mycobacteria from respiratory secretions of patients with cystic fibrosis. J. Clin. Microbiol. 31:861-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whittier, S., K. Olivier, P. Gilligan, M. Knowles, P. Della-Latta, et al. 1997. Proficiency testing of clinical microbiology laboratories using modified decontamination procedures for detection of nontuberculous mycobacteria in sputum samples from cystic fibrosis patients. J. Clin. Microbiol. 35:2706-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, Y., M. Rajagopalan, B. A. Brown, and R. J. Wallace, Jr. 1997. Randomly amplified polymorphic DNA PCR for comparison of Mycobacterium abscessus strains from nosocomial outbreaks. J. Clin. Microbiol. 35:3132-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y., M. A. Yakrus, E. A. Graviss, N. Williams-Bouyer, C. Turenne, A. Kabani, and R. J. Wallace, Jr. 2004. Pulsed-field gel electrophoresis study of Mycobacterium abscessus isolates previously affected by DNA degradation. J. Clin. Microbiol. 42:5582-5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhibang, Y., Z. BiXia, L. Qishan, C. Lihao, L. Xiangquan, and L. Huaping. 2002. Large-scale outbreak of infection with Mycobacterium chelonae subsp. abscessus after penicillin injection. J. Clin. Microbiol. 40:2626-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]