Abstract
Production of icaADBC-encoded polysaccharide intercellular adhesin, or poly-N-acetylglucosamine (PIA/PNAG), represents an important biofilm mechanism in staphylococci. We previously described a glucose-induced, ica-independent biofilm mechanism in four methicillin-resistant Staphylococcus aureus (MRSA) isolates. Here, biofilm regulation by NaCl and glucose was characterized in 114 MRSA and 98 methicillin-sensitive S. aureus (MSSA) isolates from diagnosed device-related infections. NaCl-induced biofilm development was significantly more prevalent among MSSA than MRSA isolates, and this association was independent of the isolate's genetic background as assessed by spa sequence typing. Among MSSA isolates, PIA/PNAG production correlated with biofilm development in NaCl, whereas in MRSA isolates grown in NaCl or glucose, PIA/PNAG production was not detected even though icaADBC was transcribed and regulated. Glucose-induced biofilm in MRSA was ica independent and apparently mediated by a protein adhesin(s). Experiments performed with strains that were amenable to genetic manipulation revealed that deletion of icaADBC had no effect on biofilm in a further six MRSA isolates but abolished biofilm in four MSSA isolates. Mutation of sarA abolished biofilm in seven MRSA and eight MSSA isolates. In contrast, mutation of agr in 13 MRSA and 8 MSSA isolates substantially increased biofilm (more than twofold) in only 5 of 21 (23%) isolates and had no significant impact on biofilm in the remaining 16 isolates. We conclude that biofilm development in MRSA is ica independent and involves a protein adhesin(s) regulated by SarA and Agr, whereas SarA-regulated PIA/PNAG plays a more important role in MSSA biofilm development.
The metabolic versatility of Staphylococcus aureus contributes to the impressive capacity of this pathogen to colonize and persist in a range of diverse environments. This organism can be isolated from fomites in the hospital environment, as well as from various niches in the human host, where it can exist harmlessly as a commensal, inhabiting the skin or mucous membranes, or can cause a wide variety of infections in various body sites, from superficial skin infections to deep-seated infections. S. aureus is a prominent cause of both community- and hospital-acquired infection, which is becoming increasingly difficult to treat given the ever-increasing incidence of methicillin-resistant S. aureus (MRSA) and, more recently, the emergence of glycopeptide resistance.
S. aureus possesses a wide array of virulence factors, including extracellular toxins and surface structures that facilitate tissue colonization, immune evasion, and tissue destruction. Production of many of these factors is controlled by a network of regulatory genetic loci, such as agr and sarA, which coordinate precise gene expression during different stages of infection (13). In addition to acute disease, S. aureus can cause chronic infections, many of which are mediated by the ability of this pathogen to adhere to medical devices and form biofilm. Growth of S. aureus in a biofilm requires an adaptive response by the organism. Recent transcriptome analysis identified 48 genes that were induced and 84 genes that were repressed during biofilm growth compared to planktonic growth (4).
Among staphylococci, production of a polysaccharide adhesin, termed polysaccharide intercellular adhesin (PIA) or polymeric N-acetyl-glucosamine (PNAG), by ica operon-encoded enzymes is currently the best-understood mechanism of biofilm development (34, 36). Although the majority of clinical S. aureus isolates contain the ica operon (2, 11, 17, 18), expression of the ica operon and biofilm production are tightly regulated under in vitro conditions (37). Various external signals, such as CO2 levels, anaerobicity, glucose, and osmotic stress, can alter the regulation of ica operon expression and/or biofilm development (1, 12, 37, 44). In S. aureus ATCC 35556, mutation of the ica operon impaired PIA/PNAG production and resulted in a biofilm-negative phenotype (11). In addition to the ica locus, mutation of the sarA global regulator also abolishes biofilm development in S. aureus (3, 45). In contrast, mutation of the global regulator agr has a neutral effect on biofilm development (3) or can lead to increased biofilm formation (47).
Significantly, the ica locus was recently shown to be redundant for in vitro or in vivo biofilm development in S. aureus strain UAMS-1 (4). In addition, we recently observed an absence of correlation between the activation of icaADBC expression and increased biofilm production in four MRSA isolates and that glucose-mediated biofilm development in these isolates was icaADBC independent (16). Further analysis of biofilm environmental regulation in 32 MRSA and methicillin-susceptible S. aureus (MSSA) clinical isolates revealed that MSSA isolates were more likely to form biofilm in media supplemented with NaCl, whereas MRSA and MSSA isolates were equally likely to form biofilm in glucose-supplemented medium (15). These preliminary findings suggested that significant differences may exist in the mechanisms and regulation of biofilm development in MRSA and MSSA clinical isolates.
In this study, we investigated the contribution of methicillin susceptibility to the biofilm phenotype in S. aureus. To address this question, we characterized the biofilm phenotypes of a collection of 114 MRSA and 98 MSSA clinical isolates, collected from patients with device-related infections in Beaumont Hospital, Dublin, Ireland. The regulation of ica operon expression and PIA/PNAG production by NaCl and glucose were examined in representative MRSA and MSSA isolates from this collection. The chemical composition of the extracellular matrix mediating intercellular adhesion was investigated by measuring the ability of sodium metaperiodate or proteinase K to disperse MRSA and MSSA biofilms. Phage transduction was used to construct Δica::tet, agr::tet, and sarA::tet mutations in MRSA and MSSA isolates that were amenable to genetic analysis in order to examine the role of these loci in biofilm development. To investigate the contribution of genetic background to biofilm development, the isolates were characterized by spa sequence typing. The data reveal new insights into associations between methicillin susceptibility and the ica, sarA, and agr loci in the regulation of biofilm development in MRSA and MSSA clinical isolates.
MATERIALS AND METHODS
S. aureus isolates.
Clinical S. aureus isolates were retrospectively collected from the clinical microbiology laboratory of Beaumont Hospital, Dublin, Ireland, from 1 January 2002 to 30 June 2005. Beaumont Hospital is a 720-bed tertiary referral center and contains the national neurosurgical center for the Republic of Ireland. Patient details were collected from clinical microbiology team records. Isolates were assigned as either clinically significant (e.g., causing line sepsis or bloodstream infection) or probable contaminants, based on previously published guidelines (16, 19, 49). Antibiotic susceptibility was examined by a modified Stokes’ method and by Etest (AB Biodisk, Solna, Sweden) (23). The reference strain S. aureus 8325 and its derivatives 8325-4 (cured of known prophages) and RN4220 (a chemically mutagenized restriction-deficient strain derived from 8325-4) (21, 33) were used. The previously described temperature-sensitive plasmid pSC23, carrying a Δica::tet allele (11), was used in allele replacement experiments to generate an ica deletion mutation in RN4220 (16). Strain PC6911, carrying an agrΔ::tet allele, was used as a source of an agr null mutation for transduction into clinical isolates (6, 38). Strain UAMS-240, an RN6390 derivative harboring a sarA::tet allele, was used as a source of a sarA mutation (5).
Media and growth conditions.
S. aureus strains were grown at 37°C on brain-heart infusion (BHI) medium (Oxoid) supplemented when required with chloramphenicol (10 to 20 μg/ml), erythromycin (2.5 μg/ml), and tetracycline (2.5 μg/ml). BHI broth was supplemented where indicated with 1% glucose or 4% NaCl. Bacteria were grown on Congo red agar (CRA) for the characterization of the colony morphologies of the S. aureus strains, as described previously (24).
Biofilm assays.
Semiquantitative measurements of biofilm formation were determined with Nunclon tissue culture-treated (ΔSurface) 96-well polystyrene plates (Nunc, Denmark), based on the method of Christensen et al. (7) as described by Ziebuhr et al. (50), with the following modification. Bacteria were grown in individual wells of 96-well plates at 37°C in BHI medium or BHI supplemented with 4% NaCl or 1% glucose. After 24 h of growth, the plates were washed vigorously three times with distilled H2O to remove unattached bacteria and dried for 1 hour at 60°C, as recommended by Gelosia et al. (20), prior to staining with a 0.4% crystal violet solution. Each strain was tested at least three times, and average results are presented. A biofilm-positive phenotype was defined as an optical density at 492 nm of ≥0.17.
Biofilm stability against proteinase K or sodium metaperiodate treatment was tested as described previously (35, 42).
DNA manipulations, PCR, and construction of ica, sarA, and agr deletion mutants.
Genomic and plasmid DNA purification and manipulations were performed as described previously (8-10, 24). The Δica::tet, sarA::tet, and agr::tet alleles were transferred via RN4220 to S. aureus clinical isolates by phage 80α/501 and phage 11 transduction. Transductants harboring the mutant alleles were selected on BHI agar containing 500 mg/liter sodium citrate and 2.5 μg/ml tetracycline, and the presence of the mutant alleles was confirmed by PCR analysis using primers SAdel1 (5′-TGC-AAA-TGC-CCT-TGA-TGT-AA-3′) and SALR2 (5′-GGC-GGA-AAG-TCA-GGT-TAC-AA-3′) to amplify the ica operon, primers SAsarA1 (5′-GCG-TTG-ATT-TGG-GTA-GTA-TGC-3′) and SAsarA2 (5′-TCA-CCA-AAT-TGC-GCT-AAA-CA-3′) to amplify the sarA locus, and primers SAagr1 (5′-GGG-GCT-CAC-GAC-CAT-ACT-TA-3′) and SAagr2 (5′-CGG-GGT-AGG-AAA-TTG-TAG-CA-3′) to amplify the agr locus. PCRs were carried out with Taq DNA polymerase (Invitrogen) under the following conditions: 95°C for 20 s, 53°C for 20 s, and 72°C for 3 to 6 min for 35 cycles, with 3 mM MgCl2. All oligonucleotide primers used for PCR were supplied by MWG-Biotech (Ebersberg, Germany).
RNA purification and analysis.
RNA purification, reverse transcription (RT)-PCR, and analysis of RT-PCR data were performed as described previously (8-10, 24). The constitutively expressed gyrB gene was used as an internal standard in these experiments using the primers described by Goerke et al. (22). The following primers were used to amplify the icaA transcript: icaA1, 5′-CGCACTCAATCAAGGCATTA-3′; icaA2, 5′-CCAGCAACTGTCTGACTTCG-3′. RT was performed at 50°C for 30 min, terminated at 94°C for 15 min, and followed by 21 cycles of PCR for gyrB and icaA amplification.
PIA/PNAG assays.
PIA/PNAG assays were performed as described elsewhere (27). Briefly, 5-ml overnight cultures (approximately 5 × 109 bacteria) were collected by centrifugation, resuspended in 200 to 500 μl of 0.5 M EDTA, and boiled for 5 min. The cell debris was again centrifuged, and the supernatant was treated with 200 μg proteinase K at 65°C for 1 h. The proteinase K was inactivated by boiling for 5 min, and the samples were diluted as appropriate before being applied to nitrocellulose (prewetted in Tris-buffered saline [TBS]) with a vacuum blotter. The blots were dried, rewetted in TBS, and blocked for 1 h in 1% bovine serum albumin (BSA). The primary antibody (1:5,000 dilution of rabbit anti-PIA/PNAG [a kind gift from Tomas Maira Litran and Gerald Pier] in TBS-Tween 80-0.1% BSA) was than applied to the membrane for 1 h. Horseradish peroxidase-linked anti-rabbit immunoglobulin G secondary antibody (1:5,000 dilution in TBS-Tween 80-0.1% skim milk) was then incubated with the membrane for 1 h. A chemiluminescence kit (Amersham) was used to generate light via the horseradish peroxidase-catalyzed breakdown of lumina, which was detected with a Bio-Rad Fluor-S Max charge-coupled-device camera system.
Strain typing.
Genetic background was defined by sequencing the X-repeat region of the spa gene (32) on both strands with the primers of Robinson and Enright (41). Newly discovered spa types were submitted to the eGenomics spa server (www.tools.egenomics.com). Inference of clonal complexes (CCs) based on the spa type was done by comparison to previous studies that compared multilocus sequence typing and spa typing (41) and by use of the Ridom spa server (www.spaserver.ridom.de) (25). The staphylococcal cassette chromosome mec (SCCmec) type was defined by PCR analysis of the mec and ccr genes using the primers of Ito et al. (26) and Robinson and Enright (41). SCCmec type variants were defined by multiplex PCR analysis according to the method of Oliveira and de Lencastre (39).
Statistical analyses.
Associations between methicillin susceptibility (MRSA, MSSA) and biofilm phenotypes (positive, negative) were evaluated in 2-by-2 contingency tables with odds ratios and chi-square tests by Epi Info, version 3.3.2. Stratified analyses to control for the potentially confounding effects of genetic background were done by Mantel-Haenszel tests with Epi Info, version 3.3.2.
RESULTS
Biofilm regulation in MRSA and MSSA.
Two hundred twelve S. aureus isolates (114 MRSA and 98 MSSA) implicated clinically in device-related infections were collected retrospectively from blood cultures and line tips. Previous multilocus sequence typing studies of the genetic structure of S. aureus populations have shown that most hospital-acquired MRSA strains isolated worldwide belong to one of five CCs—CC5, CC8, CC22, CC30, and CC45—which are groups of closely related clones (14, 41). These five CCs contain both MRSA and MSSA isolates. We used spa sequence typing to examine the genetic backgrounds of our isolates, and we confirmed methicillin susceptibility status by PCR of the SCCmec element. The MRSA isolates represented 16 spa types, whereas the MSSA isolates were more genetically diverse and represented 67 spa types; 33 new spa types were identified in our isolate collection (see Table S1 in the supplemental material). In total, we classified 170 of our isolates based on spa types into the five CCs that have spawned the most hospital-acquired MRSA isolates (see Table S1 in the supplemental material). Among these 170 isolates, all of the MRSA isolates with SCCmec type II were from CC8 with the exception of a single isolate from CC5, and all of the MRSA isolates with SCCmec type IV were from CC22 with the exception of a single isolate from CC45 (see Table S1 in the supplemental material). Three clones, identified by spa types 46, 382, and 781, accounted for 90% of the MRSA isolates. Thus, the findings reported here are largely within the context of these particular clones of MRSA.
Under standard laboratory conditions in BHI medium, only 8% of our 114 MRSA and 18% of our 98 MSSA isolates were capable of biofilm development. These percentages dramatically increased to 74% for MRSA and 84% for MSSA isolates when grown in BHI supplemented with 1% glucose. However, growth in the presence of 4% NaCl results in differential effects on MRSA and MSSA biofilm phenotypes. Whereas NaCl activated biofilm development in 44% of MSSA isolates, only 3% of MRSA isolates responded to NaCl. The increased propensity of MSSA isolates to form biofilm in NaCl compared to MRSA isolates was highly statistically significant (P < 0.001 [chi-square test]). Among the 170 isolates from the five CCs of hospital-acquired MRSA, the MSSA isolates were 32 times more likely than the MRSA isolates to develop a NaCl-induced biofilm (P < 0.001 [chi-square test]). Even after stratifying the analysis of these 170 isolates by their genetic backgrounds, the MSSA isolates remained eight times more likely than MRSA isolates to develop a NaCl-induced biofilm (P = 0.002 [Mantel-Haenszel test]). Thus, the association between MSSA isolates and the NaCl-induced biofilm occurs independently of the isolates’ genetic backgrounds. The glucose-induced biofilm phenotype was not associated with methicillin susceptibility in these isolates (P > 0.2 [both tests]).
Regulation of ica operon expression and PIA/PNAG production in MRSA and MSSA.
To further investigate the contribution of the ica locus and PIA/PNAG production to biofilm development in S. aureus, 9 representative MSSA isolates that were biofilm-positive in NaCl and 16 representative MRSA isolates that were biofilm positive in glucose were chosen for further study. The laboratory strain RN4220 and its isogenic Δica mutant were used as positive and negative controls, respectively.
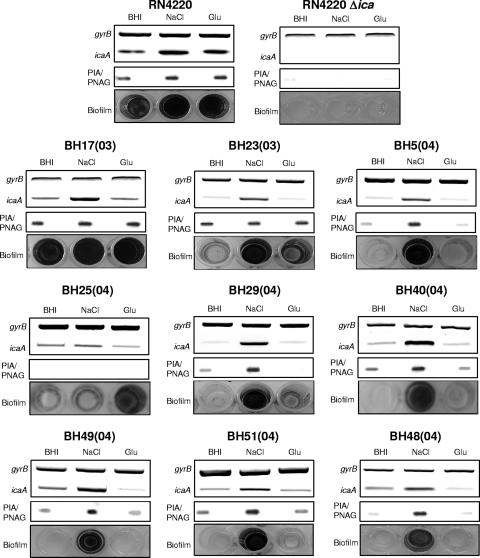
By RT-PCR, ica operon transcription was detected in all MSSA and MRSA isolates (Fig. 1 and 2). In eight of the nine MSSA clinical isolates, NaCl-induced biofilm development correlated with increased ica expression and PIA/PNAG production (Fig. 1). In one isolate, BH25(04), growth in NaCl was not associated with substantially increased ica operon expression or PIA/PNAG production (Fig. 1). Interestingly, in this strain, biofilm development was present in medium supplemented with glucose (Fig. 1). Growth of the nine MSSA isolates in glucose was not associated with activation of ica expression (Fig. 1). However, seven MSSA isolates—BH17(03), BH23(03), BH40(04), BH49(04), BH5(04), BH51(04), and BH48(04)—were found to produce PIA/PNAG under all growth conditions, including medium supplemented with glucose (Fig. 1). Taken together, these findings reveal a correlation between ica operon regulation and biofilm development in eight of the nine MSSA isolates examined. However, the correlations between ica expression and PIA/PNAG production or between PIA/PNAG production and biofilm development are less convincing, appear to be strain dependent, and suggest that levels of PIA/PNAG production alone do not always correlate with levels of biofilm formation.
FIG. 1.
Comparison of ica operon expression, PIA/PNAG production, and biofilm regulation in nine MSSA clinical isolates grown in BHI medium or in BHI supplemented with 4% NaCl or 1% glucose (Glu). Laboratory strain RN4220 and its isogenic Δica mutant were used as positive and negative controls, respectively. Transcription of gyrB (control) and icaA was measured by RT-PCR using RNA prepared from cultures grown at 37°C to an A600 of 2.0. PIA/PNAG was measured in cell extracts from overnight cultures. Biofilm formation in tissue culture-treated 96-well plates was measured three times, and representative results are shown.
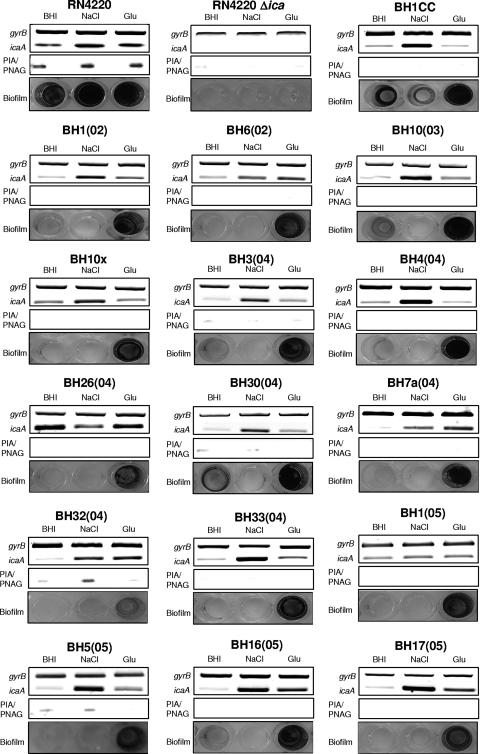
FIG. 2.
Comparison of ica operon expression, PIA/PNAG production, and biofilm regulation in 16 MRSA clinical isolates grown in BHI medium or in BHI supplemented with 4% NaCl or 1% glucose (Glu). Laboratory strain RN4220 and its isogenic Δica mutant were used as positive and negative controls, respectively. Transcription of gyrB (control) and icaA was measured by RT-PCR using RNA prepared from cultures grown at 37°C to an A600 of 2.0. PIA/PNAG was measured in cell extracts from overnight cultures. Biofilm formation in tissue culture-treated 96-well plates was measured three times, and representative results are shown.
Among the 16 MRSA isolates, growth in the presence of NaCl resulted in activation of ica transcription in 14 strains but failed to induce substantial biofilm development (Fig. 2) in any of these isolates [weak, but measurable, biofilm formation by strains BH1CC and BH32(04) was detected in medium supplemented with NaCl]. Importantly, activation of ica by NaCl in these 14 MRSA isolates was associated with measurably increased PIA/PNAG production in only 1 isolate, BH32(04) (Fig. 2). Glucose-mediated induction of biofilm formation in the 16 MRSA isolates correlated with weakly to moderately increased ica operon expression in 11 isolates: BH1(02), BH6(02), BH10(03), BH7a(04), BH32(04), BH3(04), BH30(04), BH33(04), BH5(05), BH16(05), and BH17(05) (Fig. 2). However, glucose-mediated ica activation did not correlate with increased PIA/PNAG production in any of these isolates (Fig. 2). Interestingly, ica operon transcription was more potently activated by NaCl than by glucose in all of the MRSA isolates examined except BH7a(04), BH6(02), and BH26 (04) (Fig. 2). Thus, in contrast to recent findings with S. epidermidis (8-10, 16, 30, 40) and our analysis of MSSA isolates (Fig. 1), there appears to be little correlation between ica operon regulation and biofilm formation in MRSA, suggesting that the ica operon and PIA/PNAG may not be required for biofilm development in MRSA.
Contribution of the ica locus to the biofilm phenotype in MRSA and MSSA.
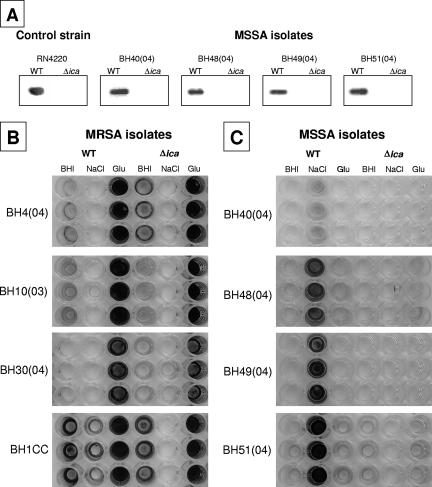
To further investigate the role of the ica locus in MRSA and MSSA biofilm development, phage transduction was used to construct Δica::tet mutations in a further six MRSA isolates and four MSSA isolates. The choice of strains and the numbers of mutants constructed were dictated by our ability to transduce the Δica::tet allele into clinical isolates. The presence of ica deletion mutations in the clinical isolates was confirmed by PCR (data not shown). In addition, PIA/PNAG-specific antiserum was used in slot blot analysis to confirm the absence of PIA/PNAG production in MSSA ica mutants grown in BHI medium supplemented with 4% NaCl (Fig. 3A). Consistent with our previous findings (16), deletion of the ica operon had no effect on the biofilm phenotype in all six MRSA isolates tested (four representative strains are shown in Fig. 3B). In contrast, deletion of the ica locus in the four MSSA isolates abolished the biofilm phenotype (Fig. 3C). These data provide further evidence for ica-independent biofilm formation in MRSA and ica-dependent biofilm formation in MSSA.
FIG. 3.
(A) Measurement of PIA/PNAG immunoreactivity in four representative wild-type (WT) MSSA clinical isolates and their corresponding Δica mutants grown in BHI medium supplemented with 4% NaCl. (B) Comparison of biofilm regulation in four representative WT MRSA clinical isolates and their corresponding Δica mutants. (C) Comparison of biofilm regulation in four WT MRSA isolates and their corresponding Δica mutants. Biofilm formation was analyzed by growing triplicate cultures in tissue culture-treated 96-well plates in BHI medium or in BHI medium supplemented with 4% NaCl or 1% glucose, followed by staining with crystal violet. Photographs of representative biofilm plates are shown.
Analysis of the extracellular matrix mediating biofilm development in MRSA and MSSA biofilms.
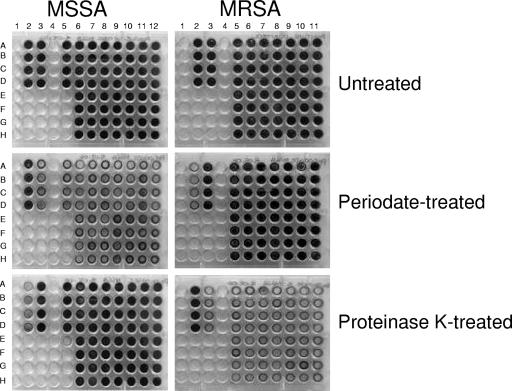
To investigate the likely chemical composition of the extracellular matrix mediating biofilm formation, we examined the ability of sodium metaperiodate and proteinase K to disperse NaCl-induced MSSA biofilms and glucose-induced MRSA biofilms. For these experiments, we chose 14 MRSA and 15 MSSA isolates capable of strong biofilm formation (A490 > 1.0 [in biofilm assays]) in media supplemented with glucose or NaCl, respectively. Sodium metaperiodate oxidation of PIA/PNAG, or indeed an alternative polysaccharide, had little effect on glucose-induced MRSA biofilms (Fig. 4). In contrast, the glucose-induced MRSA biofilms were substantially dispersed by treatment with proteinase K (Fig. 4). These data strongly suggest the involvement of a protein adhesin in glucose-mediated MRSA biofilm development and are consistent with our finding that deletion of the ica locus in 10 MRSA strains had no effect on biofilm development.
FIG. 4.
Susceptibility of glucose-induced MRSA biofilms (left) and NaCl-induced MSSA biofilms (right) to treatment with sodium metaperiodate and proteinase K. MRSA biofilms were grown in BHI medium supplemented with 1% glucose, and MSSA biofilms were grown in BHI medium supplemented with 4% NaCl for 24 h. Each strain was grown in four individual wells. The laboratory strain RN4220 (wells 2A to 2D in each plate) was used a positive control for NaCl-induced biofilm formation in MSSA. The MRSA clinical isolate BH1CC (wells 3A to 3D in each plate) was used a positive control for glucose-induced biofilm formation in MRSA. The 15 clinical MSSA isolates used in the experiment were BH6(2005) (wells 5A to 5D), BH49(2004) (wells 6A to 6D), BH51(2004) (wells 6E to 6H), BH37(2004) (wells 7A to 7D), BH48(2004) (wells 7E to 7H), BH21(2004) (wells 8A to 8D), BH29(2004) (wells 8E to 8H), BH10(2004) (wells 9A to 9D), BH18(2002) (wells 9E to 9H), BH23(2003) (wells 10A to 10D), BH27(2003) (wells 10E to 10H), BH11(2003) (wells 11A to 11D), BH17(2003) (wells 11E to 11H), BH2(2003) (wells 12A to 12D), and BH8(2003) (wells 12E to 12D). The 14 clinical MRSA isolates used in this experiment were BH26(2004) (wells 5A to 5D), BH17(2005) (wells 5E to 5D), BH4(2004) (wells 6A to 6D), BH8(2004) (wells 6E to 6H), BH23(2003) (wells 7A to 7D), BH30(2004) (wells 7E to 7H), BH10(2003) (wells 8A to 8D), BH11b(2003) (wells 8E to 8H), BH14(2002) (wells 9A to 9D), BH16(2002) (wells 9E to 9H), BH10(2002) (wells 10A to 10D), BH12(2002) (wells 10E to 10H), BH2(2002) (wells 11A to 11D), and BH6(2002) (wells 11E to 11H).
Unlike MRSA biofilms, NaCl-induced MSSA biofilms were substantially dispersed by sodium metaperiodate oxidation but were largely unaffected by treatment with proteinase K (Fig. 4), strongly suggesting the involvement of a polysaccharide adhesin in NaCl-induced MSSA biofilm development. Again, this observation is entirely consistent with our previous finding that deletion of the ica locus in four MSSA clinical isolates and the laboratory strain RN4220 abolished biofilm development (Fig. 3C) and indicates that PIA/PNAG is the polysaccharide adhesin mediating biofilm development in MSSA.
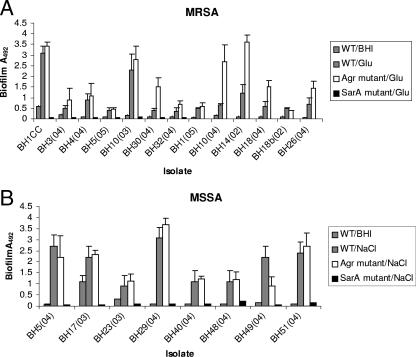
Contribution of the sarA and agr loci to biofilm development in MRSA and MSSA.
Our evidence for different biofilm mechanisms in MRSA and MSSA isolates prompted us to investigate the role of the S. aureus global virulence regulators sarA and agr, which are both known to influence the biofilm phenotype (3, 4, 45, 47).
Using phage 80α, we transduced sarA::tet and agr::tet alleles into a range of clinical isolates. Similar to our ability to construct Δica::tet alleles, the choice of strains and the numbers of mutants constructed were dictated by our ability to transduce the sarA and agr mutant alleles into clinical isolates. Among the MRSA sarA and agr mutants constructed, the majority were members of CC8 and CC22, whereas, consistent with the overall diversity of the MSSA isolates in our collection, sarA and agr mutations were constructed in MSSA isolates of more-diverse genetic backgrounds. Nevertheless, all of the MSSA and MRSA isolates in which we were able to construct sarA or agr mutations displayed biofilm phenotypes consistent with the data described above, namely, that biofilm development was induced by NaCl in the MSSA isolates whereas biofilm development was induced by glucose in the MRSA isolates. The sarA::tet allele was transduced into eight MSSA isolates and seven MRSA isolates. In all 15 sarA mutant strains, biofilm was abolished in BHI medium and BHI supplemented with NaCl (for the MSSA isolates) or glucose (for the MRSA isolates) (Fig. 5). Unlike mutation of sarA, transduction of agr::tet alleles into 13 MRSA isolates and 8 MSSA isolates resulted in either increased or no substantial difference in biofilm-forming capacity in the majority of the MRSA and MSSA isolates examined (Fig. 5). Interestingly, mutation of agr resulted in a substantial (more than twofold) increase in biofilm-forming capacity in only 5 of the 21 S. aureus isolates examined [BH30, BH10(04), BH14, BH18, and BH26]. All five of these isolates were methicillin resistant (Fig. 5), with four being members of CC22 [BH10(04), BH14, BH18, and BH26] and one being a member of CC8 (BH30). However, agr mutations in three additional CC22 MRSA isolates (BH1, BH18b, and BH26) and three CC8 MRSA isolates (BHCC, BH3, and BH4) had no substantial effect (less-than-twofold increase) on biofilm-forming capacity. Six clonal complexes were represented among the eight MSSA agr mutants constructed.
FIG. 5.
Contribution of the sarA and agr loci to biofilm regulation in clinical isolates of MRSA (A) and MSSA (B). MRSA isolates were grown in BHI medium supplemented with 1% glucose for 24 h. MSSA isolates were grown in BHI medium supplemented with 4% NaCl for 24 h. WT, wild type.
Overall, the impact of sarA and agr mutations contrasts with our observation that mutation of the ica locus does not affect biofilm development in MRSA isolates and suggests that, because of their regulatory roles in ica-dependent and ica-independent biofilm development, SarA and, to a lesser extent, Agr may be more important determinants of biofilm development in S. aureus clinical isolates than is PIA/PNAG.
DISCUSSION
Biofilm-forming capacity is now recognized as an important virulence determinant in the development of staphylococcal device-related infections. In staphylococci, the production of the exopolysaccharide PIA/PNAG by ica operon-encoded enzymes represents the most intensively studied mechanism of biofilm formation to date. Recent studies with both S. aureus and S. epidermidis have highlighted the existence of biofilm mechanisms independent of the ica operon-encoded exopolysaccharide PIA/PNAG (4, 16, 42). Nevertheless, it is interesting to note that although the ica locus is more likely to be associated with clinically significant S. epidermidis isolates than with carriage isolates, in S. aureus the vast majority of isolates appear to carry this gene cluster (15, 31). Clearly the contribution of the ica locus to biofilm development in S. aureus is complex and is likely to be both strain and environment dependent. We previously reported the existence of a possible correlation between methicillin susceptibility and the environmental regulation of biofilm development in clinical S. aureus isolates and that icaADBC-independent biofilm formation was possible in four clinical MRSA isolates (15, 16).
In this study, our findings provide further evidence for differential mechanisms of biofilm development in MSSA and MRSA clinical isolates. We examined the environmental regulation of ica operon expression, PIA/PNAG production, and biofilm formation in MRSA and MSSA isolates in media supplemented with NaCl or glucose. Among MSSA isolates, we observed strain-to-strain variation in terms of the correlation between ica operon transcriptional regulation and PIA/PNAG production. Nevertheless, a strong correlation was evident between PIA/PNAG production and biofilm development in MSSA isolates. In contrast, although the ica operon was expressed and subject to environmental regulation in MRSA isolates, no correlation existed between ica operon transcriptional activity and PIA/PNAG production or biofilm development.
Biofilm development was significantly induced by glucose in both MRSA and MSSA isolates. In contrast, NaCl was significantly more likely to induce biofilm formation in MSSA isolates than in MRSA isolates, and this effect was independent of an isolate's genetic background. NaCl is a known activator of ica operon transcription (16, 40) and, consistent with this biofilm development in MSSA isolates, is apparently icaADBC-dependent and involves the production of PIA/PNAG. Biofilm development in MRSA isolates is primarily glucose induced, is ica independent, and apparently involves a protein adhesin. In general, these experiments suggest the involvement of a protein adhesin in glucose-induced MRSA biofilm development and a polysaccharide adhesin in NaCl-induced MSSA biofilm development. However an alternative explanation can also be proposed, namely, that glucose-induced biofilm in S. aureus (either MSSA or MRSA) is mediated by a protein adhesin and that NaCl-induced biofilm in S. aureus (either MRSA or MSSA) is mediated by a polysaccharide adhesin. Interestingly, dispersion assay experiments with three MSSA isolates that formed strong biofilm in glucose revealed that one biofilm was dispersed with proteinase K and the second was partially dispersed by periodate only, while the third was not affected by either periodate or proteinase K (our unpublished data). We are currently conducting further investigations into the nature of glucose-induced MSSA biofilms and NaCl-induced MRSA biofilms. In any event, these data reveal a previously unknown level of complexity in terms of both biofilm mechanisms and regulation in S. aureus.
Interestingly, mutation of the global virulence regulator sarA abolished biofilm in 7 MSSA and 10 MRSA isolates examined in this study. These findings suggest that sarA encodes a master regulator controlling at least two independent mechanisms of biofilm development in S. aureus. Consistent with this conclusion, SarA is known to repress production of four major extracellular proteases in S. aureus (SspA, SspB, Aur, and ScpA), which results in reduced levels of cell-bound fibronectin-binding proteins and protein A (29). Hence, the putative protein adhesin(s) mediating ica-independent biofilm in MRSA may be modified or degraded in a SarA mutant. It is also interesting to note that different levels of sarA expression in clinical isolates of S. aureus have been linked to variations in extracellular protease production (28) and that SarA can directly and positively regulate levels of fnbA transcription (5, 48). In terms of ica-dependent biofilm in MSSA, SarA has previously been shown to positively regulate ica operon expression and PIA/PNAG production (45).
Unlike SarA, mutations in agr either enhanced biofilm development or did not significantly alter biofilm-forming capacity in the majority of MRSA and MSSA isolates examined. This finding is broadly consistent with previous studies that indicated that mutation of agr can enhance S. aureus biofilm development (3, 47). However. agr mutations resulted in a substantial (more-than-twofold) increase in biofilm-forming capacity in only 5 of 21 S. aureus isolates examined. All five of these isolates were MRSA, four from CC22 and one from CC8. Because agr mutations in three additional CC22 and three CC8 MRSA isolates did not significantly affect biofilm-forming capacity, it is difficult to attribute strain-to-strain differences in the impact of agr mutations on the biofilm phenotype to different genetic backgrounds.
Given that we observed significant differences in the basal levels of biofilm produced by biofilm-positive clinical S. aureus isolates, it seems possible that the regulatory pathways controlling biofilm adhesins vary between strains. In this context, the significantly different biofilm phenotypes associated with agr mutations in clinical isolates suggest that strain-to-strain differences in the pathways controlling the regulation of putative surface protein adhesins or PIA/PNAG are perhaps more important than the potential impact of decreased levels of extracellular proteases (43) or the absence of the agr-encoded δ toxin (which has surfactant properties) (47) in agr mutants. Interestingly, in contrast to SarA, Agr positively regulates expression of the four major extracellular proteases (43). Consistent with this, all five clinical isolates in which agr mutation substantially (more than twofold) increased biofilm-forming capacity were MRSA isolates. In contrast, previous studies have indicated that mutation of agr does not result in significantly altered icaADBC expression or PIA/PNAG production (46); consistent with this, mutation of agr did not result in substantially (more-than-twofold) increased biofilm-forming capacity in any of the MSSA isolates examined.
These data raise the intriguing question of why the ica locus is maintained, expressed, and regulated in MRSA isolates. Perhaps the acquisition of resistance to multiple antibiotics that target the cell wall inadvertently results in impaired biosynthesis or export of PIA/PNAG. Such changes in the cell surface of MRSA strains may also be accompanied by the unintended redeployment of a cell surface protein(s) not normally involved in adherence to inanimate surfaces on naked (as opposed to surfaces coated with host proteins) polystyrene or other biomaterials. Given that many S. aureus cell wall-anchored proteins are involved in binding to host matrix proteins, the interesting possibility exists that in the absence of PIA/PNAG, one or more of these adhesins may also play a role in biofilm development in MRSA. In any event, our evidence for different mechanisms of biofilm development in MSSA and MRSA isolates is likely to be significant in our understanding of the pathogenesis of S. aureus device-related infections involving biofilms and for the development of novel therapeutics targeting this important staphylococcal phenotype.
Supplementary Material
Acknowledgments
This study was funded by a Clinical Research Training Fellowship from the Health Research Board (Ireland) to E. O'Neill, grants from the Hospital Infection Society (United Kingdom) and the Health Research Board to J. P. O'Gara, and institutional funds from New York Medical College and a grant from the American Heart Association to D. A. Robinson.
We are grateful to P. D. Fey and T. J. Foster for generously providing phage 80α, phage 11, and phage 85; S. Foster for strain PC6911, carrying an agrΔ::tet allele; M. Smeltzer for strain UAMS-240, carrying a sarA::tet allele; and T. Maira Litran and G. B. Pier for rabbit anti-PIA/PNAG serum. Plasmid pSC23, used in the construction of the Δica::tet allele, was a kind gift from S. Cramton and F. Gotz. We thank S. O'Donnell and L. Holland for experimental advice throughout the study, H. Grundmann for helpful comments on our findings, and T. J. Foster for critical reading of the manuscript.
Footnotes
Published ahead of print on 28 February 2007.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Ammendolia, M. G., R. Di Rosa, L. Montanaro, C. R. Arciola, and L. Baldassarri. 1999. Slime production and expression of the slime-associated antigen by staphylococcal clinical isolates. J. Clin. Microbiol. 37:3235-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arciola, C. R., L. Baldassarri, and L. Montanaro. 2001. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 39:2151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beenken, K. E., J. S. Blevins, and M. S. Smeltzer. 2003. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect. Immun. 71:4206-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blevins, J. S., K. E. Beenken, M. O. Elasri, B. K. Hurlburt, and M. S. Smeltzer. 2002. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect. Immun. 70:470-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan, P. F., and S. J. Foster. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232-6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, G. D., W. A. Simpson, J. J. Younger, L. M. Baddour, F. F. Barrett, D. M. Melton, and E. H. Beachey. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. icaR encodes a transcriptional repressor involved in environmental regulation of ica operon expression and biofilm formation in Staphylococcus epidermidis. J. Bacteriol. 184:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2004. Inactivations of rsbU and sarA by IS256 represent novel mechanisms of biofilm phenotypic variation in Staphylococcus epidermidis. J. Bacteriol. 186:6208-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conlon, K. M., H. Humphreys, and J. P. O'Gara. 2002. Regulation of icaR gene expression in Staphylococcus epidermidis. FEMS Microbiol. Lett. 216:173-179. [DOI] [PubMed] [Google Scholar]
- 11.Cramton, S. E., C. Gerke, N. F. Schnell, W. W. Nichols, and F. Gotz. 1999. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 67:5427-5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cramton, S. E., M. Ulrich, F. Gotz, and G. Doring. 2001. Anaerobic conditions induce expression of polysaccharide intercellular adhesin in Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 69:4079-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enright, M. C., D. A. Robinson, G. Randle, E. J. Feil, H. Grundmann, and B. G. Spratt. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. USA 99:7687-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2006. Environmental regulation of biofilm development in methicillin-resistant and methicillin-susceptible Staphylococcus aureus clinical isolates. J. Hosp. Infect. 62:120-122. [DOI] [PubMed] [Google Scholar]
- 16.Fitzpatrick, F., H. Humphreys, and J. P. O'Gara. 2005. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J. Clin. Microbiol. 43:1973-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler, V. G., Jr., P. D. Fey, L. B. Reller, A. L. Chamis, G. R. Corey, and M. E. Rupp. 2001. The intercellular adhesin locus ica is present in clinical isolates of Staphylococcus aureus from bacteremic patients with infected and uninfected prosthetic joints. Med. Microbiol. Immunol. (Berlin) 189:127-131. [DOI] [PubMed] [Google Scholar]
- 18.Frank, K. L., A. D. Hanssen, and R. Patel. 2004. icaA is not a useful diagnostic marker for prosthetic joint infection. J. Clin. Microbiol. 42:4846-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garner, J. S., W. R. Jarvis, T. G. Emori, T. C. Horan, and J. M. Hughes. 1988. CDC definitions for nosocomial infections. Am. J. Infect. Control. 16:128-140. [DOI] [PubMed] [Google Scholar]
- 20.Gelosia, A., L. Baldassarri, M. Deighton, and T. Van Nguyen. 2001. Phenotypic and genotypic markers of Staphylococcus epidermidis virulence. Clin. Microbiol. Infect. 7:193-199. [DOI] [PubMed] [Google Scholar]
- 21.Gertz, S., S. Engelmann, R. Schmid, K. Ohlsen, J. Hacker, and M. Hecker. 1999. Regulation of sigmaB-dependent transcription of sigB and asp23 in two different Staphylococcus aureus strains. Mol. Gen. Genet. 261:558-566. [DOI] [PubMed] [Google Scholar]
- 22.Goerke, C., S. Campana, M. G. Bayer, G. Doring, K. Botzenhart, and C. Wolz. 2000. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect. Immun. 68:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosden, P. E., J. M. Andrews, K. E. Bowker, H. A. Holt, A. P. MacGowan, D. S. Reeves, J. Sunderland, and R. Wise. 1998. Comparison of the modified Stokes’ method of susceptibility testing with results obtained using MIC methods and British Society of Antimicrobial Chemotherapy breakpoints. J. Antimicrob. Chemother. 42:161-169. [DOI] [PubMed] [Google Scholar]
- 24.Handke, L. D., K. M. Conlon, S. R. Slater, S. Elbaruni, F. Fitzpatrick, H. Humphreys, W. P. Giles, M. E. Rupp, P. D. Fey, and J. P. O'Gara. 2004. Genotypic and phenotypic analysis of biofilm phenotypic variation in multiple Staphylococcus epidermidis isolates. J. Med. Microbiol. 53:367-374. [DOI] [PubMed] [Google Scholar]
- 25.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito, T., Y. Katayama, K. Asada, N. Mori, K. Tsutsumimoto, C. Tiensasitorn, and K. Hiramatsu. 2001. Structural comparison of three types of staphylococcal cassette chromosome mec integrated in the chromosome in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 45:1323-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jefferson, K. K., D. B. Pier, D. A. Goldmann, and G. B. Pier. 2004. The teicoplanin-associated locus regulator (TcaR) and the intercellular adhesin locus regulator (IcaR) are transcriptional inhibitors of the ica locus in Staphylococcus aureus. J. Bacteriol. 186:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karlsson, A., and S. Arvidson. 2002. Variation in extracellular protease production among clinical isolates of Staphylococcus aureus due to different levels of expression of the protease repressor sarA. Infect. Immun. 70:4239-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlsson, A., P. Saravia-Otten, K. Tegmark, E. Morfeldt, and S. Arvidson. 2001. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect. Immun. 69:4742-4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knobloch, J. K., K. Bartscht, A. Sabottke, H. Rohde, H. H. Feucht, and D. Mack. 2001. Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J. Bacteriol. 183:2624-2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knobloch, J. K., M. A. Horstkotte, H. Rohde, and D. Mack. 2002. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med. Microbiol. Immunol. 191:101-106. [DOI] [PubMed] [Google Scholar]
- 32.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kullik, I., P. Giachino, and T. Fuchs. 1998. Deletion of the alternative sigma factor sigmaB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mack, D., W. Fischer, A. Krokotsch, K. Leopold, R. Hartmann, H. Egge, and R. Laufs. 1996. The intercellular adhesin involved in biofilm accumulation of Staphylococcus epidermidis is a linear β-1,6-linked glucosaminoglycan: purification and structural analysis. J. Bacteriol. 178:175-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mack, D., N. Siemssen, and R. Laufs. 1992. Parallel induction by glucose of adherence and a polysaccharide antigen specific for plastic-adherent Staphylococcus epidermidis: evidence for functional relation to intercellular adhesion. Infect. Immun. 60:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKenney, D., J. Hubner, E. Muller, Y. Wang, D. A. Goldmann, and G. B. Pier. 1998. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 66:4711-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenney, D., K. L. Pouliot, Y. Wang, V. Murthy, M. Ulrich, G. Doring, J. C. Lee, D. A. Goldmann, and G. B. Pier. 1999. Broadly protective vaccine for Staphylococcus aureus based on an in vivo-expressed antigen. Science 284:1523-1527. [DOI] [PubMed] [Google Scholar]
- 38.Novick, R. P., H. F. Ross, S. J. Projan, J. Kornblum, B. Kreiswirth, and S. Moghazeh. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967-3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira, D. C., and H. de Lencastre. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rachid, S., K. Ohlsen, W. Witte, J. Hacker, and W. Ziebuhr. 2000. Effect of subinhibitory antibiotic concentrations on polysaccharide intercellular adhesin expression in biofilm-forming Staphylococcus epidermidis. Antimicrob. Agents Chemother. 44:3357-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson, D. A., and M. C. Enright. 2003. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:3926-3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rohde, H., C. Burdelski, K. Bartscht, M. Hussain, F. Buck, M. A. Horstkotte, J. K. Knobloch, C. Heilmann, M. Herrmann, and D. Mack. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883-1895. [DOI] [PubMed] [Google Scholar]
- 43.Shaw, L., E. Golonka, J. Potempa, and S. J. Foster. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150:217-228. [DOI] [PubMed] [Google Scholar]
- 44.Stepanovic, S., N. Djukic, V. Djordjevic, and S. Djukic. 2003. Influence of the incubation atmosphere on the production of biofilm by staphylococci. Clin. Microbiol. Infect. 9:955-958. [DOI] [PubMed] [Google Scholar]
- 45.Valle, J., A. Toledo-Arana, C. Berasain, J. M. Ghigo, B. Amorena, J. R. Penades, and I. Lasa. 2003. SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus. Mol. Microbiol. 48:1075-1087. [DOI] [PubMed] [Google Scholar]
- 46.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 47.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 48.Wolz, C., P. Pohlmann-Dietze, A. Steinhuber, Y. T. Chien, A. Manna, W. van Wamel, and A. Cheung. 2000. Agr-independent regulation of fibronectin-binding protein(s) by the regulatory locus sar in Staphylococcus aureus. Mol. Microbiol. 36:230-243. [DOI] [PubMed] [Google Scholar]
- 49.Yokoe, D. S., J. Anderson, R. Chambers, M. Connor, R. Finberg, C. Hopkins, D. Lichtenberg, S. Marino, D. McLaughlin, E. O'Rourke, M. Samore, K. Sands, J. Strymish, E. Tamplin, N. Vallonde, and R. Platt. 1998. Simplified surveillance for nosocomial bloodstream infections. Infect. Control Hosp. Epidemiol. 19:657-660. [DOI] [PubMed] [Google Scholar]
- 50.Ziebuhr, W., C. Heilmann, F. Gotz, P. Meyer, K. Wilms, E. Straube, and J. Hacker. 1997. Detection of the intercellular adhesion gene cluster (ica) and phase variation in Staphylococcus epidermidis blood culture strains and mucosal isolates. Infect. Immun. 65:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.