Abstract
A multilocus sequence typing (MLST) scheme was devised for Aspergillus fumigatus. The system involved sequencing seven gene fragments and was applied to a panel of 100 isolates of A. fumigatus from diverse sources. Thirty different sequence types were found among the 100 isolates, and 93% of the isolates differed from the other isolates by only one allele sequence, forming a single clonal cluster as indicated by the eBURST algorithm. The discriminatory power of the MLST method was only 0.93. These results strongly indicate that A. fumigatus is a species of a relatively recent origin, with low levels of sequence dissimilarity. Typing methods based on variable numbers of tandem repeats offer higher levels of strain discrimination. Mating type data for the 100 isolates showed that 71 isolates were type MAT1-2 and 29 isolates were MAT1-1.
Differentiation between strains of the same microbial species can be accomplished by multilocus sequence typing (MLST). This method compares nucleotide polymorphisms within regions of five to seven genes, traditionally housekeeping genes, which are under selective pressure to retain function. MLST was developed to facilitate studies of epidemiology and population structure in bacterial populations (32). Polymorphisms giving rise to allelic variants are recorded as bar codes of integers which together constitute a strain sequence type (ST). The digital nature of MLST makes it globally portable so that data can be compiled from multiple contributors. Active MLST schemes for several microorganisms are publicly available at http://www.mlst.net/, where sequence data for MLST alleles can be uploaded to ascertain allele genotypes and strain STs. Recently, MLST was used to investigate populations of human pathogenic fungi, including Candida albicans (7), Candida glabrata (16), Candida tropicalis (48), Candida krusei (22), Cryptococcus neoformans var. gattii (20), Cryptococcus neoformans var. grubii (30), Histoplasma capsulatum (23), and Coccidioides immitis (25).
Aspergillus fumigatus is a saprotrophic mold fungus commonly found in soil enriched with organic material (49). This fungus produces huge numbers of airborne spores, which are found ubiquitously in the environment, including the air column (15). Despite having its primary niche in organic material in soil, A. fumigatus has become a major pathogenic organism in humans, coincident with compromised or suppressed host immunity (6). In the immunocompromised host, inhaled spores can initiate serious invasive aspergillosis, a condition carrying a prognosis of at least 50% mortality even when antifungal drugs are administered (29). Vulnerable individuals cannot be completely protected from airborne spores; even HEPA-filtered air in bone marrow transplant units may contain four Aspergillus spores per m3, which may be brought in by the patient or staff (28). Ascertaining relationships between the epidemiology of aspergillosis and the population structure of A. fumigatus is therefore desirable, and attempts have been made using a variety of genotyping methods, although with little success in discerning population structure within the species (52).
A search of the incompletely sequenced A. fumigatus genome revealed some of the genetic elements that may permit a sexual cycle (38). The sexual genetic elements in A. fumigatus constitute a putative heterothallic system. The original genome-sequencing strain AF293 has the high-mobility-group domain protein at the MAT locus (36). A survey of 290 isolates revealed distributions of strains comprising either the high-mobility-group protein (MAT1-2) or the complementary alpha box domain protein (MAT1-1) at the MAT locus at either 57% or 43%, respectively (37). In the present study, we describe the development of a seven-locus MLST scheme for A. fumigatus and discuss the findings from MLST analysis of 100 clinical and environmental isolates. In addition, we have determined the distribution of sexual idiomorphs among our collection. The results show a low level of sequence variation between most isolates of A. fumigatus, suggesting a relatively recent evolutionary origin for the species.
MATERIALS AND METHODS
Isolates.
The 100 isolates of A. fumigatus used in this study were sourced from both clinical and environmental settings (Table 1). They were taken from our own stock collection and provided by the kindness of collaborators. Each isolate originated from a separate source except for IHEM20373, which was obtained from a culture collection but which was found, after it had been typed, to be from the same source as J960330. The isolate originally received as TX2684 was found to consist of colonies of opposite mating types, which were typed separately by MLST as TX2684A and TX2684B. For four isolates, no information concerning the origins was known. Among the remaining 96, 46 came from the United Kingdom, 19 from the United States, 17 from Australia, 8 from France, and 2 each from Belgium, Canada, and The Netherlands (Table 1). Fifty-six isolates had been originally cultured since 2000, and 36 were cultured in the 1990s, 1 in the 1980s, and 2 in the 1970s. Eighteen isolates had previously been typed by PCR of genes encoding the intergenic spacer regions of the large and small ribosomal subunits (41). The fungi were stored at −80°C as spore suspensions in YPG-glycerol (1:1) (1% yeast extract [Difco, Sparks, MD], 2% Bacto peptone [Difco], 2% glucose [Fisher, Loughborough, United Kingdom], 50% glycerol [Fisher]) and then streaked onto Sabouraud agar and grown at 35°C until conidia formed.
TABLE 1.
List of 100 A. fumigatus isolates typed by MLST
| Strain | Geographical source | Other detailsa | Decade isolated | ST | UPGMA subgroup | Mating type |
|---|---|---|---|---|---|---|
| 81/001 | United Kingdom | 1980 | 1 | B | MAT1-1 | |
| AM2002/0066 | United Kingdom | Sputum; patient with CF | 2000 | 2 | A | MAT1-2 |
| TX01-2819 | United States | 2000 | 2 | A | MAT1-2 | |
| WC04-200434 | Australia | Sputum isolate | 2000 | 3 | A | MAT1-2 |
| J931199 | France | 1990 | 3 | A | MAT1-2 | |
| J932614 | France | 1990 | 3 | A | MAT1-2 | |
| WC04-200492 | Australia | BAL isolate | 2000 | 4 | A | MAT1-2 |
| WC04-201352 | Australia | Sputum isolate | 2000 | 4 | A | MAT1-2 |
| AF23 | United Kingdom | Environment BMT patient; DNA type H (41) | 1990 | 5 | A | MAT1-1 |
| AF22 | United Kingdom | Environment BMT patient; DNA type H (41) | 1990 | 5 | A | MAT1-1 |
| J940362 | Belgium | 1990 | 5 | A | MAT1-1 | |
| AM2004/0051 | United Kingdom | BAL isolate | 2000 | 5 | A | MAT1-1 |
| WC04-201446 | Australia | Rhinitis isolate | 2000 | 5 | A | MAT1-1 |
| TX01-2725 | United States | 2000 | 5 | A | MAT1-1 | |
| WC04-202908 | Australia | Isolate from mitral valve tissue | 2000 | 5 | A | MAT1-2 |
| MGR01-2507 | United States | 2000 | 5 | A | MAT1-2 | |
| MGR01-2705 | United States | 2000 | 5 | A | MAT1-2 | |
| AM2004/0044 | United Kingdom | Sputum; patient with CF | 2000 | 5 | A | MAT1-2 |
| AM2004/0049 | United Kingdom | Sputum; patient with CF | 2000 | 5 | A | MAT1-2 |
| AM2004/0055 | United Kingdom | Sputum; patient with CF | 2000 | 5 | A | MAT1-2 |
| WC04-201208 | Australia | Sputum isolate | 2000 | 5 | A | MAT1-2 |
| WC03-202715 | Australia | BAL isolate | 2000 | 5 | A | MAT1-2 |
| WC02-202431 | Australia | BAL isolate | 2000 | 5 | A | MAT1-2 |
| J980659/3 | Belgium | Isolated from a toenail sample | 1990 | 5 | A | MAT1-2 |
| TX01-2837 | United States | 2000 | 5 | A | MAT1-2 | |
| IHEM20452 | Canada | Sputum isolate | 1990 | 5 | A | MAT1-2 |
| TX01-2841 | United States | 2000 | 6 | B | MAT1-2 | |
| AM2004/0054 | United Kingdom | Isolated from an ear swab | 2000 | 7 | A | MAT1-1 |
| TX01-2855 | United States | 2000 | 7 | A | MAT1-1 | |
| AF04 | United Kingdom | Environment BMT patient; DNA type F (41) | 1990 | 7 | A | MAT1-2 |
| AF11 | United Kingdom | Patient 3; nasal swab; DNA type H (41) | 1990 | 7 | A | MAT1-2 |
| AF16 | United Kingdom | Patient 6; pericardial fluid; DNA type H (41) | 1990 | 7 | A | MAT1-2 |
| AF02 | United Kingdom | Environmental isolate; DNA type F (41) | 1990 | 7 | A | MAT1-2 |
| AF12 | United Kingdom | Patient 1; nasal swab; DNA type H (41) | 1990 | 7 | A | MAT1-2 |
| WC02-202494 | Australia | 2000 | 7 | A | MAT1-2 | |
| AF06 | United Kingdom | Environmental isolate; DNA type F (41) | 1990 | 7 | A | MAT1-2 |
| AM2002/0063 | United Kingdom | Isolated from aspergilloma | 2000 | 7 | A | MAT1-2 |
| AM2002/0065 | United Kingdom | Sputum, patient with ABPA | 2000 | 7 | A | MAT1-2 |
| AF05 | United Kingdom | Environmental isolate; DNA type F (41) | 1990 | 7 | A | MAT1-2 |
| AF07 | United Kingdom | Environment BMT patient; DNA type F (41) | 1990 | 8 | MAT1-1 | |
| AF70 | United Kingdom | Environmental isolate; DNA type A (41) | 1990 | 8 | MAT1-1 | |
| K4437 | 8 | MAT1-2 | ||||
| AF19 | United Kingdom | Environmental isolate; DNA type H (41) | 1990 | 9 | B | MAT1-1 |
| AF69 | United Kingdom | Environmental isolate; DNA type A (41) | 1990 | 9 | B | MAT1-1 |
| AM2004/0056 | United Kingdom | Sputum isolate | 2000 | 9 | B | MAT1-1 |
| J941369 | France | 1990 | 9 | B | MAT1-1 | |
| AF60 | United Kingdom | Environmental isolate; DNA type A (41) | 1990 | 9 | B | MAT1-2 |
| AM2002/0067 | United Kingdom | Patient with IPA | 2000 | 9 | B | MAT1-2 |
| AM2004/0043 | United Kingdom | Sputum isolate | 2000 | 9 | B | MAT1-2 |
| R3317 | United States | 9 | B | MAT1-2 | ||
| AF20 | United Kingdom | Environmental isolate; DNA type H (41) | 1990 | 10 | B | MAT1-1 |
| 74/037 | United Kingdom | Laboratory contaminant | 1970 | 11 | A | MAT1-1 |
| AF71 | United Kingdom | Environmental isolate; DNA type A (41) | 1990 | 11 | A | MAT1-2 |
| AM2004/0048 | United Kingdom | Sputum; patient with CF | 2000 | 11 | A | MAT1-2 |
| WC04-202367 | Australia | BAL isolate | 2000 | 11 | A | MAT1-2 |
| J960158 | France | 1990 | 11 | A | MAT1-2 | |
| AF78 | United Kingdom | Environmental isolate; DNA type E (41) | 1990 | 12 | B | MAT1-2 |
| AM2004/0053 | United Kingdom | Isolated from a throat swab | 2000 | 12 | B | MAT1-2 |
| AF21 | United Kingdom | Environmental isolate; DNA type H (41) | 1990 | 13 | A | MAT1-1 |
| 73/063 | United Kingdom | Isolated from a lymph node; postmortem | 1970 | 14 | B | MAT1-1 |
| MCR01-2529 | United States | 2000 | 14 | B | MAT1-1 | |
| MGR01-2556 | United States | 2000 | 14 | B | MAT1-1 | |
| MGR02-40 | United States | 2000 | 14 | B | MAT1-2 | |
| AM2004/0052 | United Kingdom | 2000 | 14 | B | MAT1-2 | |
| TX01-2684B | United States | 2000 | 14 | B | MAT1-2 | |
| TX01-2711 | United States | 2000 | 14 | B | MAT1-2 | |
| J950637 | France | 1990 | 14 | B | MAT1-2 | |
| AM2002/0062 | United Kingdom | Sputum; patient with ABPA | 2000 | 15 | A | MAT1-2 |
| J960330 | United Kingdom | NCPF7099; patient with invasive aspergillosis | 1990 | 16 | A | MAT1-2 |
| WC02-203746 | Australia | Bronchial washing isolate | 2000 | 16 | A | MAT1-2 |
| TX01-2748 | United States | 2000 | 16 | A | MAT1-2 | |
| IHEM20373 | United Kingdom | Originally J960330 but obtained from different culture collection | 1990 | 16 | A | MAT1-2 |
| J970377 | Netherlands | 1990 | 17 | MAT1-2 | ||
| J970466 | Netherlands | 1990 | 18 | MAT1-2 | ||
| MGR01-2659 | United States | 2000 | 19 | B | MAT1-2 | |
| WC04-201218 | Australia | Isolated from mitral valve tissue | 2000 | 19 | B | MAT1-2 |
| WC02-200231 | Australia | Isolated from leg tissue | 2000 | 20 | A | MAT1-1 |
| J960181 | United Kingdom | AF90 (13) | 1990 | 20 | A | MAT1-1 |
| J960180 | United Kingdom | AF91 (13) | 1990 | 20 | A | MAT1-1 |
| AM2004/0040 | United Kingdom | Sputum isolate | 2000 | 20 | A | MAT1-2 |
| AM2004/0042 | United Kingdom | Sputum isolate | 2000 | 20 | A | MAT1-2 |
| AM2004/0047 | United Kingdom | Sputum; patient with CF | 2000 | 20 | A | MAT1-2 |
| TX01-2898 | United States | 2000 | 20 | A | MAT1-2 | |
| AM2004/0045 | United Kingdom | Cough swab; patient with CF | 2000 | 21 | B | MAT1-2 |
| AM2004/0046 | United Kingdom | 2000 | 22 | MAT1-2 | ||
| AM2004/0057 | United Kingdom | Isolated from an ear swab | 2000 | 23 | B | MAT1-2 |
| WC04-202056 | Australia | Isolated from right maxillary tissue | 2000 | 24 | A | MAT1-2 |
| WC03-202903 | Australia | 2000 | 24 | A | MAT1-2 | |
| WC04-201263 | Australia | 2000 | 24 | A | MAT1-2 | |
| WC04-201640 | Australia | Sputum isolate | 2000 | 24 | A | MAT1-2 |
| J941185 | France | 1990 | 24 | A | MAT1-2 | |
| TX01-2825 | United States | 2000 | 25 | A | MAT1-1 | |
| J941218 | France | 1990 | 25 | A | MAT1-1 | |
| IHEM20368 | France | 1990 | 25 | A | MAT1-1 | |
| IHEM20453 | Canada | BAL isolate | 1990 | 26 | B | MAT1-1 |
| TX01-2656 | United States | 2000 | 26 | B | MAT1-2 | |
| AF05-55308 | 2000 | 27 | B | MAT1-2 | ||
| B42928 | 28 | B | MAT1-2 | |||
| B64620 | 29 | A | MAT1-2 | |||
| TX01-2684A | United States | 30 | A | MAT1-1 |
ABPA, allergic bronchopulmonary aspergillosis; BAL, bronchoalveolar lavage; BMT, bone marrow transplantation; CF, cystic fibrosis; IPA, invasive pulmonary aspergillosis.
The identity of the isolates as A. fumigatus and not a closely related species, such as Aspergillus lentulus or Aspergillus udagawae, was confirmed by PCR and StyI restriction digestion as described elsewhere (3).
DNA extraction.
Mycelia were grown from conidial inocula in 20 ml of liquid YPG with shaking at 200 rpm at 30°C for 3 days. The mycelia were strained to remove the supernatant, and then 0.3-g glass beads (0.45- to 0.52-mm diameters; Sigma, St. Louis, MO), 300 μl extraction buffer (100 mM Tris-HCl, pH 8, 100 mM NaCl, 2% Triton X-100, 1% sodium dodecyl sulfate, 1 mM EDTA), and 300 μl phenol-chloroform (1:1) were added. The mycelia were disrupted by vortexing for 30 min. To the lysate, 200 μl TE (10 mM Tris-HCl, pH 8, 1 mM EDTA) was added, and insoluble debris was removed in a bench-top centrifuge run at full speed for 5 min. The DNA in the aqueous phase was extracted a second time by adding an equal volume of phenol-chloroform (1:1), vortexing it for 30 s, and repeating the centrifugation step. Nucleic acid from the aqueous phase was precipitated with 1 ml ethanol and pelleted as before. The pellet was resuspended in TE containing 250 μg ml−1 RNase (Sigma) and incubated for 30 min at 37°C. DNA was precipitated with 1 ml isopropanol and 10 μl 3 M sodium acetate. Following centrifugation for 10 min, DNA pellets were dried and resuspended in 50 μl water.
Typing of sexual idiomorphs.
MAT1-1 and MAT1-2 sexual idiomorphs were differentiated by PCR as described previously (37). Briefly, a multiplex PCR was performed with reaction volumes of 25 μl containing 1 μl of genomic DNA as the template, 5 μl 5× GoTaq buffer (Promega, Southampton, United Kingdom), 2 mM MgCl2 (Promega), 200 μM deoxynucleoside triphosphate mix (Invitrogen, Paisley, United Kingdom), and 2.5 units GoTaq DNA polymerase (Promega). The reaction mixture contained 0.64 μM of reverse primer AFM3 (5′-CGGAAATCTGATGTCGCCACG-3′), which is common to both idiomorphs, and 0.32 μM each of forward primer AFM1, which is specific to MAT-1 (5′-CCTTGACGCGATGGGGTGG-3′), and forward primer AFM2, which is specific to MAT-2 (5′-CGCTCCTCATCAGAACAACTCG-3′). PCR was performed with a thermocycler (TC-412; Techne, Cambridge, United Kingdom) programmed as follows: 95°C for 5 min; 35 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min; and a final elongation step at 72°C for 5 min.
Selection of gene fragments for MLST scheme.
A selection of 27 A. fumigatus genes from 0.7 to 3.3 kb was sequenced across a set of 12 isolates chosen to maximize the diversities of geographical sources, anatomical properties, and dates of isolation and therefore increase the likelihood of representation of diverse genotypes. Primers for PCR amplification and sequencing of the seven fragments were designed from sequences taken from GenBank (http://www.ncbi.nlm.nih.gov/) and A. fumigatus GeneDB (http://www.genedb.org/genedb/asp/). Results from these pilot sequences were used to determine a final panel of seven gene fragments that gave the highest discrimination for MLST analysis ( Table 2). In accordance with feasible automated-sequencing run length, fragments of up to 590 bp containing the most discriminatory single-nucleotide polymorphisms (SNPs) were identified.
TABLE 2.
Gene fragments and primers used for MLST
| Gene fragment | Gene product | GenBank accession no. | Primersb | Amplicon size (bp) | Sequencing result
|
Size of sequenced fragment (bp) | |
|---|---|---|---|---|---|---|---|
| 5′ | 3′ | ||||||
| ANXC4 | Annexin | AY598940 | F, 5′-GCGAGATAGCAACACTTCAGT-3′ | 729 | GGTCGCCA | GGCTTCGT | 400 |
| R, 5′-GGATACTGTTGCCCTAGATTTG-3′ | |||||||
| BGT1 | Beta-1,3-glucanosyl transferase | AF38596 | F, 5′-GATCGGTTGCCAGTCTTTGA-3′ | 811 | GTCTATTC | AAACGAGG | 500 |
| R, 5′-AATGGACGCAGAATGAAACT-3′ | |||||||
| CAT1 | Catalase | XM_743457 | F, 5′-AGCTCAACCGTCGTGAT-3′ | 754 | GACGTCCG | ACTCGGCC | 590 |
| R, 5′-TGCCATGCCCAGACATA-3′ | |||||||
| LIP | Lipase | AfA24A6.090ca | F, 5′-CGCCTCACTTCTCCTCA-3′ | 802 | GGTTTGGC | CGCGAAGG | 503 |
| R, 5′-TGCGAAATGGCTGACG-3′ | |||||||
| MAT1-2 | Mating type protein | XM_746497 | F, 5′-ACACCCTTCGACTTTCCA-3′ | 360 | CTCCCAAG | CGGGCTGC | 135 |
| R, 5′-GATCGCATAGTGTGAGTGG-3′ | |||||||
| SODB | Superoxide dismutase | XM_746579 | F, 5′-GCTCCAAGAGCTGCTAC-3′ | 673 | GATTATGG | TGACGCCT | 510 |
| R, 5′-AAGCGCTTCTCCACAGC-3′ | |||||||
| ZRF2 | Zinc transporter | AY611772 | F, 5′-CTCATCCAAGCTTGTTTCC-3′ | 639 | GCCTTGAT | TTCGTTGC | 400 |
| R, 5′-GTACCGCATCACCATCAA-3′ | |||||||
The sequence of LIP is not deposited in GenBank but can be found in the A. fumigatus GeneDB (http://www.genedb.org/genedb/asp/index.jsp).
F, forward primer; R, reverse primer.
Sequencing of MLST fragments.
Gene fragments were amplified by PCRs in 96-well microdilution plates to amplify gene fragments of <1 kb with the primers listed in Table 2. Reaction volumes of 50 μl contained 2 μl of genomic DNA as a template, 10 μl 5× Green GoTaq buffer (Promega), 1.5 mM MgCl2 (Promega), 100 μM deoxynucleoside triphosphate mix (Invitrogen), 0.2 μM each of forward and reverse primers (Invitrogen), and 5 U GoTaq DNA polymerase (Promega). PCR was performed with a TC-412 thermocycler (Techne, Cambridge, United Kingdom), with a cycle program of 94°C for 5 min; 35 cycles of 94°C for 1 min, 50°C for 1 min, and 68°C for 1 min; and a final elongation step at 68°C for 10 min. Amplified DNA fragments were purified by combining PCR mixtures with 60 μl of 20% polyethylene glycol 8000 (Sigma) and 2.5 M NaCl solution for 30 min at room temperature and then centrifuging 96-well plates at 2,254 × g for 60 min. The plates were inverted onto blotting paper to remove the supernatant. The DNA pellets were washed with 150 μl 70% ethanol and centrifuged for 10 min, and the supernatant was removed by blotting. Plates inverted on tissue paper were centrifuged for 1 min at 500 × g to remove residual ethanol. DNA was resuspended in 50 μl distilled H2O for use as template DNA for subsequent sequencing reactions. Reaction volumes of 5 μl were set up in 96-well plates and contained 1 μl of purified gene fragment, 2 μl of 0.67 μM forward primer, 0.25 μl Big Dye (Applied Biosystems, Warrington, United Kingdom), 0.875 μl 5× buffer (Applied Biosystems), and 0.875 μl distilled H2O. Reaction mixtures were prepared as described above with the reverse primer. Sequencing PCR was performed with 25 cycles of 96°C for 10 s, 52°C for 5 s, and 60°C for 2 min. DNA was purified by addition of 50 μl of a 250:1 solution of ethanol-sodium acetate and incubation at room temperature for 45 min. DNA was pelleted and washed in 96-well plates as described above. Automated DNA sequencing was performed at the Department of Zoology, University of Oxford, United Kingdom. Forward and reverse DNA sequence chromatograms were analyzed with DNASTAR software to identify interstrain SNPs.
Statistical analysis of MLST data.
MLST sequence data were analyzed to determine relationships between the 100 strains. Concatenated SNPs for all MLST alleles of each strain were input with MEGA 3.0 software (26) to generate an unweighted-pair group method with arithmetic averages (UPGMA) dendrogram based on p-distance with 1,000 bootstrap replications. eBURST version 3 (http://eburst.mlst.net/) was used to ascertain clonal cluster relationships between isolates on the basis of single allele differences, regardless of numbers of SNP differences between alleles.
RESULTS
A. fumigatus strain differentiation by MLST.
A total of 27 gene fragments was sequenced for a set of 12 isolates, expected to be genetically diverse, to determine an optimal panel of gene fragments for MLST. Initially, housekeeping genes were considered potential MLST fragment candidates. These are defined as genes for which the ratio of nonsynonymous/synonymous amino acid changes resulting from SNPs is <1. Traditionally, only such genes were used in bacterial schemes when MLST was first devised (32). A. fumigatus housekeeping genes showed such low interstrain sequence variability that we eventually included nonhousekeeping genes to obtain a higher frequency of interisolate SNPs. The 20 genes that were considered but not chosen for the final selection were AREA, CALN, CSN, CPCA, CRNA, FACC, HIS3, HSP70, LEU2, MAN70, MANA, NIAD, NIIA, PEP, PREA, PREB, SOK, STEA, STUA, and TRPC.
Seven MLST fragments that represented the minimum set needed to differentiate the panel of 12 isolates were selected (Table 2). This MLST scheme was used to type 100 A. fumigatus isolates, including both clinical and environmental specimens, from our culture collection (Table 1). The polymorphic sites in the MLST fragments for A. fumigatus are shown in Table 3. Details of the MLST system can be found at http://pubmlst.org/afumigatus/. Among the 100 strains analyzed, we distinguished 30 STs (Table 1), giving our scheme a discriminatory index of 0.93 (21). ST 5 was the most common (Table 1), with 18 isolates, followed by ST 7 (12 isolates), ST 14 (8 isolates), and ST 20 (7 isolates). For 19 STs, only one or two examples each were found among the isolates tested (Table 1).
TABLE 3.
Numbers of polymorphic sites within MLST fragments and noncoding sequences and ratios of nonsynonymous to synonymous and nontrivial to trivial amino substitutions resulting from nucleotide changes at polymorphic sites
| MLST fragment | No. of polymorphisms
|
Ratio of amino acid substitutions
|
||
|---|---|---|---|---|
| Total | In noncoding sequences | Nonsynonymous to synonymous | Nontrivial to triviala | |
| ANXC4 | 8 | 0 | 3 | 0.6 |
| BGT1 | 7 | 1 | 2 | 0.5 |
| CAT1 | 8 | 3 | 0.67 | 0.25 |
| LIP | 8 | 0 | 1.67 | 0.33 |
| MAT1-2 | 3 | 0 | 3 | 2 |
| SODB | 3 | 0 | 0 | 0 |
| ZRF2 | 4 | 0 | 4 | 1 |
A trivial substitution is defined as one that does not alter side chain polarity or acidity/basicity; a nontrivial substitution involves a change of one or both properties.
Polymorphic sites were found predominantly in coding sequences; however, some were found in introns that were discovered by comparison of our direct sequence data with cDNA sequences in GenBank (Table 3). The BGT1 fragment used for MLST contained a 71-bp intron without polymorphic sites. The CAT1 fragment contained two introns of 66 bp and 105 bp, which included, respectively, one and two of the eight SNPs for this gene. One SNP was upstream of the start codon in the BGT1 fragment (Table 3). MLST can be based on coding and noncoding sequences because the technology relies upon the digital information of genomic DNA sequences and not expressional information. The amino acid substitutions for polymorphisms within the coding sequence were determined. The ratio of nonsynonymous to synonymous amino acid substitutions was determined for each MLST fragment (Table 3) and found to be >1 for five of the seven genes. Therefore, to determine the functionally relevant substitutions, the ratio of substitutions that maintained amino acid side chain polarity and acidity/basicity to those that did not was also calculated. These ratios were <1 for five gene fragments, which we consider acceptably low (Table 3).
Determination of distribution of sexual idiomorphs MAT1-1 and MAT1-2.
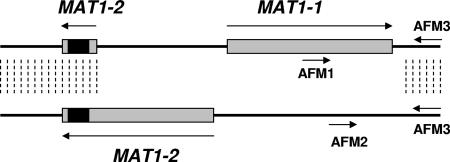
In addition to applying the MLST scheme to genotype our A. fumigatus collection, we determined the mating type for each strain, using a multiplex PCR with primers AFM1, AFM2, and AFM3 (Fig. 1), as described previously (37). The locus-specific amplicon of the sexual idiomorph MAT1-1 is 834 bp, and that of MAT1-2 is 438 bp. It should be noted that the MAT1-2 fragment sequenced in the MLST scheme can be amplified from all A. fumigatus isolates because both MAT loci carry the 3′ MAT1-2 coding region, within which lies the 135-bp MLST fragment (Fig. 1). The position of the MLST MAT1-2 fragment is from nucleotides +1106 to +1240 of MAT1-2. Of the 100 A. fumigatus strains that we tested, 29% were sexual idiomorph MAT1-1 and 71% were MAT1-2, a ratio of approximately 1:2. There was no association between mating type and ST, since both mating types were represented among the most populous STs.
FIG. 1.
The MAT locus in A. fumigatus. Both sexual idiomorphs are shown aligned together as described by Paoletti et al. (37). The upper sequence has only the 3′ portion of the MAT1-2 ORF and the complete MAT1-1 ORF. The lower sequence has the complete MAT1-2 ORF. Dashed lines indicate areas of sequence homology between both idiomorphs. Primers AFM1, AFM2, and AFM3, used for the multiplex PCR to distinguish the sexual idiomorph genotypes, are indicated. The black box indicates the location of MLST fragment MAT1-2.
A. fumigatus phylogenetics.
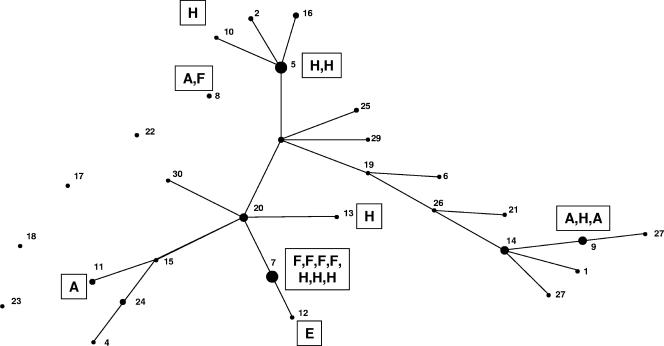
An eBURST analysis of the MLST data, which constructs a tree of STs joined to indicate pairs that differ in only one of the seven alleles sequenced, is shown in Fig. 2. All but 5 of the 30 STs formed a single clonal cluster of related isolates, with ST 20 as the putative founding isolate, according to eBURST. Figure 2 shows the genotype assignments of the 18 isolates that had been typed in a previous study on the basis of intergenic spacer regions in ribosome-encoding DNA (41). It is obvious that no relation could be found between the positions of STs on the branches of the eBURST clonal cluster and previously determined genotypes.
FIG. 2.
Clonal cluster and five single STs generated by eBURST analysis of MLST data, representing all 30 STs found from 100 A. fumigatus isolates. The putative founder of the complex is ST 20. The diameter of each circle represents the number of members within each ST group. Connecting lines are of arbitrary lengths but indicate that STs linked by a single line are different only at one allele out of seven. The boxed letters indicate genotypes assigned in a previous study (39).
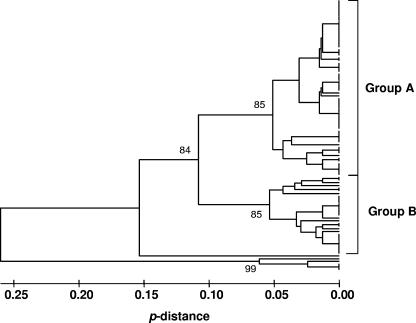
A UPGMA dendrogram of the MLST data, based on p-distance (Fig. 3), separated four of the five isolates that were singletons in the eBURST snapshot (Fig. 2) as distant relatives of the rest. The UPGMA dendrogram divided the isolates along lines similar to those for eBURST, but with ST 23, the fifth eBURST singleton, included with the main cluster in this analysis (Fig. 3). Isolates with STs 8, 17, 18, and 22 remained well distanced from other isolates. The main set of isolates could be differentiated into two subgroups by UPGMA, with strong bootstrap support (Fig. 3). The subgroup assignments of the 100 isolates are listed in Table 1, together with other genotype details. No significant association was found between the subgroups and the geographic origins or mating types of the isolates.
FIG. 3.
UPGMA dendrogram for 100 A. fumigatus isolates sequenced for MLST. The large cluster, which contained 94% of the isolates, was subdivided into two groups (indicated at the right) based on the high bootstrap values at the cluster nodes (numbers shown on the dendrogram).
DISCUSSION
The publication of the A. fumigatus genome sequence (36) is a driver for molecular genetic studies and has already helped correct taxonomic assignments based on phenotypic traits. Phylogenetic analysis revealed a sibling species of A. fumigatus, designated A. lentulus, in a study that compared sequences at five loci in slow-sporulating variants with those of Af293 (2), the A. fumigatus strain used for whole-genome sequencing (36). Sequence comparison based on part of the β-tubulin gene and 18S rRNA revealed that phenotypically atypical strains previously thought to be A. fumigatus may be a separate, more recently evolved species (24). Cryptic speciation within A. fumigatus was revealed following a microsatellite analysis of 63 isolates (39). In a recent study, misidentified A. fumigatus isolates were reclassified as A. lentulus and A. udagawae on the basis of restriction fragment length polymorphisms (RFLP) (3). None of our 100 isolates was slow to form conidia, the main phenotypic differentiator for A. lentulus and A. udagawae, and PCR/StyI testing confirmed that the isolates were A. fumigatus. In our experience with MLST analysis of many fungal species, we have never encountered examples in which isolates of another species gave identical PCR products with all the MLST genes when set up for sequencing reactions. This information therefore serves as a double check on species identity.
Comparison of nucleotide sequences of organisms is the most unequivocal method by which strains of any microorganism can be differentiated (50). We therefore devised a seven-gene MLST scheme to genotype A. fumigatus. Housekeeping genes possessed very low numbers of polymorphisms in this species and therefore were not useful for designing an MLST scheme. The infrequency with which we found polymorphisms within many genes scattered throughout the genome is a general indicator that A. fumigatus has low interstrain variation in its genome, suggesting recent evolution relative to that of other fungal species. MLST is, in effect, the successor to multilocus enzyme electrophoresis (MLEE). MLEE has previously been applied to A. fumigatus. Rodriguez et al. (42) found 48 electrophoretic types from 91 isolates tested with 12 polymorphic loci. Bertout et al. (5) differentiated eight electrophoretic types from 50 isolates based on seven loci. These MLEE results therefore support our finding from MLST that the level of interstrain variation for coding regions of genes is low. Sequence-specific DNA primer analysis distinguished 22 genotypes from 51 isolates, with a discriminatory power of 0.96 (34), and another sequence-specific DNA primer study found 19 genotypes in 81 isolates (46). Once again, typing systems based on sequence differentials failed to show high discriminatory power with A. fumigatus.
The total number of polymorphic sites from all seven MLST fragments in our system was 41 (Table 3); therefore, 1.35% of the 3,038 nucleotides from seven MLST fragments have polymorphisms. This “SNP return” is much lower than that for MLST with other pathogenic fungi, such as C. albicans, presently with 172 SNPs (6.0%) among the 2,883 bases sequenced; C. tropicalis, with 169 SNPs (6.3%) among the 2,677 bases sequenced; and C. glabrata, with 122 SNPs (3.7%) among the 3,345 bases sequenced (results from our own current databases for these species). The 1.35% SNP return for A. fumigatus is comparable with the polymorphic site rate of 1.6% reported in a study that compared three intergenic sequences in strains of A. fumigatus and the closely related taxa Neosartorya fischeri and Neosartorya spinosa, both of which fall within the subgenus Fumigati subgroup Fumigati (44). Normally, there is an expectation that intergenic sequences should have greater variability than the coding regions which constitute the majority of our MLST scheme. Rydholm et al. (44) found that the two Neosartorya species had greater interstrain variation than A. fumigatus. The consistently low typeability of A. fumigatus based on DNA sequences of coding and intergenic regions, combined with the observation that 93% of the isolates in our panel fell into a single clonal cluster by eBURST analysis (Fig. 2), reemphasizes the conclusion that A. fumigatus must be a recently evolved species and has therefore accumulated fewer mutations.
The fragments of approximately 500 bp used for MLST were chosen primarily because they contained the highest possible numbers of SNPs. They do not represent entire open reading frames (ORFs) and, in the present study, include introns and noncoding regions upstream of ORFs. For CAT1, Calera et al. previously described five introns in the ORF, with sizes of 66, 49, 85, 56, and 59 bp (from the 5′ to the 3′ end) (9). Our MLST fragment includes the first two of these introns, but the sizes were 66 and 105 bp, as deduced by comparison of GenBank mRNA data (sequence XM 743457) with our own sequence data. Similarly, for BGT1, the first of three introns previously described for this gene (35) was included in our MLST fragment, but comparison of the GenBank mRNA sequence (XM 747418) with our own data indicated an intron of 71 bp instead of the 42 bp originally reported (35).
Earlier approaches to A. fumigatus strain differentiation included immunoblot fingerprinting (8), RFLP typing (10), randomly amplified polymorphic DNA typing (31, 33, 40), and DNA fingerprinting by Southern blot analysis and probe hybridization (18). The discriminatory power of each of these approaches was superior to that of our MLST, but in none of the studies did the isolate population yield more than two-thirds of its number as different types. (MLST and its analogous approaches yielded different types amounting to approximately one-third of the number of isolates tested.) By comparison, C. albicans MLST generated 351 strain types from a panel of 416 isolates, so 84% of the test population emerged as different types. The discriminatory power of typing by amplified fragment length polymorphisms was not assessed (53), but the data suggest that this approach is at least as effective as RFLP or randomly amplified polymorphic DNA typing. By far, the most successful typing approach for A. fumigatus has been microsatellite, or short tandem repeat, analysis. This method has been used successfully by several investigators (4, 14, 19, 43, 51) and has been shown to have a discriminatory power of 0.99 and better (4, 14).
To achieve finer discrimination of A. fumigatus strains, some authors have used different methods in combination (1, 42, 45). However, applying multiple tests makes the task of typing A. fumigatus more laborious, and some typing methods may be compromised in their accuracy, for example, because interpretation of band sizes is open to error. The advantages of MLST as a typing approach are its portability (sequencing can be done anywhere, with identical results for identical DNA samples) and its archiveability (results can be stored in a central web database). Our study shows that MLST is a practical proposition for typing A. fumigatus, but for high-level strain discrimination, microsatellite typing is considerably more effective. This approach, like MLST, also offers portability and archiveability. However, its application to population genetic studies is limited by homoplasy arising from highly mutable sequences.
The A. fumigatus genome sequence project has revealed genetic elements for sexual behavior (36-38), opening the possibility that sexual recombination may occur. No observations of mating or meiosis have so far been made for A. fumigatus; therefore, this species will remain classified as an asexual haploid fungus for the time being. The MAT1-2 idiomorph was more commonly found than the MAT1-1 idiomorph (71% versus 29%) in our panel of isolates. Other studies also found MAT1-2 to be the dominant mating type: 57% versus 43% of 290 isolates (37) and 55% versus 45% of 102 isolates (44). The predominance of the MAT1-2 idiomorph may indicate a shift from the 1:1 ratio that is consistent with sexual reproduction. There is some evidence that recombination has occurred in the past for A. fumigatus (37); however, other studies (44) have found the same lack of variation between isolates from all parts of the world, indicating a predominantly clonal mode of reproduction. Compared with Candida albicans, also considered a largely clonal species, which generates 18 eBURST clonal clusters out of 416 isolates (47), there is very strong evidence for A. fumigatus both as a clonally reproducing organism and as a species of a relatively recent origin.
A major application of any microbial strain typing system is to elucidate the epidemiology of infection. The many clinical forms of aspergillosis result from inhalation of A. fumigatus from environmental sources, yet research attempting to make positive associations between environmental isolates and isolates infecting patients has not easily made such links, regardless of the typing method used. While some investigators have found examples of indistinguishable A. fumigatus isolates in some individual patients and their hospital environments (10, 19, 33), others have reported multiple strain types in the same patient and even the same sample (10, 11, 33). The best evidence for common types in infecting and environmental isolates comes more from statistical clustering of strain types (10, 17, 19, 53) than from unequivocal evidence of an indistinguishable strain type in a clinical sample and a sample, say, from the patient's ward. One study even drew the conclusion of a negative association between clinical and environmental isolates (43). By contrast, two studies found indistinguishable isolates in several patients, a situation compatible with cross-infection with a common strain type (12, 27); the finding is also more likely to arise in studies based on typing methods with low discriminatory power.
We cross referenced our own data with those from another study, because 18 isolates were genotyped in both studies (41). MLST could further resolve strains that had previously been classified as the same type, with up to seven allelic differences found (41). However, there was no cross-association between types determined by the two methods (Fig. 2). We have no explanation for why the results from ribosomal DNA typing do not match those from MLST.
MLST is a useful tool for differentiating isolates in an unambiguous manner. However, the inherent lack of sequence variability between A. fumigatus isolates probably restricts the value of MLST to preliminary screening in situations such as putative endemic outbreaks. High-level strain discrimination for A. fumigatus is better served by microsatellite typing.
Acknowledgments
This work was supported by the Wellcome Trust (grant 074898).
We gratefully thank our colleagues who supplied us with A. fumigatus isolates for typing: David H. Ellis, Annette Fothergill, Elizabeth M. Johnson, Nicole Nolard, and Francoise Symoens.
Footnotes
Published ahead of print on 21 March 2007.
REFERENCES
- 1.Anderson, M. J., K. Gull, and D. W. Denning. 1996. Molecular typing by random amplification of polymorphic DNA and M13 Southern hybridization of related paired isolates of Aspergillus fumigatus. J. Clin. Microbiol. 34:87-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balajee, S. A., J. L. Gribskov, E. Hanley, D. Nickle, and K. A. Marr. 2005. Aspergillus lentulus sp. nov., a new sibling species of A. fumigatus. Eukaryot. Cell 4:625-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balajee, S. A., D. Nickle, J. Varga, and K. A. Marr. 2006. Molecular studies reveal frequent misidentification of Aspergillus fumigatus by morphotyping. Eukaryot. Cell 5:1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bart-Delabesse, E., J. Sarfati, J.-P. Debeaupuis, W. Van Leeuwen, A. Van Belkum, S. Bretagne, and J.-P. Latge. 2001. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates. J. Clin. Microbiol. 39:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertout, S., F. Renaud, T. De Meeüs, M.-A. Piens, B. Lebeau, M.-A. Viviani, M. Mallié, J.-M. Bastide, and the EBGA Network. 2000. Multilocus enzyme electrophoresis analysis of Aspergillus fumigatus strains isolated from the first clinical sample from patients with invasive aspergillosis. J. Med. Microbiol. 49:375-381. [DOI] [PubMed] [Google Scholar]
- 6.Bodey, G. P., and S. Vartivarian. 1989. Aspergillosis. Eur. J. Clin. Microbiol. Infect. Dis. 8:413-437. [DOI] [PubMed] [Google Scholar]
- 7.Bougnoux, M. E., A. Tavanti, C. Bouchier, N. A. R. Gow, A. Magnier, A. D. Davidson, M. C. J. Maiden, C. d'Enfert, and F. C. Odds. 2003. Collaborative consensus for optimized multilocus sequence typing of Candida albicans. J. Clin. Microbiol. 41:5265-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burnie, J. P., R. C. Matthews, I. Clark, and L. J. Milne. 1989. Immunoblot fingerprinting Aspergillus fumigatus. J. Immunol. Methods 118:179-186. [DOI] [PubMed] [Google Scholar]
- 9.Calera, J. A., S. Paris, M. Monod, A. J. Hamilton, J. P. Debeaupuis, M. Diaquin, R. Lopez-Medrano, F. Leal, and J.-P. Latgé. 1997. Cloning and disruption of the antigenic catalase gene of Aspergillus fumigatus. Infect. Immun. 65:4718-4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chazalet, V., J.-P. Debeaupuis, J. Sarfati, J. Lortholary, P. Ribaud, P. Shah, M. Cornet, H. V. Thein, E. Gluckman, G. Brücker, and J.-P. Latge. 1998. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J. Clin. Microbiol. 36:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cimon, B., F. Symoens, R. Zouhair, D. Chabasse, N. Nolard, A Defontaine, and J.-P. Bouchara. 2001. Molecular epidemiology of airway colonization by Aspergillus fumigatus in cyctic fibrosis patients. J. Med. Mycol. 50:367-374. [DOI] [PubMed] [Google Scholar]
- 12.Debeaupuis, J.-P., J. Sarfati, V. Chazalet, and J.-P. Latgé. 1997. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect. Immun. 65:3080-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Valk, H. A., J. F. G. M. Meis, I. M. Curfs, K. Muehlethaler, J. W. Mouton, and C. H. W. Klaassen. 2005. Use of a novel panel of nine short tandem repeats for exact and high-resolution fingerprinting of Aspergillus fumigatus isolates. J. Clin. Microbiol. 43:4112-4120, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Vries, G. A. 1960. Aspergillus fumigatus and actinomycetes in air. Temperature and pH as selective factors in the analysis of mycoflora of the air by means of the culture plate technique. Acta Allergol. 15:99-106. [PubMed] [Google Scholar]
- 16.Dodgson, A. R., C. Pujol, D. W. Denning, D. R. Soll, and A. J. Fox. 2003. Multilocus sequence typing of Candida glabrata reveals geographically enriched clades. J. Clin. Microbiol. 41:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girardin, H., J. Sarfati, H. Kobayashi, J. P. Bouchara, and J.-P. Latgé. 1994. Use of DNA moderately repetitive sequence to type Aspergillus fumigatus isolates from aspergilloma patients. J. Infect. Dis. 169:683-685. [DOI] [PubMed] [Google Scholar]
- 18.Girardin, H., J. Sarfati, F. Traoré, J. Dupouy Camet, F. Derouin, and J.-P. Latgé. 1994. Molecular epidemiology of nosocomial invasive aspergillosis. J. Clin. Microbiol. 32:684-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guarro, J., M. Sole, R. Castany, J. Cano, A. Teixido, I. Pujol, J. Gene, A. Castro, and P. Sarda. 2005. Use of random amplified microsatellites to type isolates from an outbreak of nosocomial aspergillosis in a general medical ward. Med. Mycol. 43:365-371. [DOI] [PubMed] [Google Scholar]
- 20.Halliday, C. L., and D. A. Carter. 2003. Clonal reproduction and limited dispersal in an environmental population of Cryptococcus neoformans var. gattii isolates from Australia. J. Clin. Microbiol. 41:703-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hunter, P. R. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobsen, M. D., N. A. R. Gow, M. C. J. Maiden, D. J. Shaw, and F. C. Odds. 2007. Strain typing and determination of population structure of Candida krusei by multilocus sequence typing. J. Clin. Microbiol. 45:317-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasuga, T., J. W. Taylor, and T. J. White. 1999. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus Histoplasma capsulatum Darling. J. Clin. Microbiol. 37:653-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Katz, M. E., A. M. Dougall, K. Weeks, and B. F. Cheetham. 2005. Multiple genetically distinct groups revealed among clinical isolates identified as atypical Aspergillus fumigatus. J. Clin. Microbiol. 43:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koufopanou, V., A. Burt, and J. W. Taylor. 1997. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. USA 94:5478-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 27.Lasker, B. A. 2002. Evaluation of performance of four genotypic methods for studying the genetic epidemiology of Aspergillus fumigatus isolates. J. Clin. Microbiol. 40:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leenders, A. C. A. P., A. van Belkum, M. Behrendt, A. Luijendijk, and H. A. Verbrugh. 1999. Density and molecular epidemiology of Aspergillus in air and relationship to outbreaks of Aspergillus infection. J. Clin. Microbiol. 37:1752-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, S. J., J. Schranz, and S. M. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358-366. [DOI] [PubMed] [Google Scholar]
- 30.Litvintseva, A. P., R. Thakur, R. Vilgalys, and T. G. Mitchell. 2006. Multilocus sequence typing reveals three genetic subpopulations of Cryptococcus neoformans var. grubii (serotype A), including a unique population in Botswana. Genetics 172:2223-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loudon, K., J. Burnie, A. Coke, and R. Matthews. 1993. Application of polymerase chain reaction to fingerprinting Aspergillus fumigatus by random amplification of polymorphic DNA. J. Clin. Microbiol. 31:1117-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maiden, M. C. J., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Menotti, J., J. Waller, O. Meunier, V. Letscher-Bru, R. Herbrecht, and E. Candolfi. 2005. Epidemiological study of invasive pulmonary aspergillosis in a haematology unit by molecular typing of environmental and patient isolates of Aspergillus fumigatus. J. Hosp. Infect. 60:61-68. [DOI] [PubMed] [Google Scholar]
- 34.Mondon, P., M. P. Brenier, E. Coursange, B. Lebeau, P. Ambroisethomas, and R. Grillot. 1997. Molecular typing of Aspergillus fumigatus strains by sequence-specific DNA primer (SSDP) analysis. FEMS Immunol. Med. Microbiol. 17:95-102. [DOI] [PubMed] [Google Scholar]
- 35.Mouyna, I., R. P. Hartland, T. Fontaine, M. Diaquin, C. Simenel, M. Delepierre, B. Henrissat, and J.-P. Latgé. 1998. A 1,3-β-glucanosyltransferase isolated from the cell wall of Aspergillus fumigatus is a homologue of the yeast Bgl2p. Microbiology 144:3171-3180.9846753 [Google Scholar]
- 36.Nierman, W. C., A. Pain, M. J. Anderson, J. R. Wortman, H. S. Kim, J. Arroyo, M. Berriman, K. Abe, D. B. Archer, C. Bermejo, J. Bennett, P. Bowyer, D. Chen, M. Collins, R. Coulsen, R. Davies, P. S. Dyer, M. Farman, N. Fedorova, N. Fedorova, T. V. Feldblyum, R. Fischer, N. Fosker, A. Fraser, J. L. Garcia, M. J. Garcia, A. Goble, G. H. Goldman, K. Gomi, S. Griffith-Jones, R. Gwilliam, B. Haas, H. Haas, D. Harris, H. Horiuchi, J. Huang, S. Humphray, J. Jimenez, N. Keller, H. Khouri, K. Kitamoto, T. Kobayashi, S. Konzack, R. Kulkarni, T. Kumagai, A. Lafton, J. P. Latge, W. X. Li, A. Lord, W. H. Majoros, G. S. May, B. L. Miller, Y. Mohamoud, M. Molina, M. Monod, I. Mouyna, S. Mulligan, L. Murphy, S. O'Neil, I. Paulsen, M. A. Penalva, M. Pertea, C. Price, B. L. Pritchard, M. A. Quail, E. Rabbinowitsch, N. Rawlins, M. A. Rajandream, U. Reichard, H. Renauld, G. D. Robson, S. R. de Cordoba, J. M. Rodriguez-Pena, C. M. Ronning, S. Rutter, S. L. Salzberg, M. Sanchez, J. C. Sanchez-Ferrero, D. Saunders, K. Seeger, R. Squares, S. Squares, M. Takeuchi, F. Tekaia, G. Turner, C. R. V. de Aldana, J. Weidman, O. White, J. Woodward, J. H. Yu, C. Fraser, J. E. Galagan, K. Asai, M. Machida, N. Hall, B. Barrell, and D. W. Denning. 2005. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature 438:1151-1156. [DOI] [PubMed] [Google Scholar]
- 37.Paoletti, M., C. Rydholm, E. U. Schwier, M. J. Anderson, G. Szakacs, F. Lutzoni, J. P. Debeaupuis, J. P. Latgé, D. W. Denning, and P. S. Dyer. 2005. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 15:1242-1248. [DOI] [PubMed] [Google Scholar]
- 38.Poggeler, S. 2002. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 42:153-160. [DOI] [PubMed] [Google Scholar]
- 39.Pringle, A., D. M. Baker, J. L. Platt, J. P. Wares, J. P. Latge, and J. W. Taylor. 2005. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution 59:1886-1899. [PubMed] [Google Scholar]
- 40.Raclasky, V., J. Trtkova, L. Ruskova, V. Buchta, R. Bolehovska, M. Vackova, and P. Hamal. 2006. Primer R108 performs best in the RAPD strain typing of three Aspergillus species frequently isolated from patients. Folia Microbiol. 51:136-140. [DOI] [PubMed] [Google Scholar]
- 41.Radford, S. A., E. M. Johnson, J. P. Leeming, M. R. Millar, J. M. Cornish, A. B. M. Foot, and D. W. Warnock. 1998. Molecular epidemiological study of Aspergillus fumigatus in a bone marrow transplantation unit by PCR amplification of ribosomal intergenic spacer sequences. J. Clin. Microbiol. 36:1294-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodriguez, E., F. Symoens, P. Mondon, M. Mallie, M. A. Piens, B. Lebeau, A. M. Tortorano, F. Chaib, A. Carlotti, J. Villard, M. A. Viviani, F. Chapuis, N. Nolard, R. Grillot, and J. M. Bastide. 1999. Combination of three typing methods for the molecular epidemiology of Aspergillus fumigatus infections. J. Med. Microbiol. 48:181-194. [DOI] [PubMed] [Google Scholar]
- 43.Rosehart, K., M. H. Richards, and M. J. Bidochka. 2002. Microsatellite analysis of environmental and clinical isolates of the opportunist fungal pathogen Aspergillus fumigatus. J. Med. Microbiol. 51:1128-1134. [DOI] [PubMed] [Google Scholar]
- 44.Rydholm, C., G. Szakacs, and F. Lutzoni. 2006. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryot. Cell 5:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semighini, C. P., G. Delmas, S. Park, D. Amstrong, D. Perlin, and G. H. Goldman. 2001. New restriction fragment length polymorphism (RFLP) markers for Aspergillus fumigatus. FEMS Immunol. Med. Microbiol. 31:15-19. [DOI] [PubMed] [Google Scholar]
- 46.Symoens, F., J. Burnod, B. Lebeau, M. A. Viviani, M. A. Piens, A. M. Tortorano, N. Nolard, F. Chapuis, and R. Grillot. 2002. Hospital-acquired Aspergillus fumigatus infection: can molecular typing methods identify an environmental source? J. Hosp. Infect. 52:60-67. [DOI] [PubMed] [Google Scholar]
- 47.Tavanti, A., A. D. Davidson, M. J. Fordyce, N. A. R. Gow, M. C. J. Maiden, and F. C. Odds. 2005. Population structure and properties of Candida albicans, as determined by multilocus sequence typing. J. Clin. Microbiol. 43:5601-5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tavanti, A., A. D. Davidson, E. M. Johnson, M. C. J. Maiden, D. J. Shaw, N. A. R. Gow, and F. C. Odds. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 43:5593-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thom, C., and K. B. Raper. 1945. A manual of the aspergilli. Waverly Press, Baltimore, MD.
- 50.Urwin, R., and M. C. J. Maiden. 2003. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. 11:479-487. [DOI] [PubMed] [Google Scholar]
- 51.van Belkum, A., W. Quint, P. De, W. Melchers, and J. Meis. 1993. Typing of Aspergillus species and Aspergillus fumigatus isolates by interrepeat polymerase chain reaction. J. Clin. Microbiol. 31:2502-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Varga, J., and B. Tóth. 2003. Genetic variability and reproductive mode of Aspergillus fumigatus. Infect. Genet. Evol. 3:3-17. [DOI] [PubMed] [Google Scholar]
- 53.Warris, A., C. H. W. Klaassen, J. F. G. M. Meis, M. T. de Ruiter, H. A. de Valk, T. G. Abrahamsen, P. Gaustad, and P. E. Verweij. 2003. Molecular epidemiology of Aspergillus fumigatus isolates recovered from water, air, and patients shows two clusters of genetically distinct strains. J. Clin. Microbiol. 41:4101-4106. [DOI] [PMC free article] [PubMed] [Google Scholar]