Abstract
This study aimed to reconstruct the evolutionary history of Beijing strains of Mycobacterium tuberculosis and to test the hypothesis that evolution has influenced the ability of the Beijing strains within the different Beijing sublineages to spread and cause disease. A PCR-based method was used to analyze the genome structure of 40 different loci in 325 Beijing isolates collected from new and retreatment tuberculosis patients from an urban setting and 270 Beijing isolates collected from high-risk tuberculosis patients from a rural setting in the Western Cape, South Africa. The resulting data were subjected to phylogenetic analysis using the neighbor joining algorithm. Phylogenetic reconstructions were highly congruent with the “gold standard” phylogenetic tree based on synonymous single-nucleotide polymorphisms, thereby allowing a prediction of the order in which the evolutionary events had occurred. A total of seven independently evolving Beijing sublineages were identified. Analysis of epidemiological data in relation to the Beijing sublineage suggested an association between recent evolutionary change and frequency of occurrence in an urban population (P < 0.001) as well as in the rural population (P < 0.001). This concept was further supported by an association between more recently evolved Beijing strains and an increased ability to transmit and to cause disease (odds ratio, 5.82; 95% confidence interval, 3.13 to 10.82 [P < 0.001]). An association between Beijing sublineage and demographic and clinical parameters and drug resistance could not be demonstrated. From these data, we suggest that the pathogenic characteristics of Beijing strains are not conserved but rather that strains within individual lineages have evolved unique pathogenic characteristics.
From molecular epidemiological data, it has been hypothesized that Beijing strains of Mycobacterium tuberculosis have evolved unique properties (10), including the abilities to evade the protective effect of Mycobacterium bovis BCG vaccination (23), to disseminate more efficiently than non-Beijing strains (4), and to acquire drug resistance more frequently as the consequence of single-nucleotide polymorphisms (SNPs) in the mismatch repair genes (15). In some studies, disease caused by a Beijing strain has been associated with treatment relapse (16), while in another study a febrile response was noted during the earlier stages of treatment of tuberculosis caused by a Beijing strain (21). However, studies of chest radiography data have not been able to consistently demonstrate differences between the pathogenic characteristics of Beijing and non-Beijing strains (5, 7, 17). It is currently not known whether these results can be explained by different pathogenic characteristics among different Beijing strains, as a recent study has shown that extrathoracic tuberculosis was associated with only a subset of Beijing strains (12).
M. tuberculosis strains with a Beijing genotype are members of principal genetic group 1 and are characterized by a distinct spoligotype pattern, closely related IS6110 DNA fingerprints, identical variable-number tandem repeat sequences, and an IS6110 insertion in the NTF 1 region (Rv0001:Rv0002) (10). Comparative genomics have shown that Beijing strains have evolved through mechanisms including IS6110 transposition (2), SNPs in the mismatch repair genes (15), deletion of chromosomal domains (regions of difference [RDs]) (19), and synonymous SNPs (sSNPs) (9). An evolutionary scenario based on mutations in the mismatch repair genes mutT2 and mutT4 and ogt genes has suggested the absence of these mutations in the ancestral Beijing strains (15). Subsequent mutations in the mutT4 gene (codon 48) and ogt gene (codon 37) or ogt and mutT2 genes (codons 12 and 58, respectively) generated two independently evolving subgroups (15). Mapping of the RD domains showed that the Beijing strain family is a monophyletic clade, with four distinct evolutionary branches defined by the RD105, RD181, RD150, and RD142 deletions, respectively (19). Phylogenetic analysis according to sSNPs supported the monophyletic clade structure and suggested 11 Beijing sequence types (STs) (9). However, the order in which all of the above evolutionary events occurred remained to be determined.
In this study we aimed to reconstruct the evolutionary history of Beijing strains from the Western Cape, South Africa, through the analysis of the genomic structure of 40 different loci. This phylogeny was used to test the hypothesis that evolution has influenced the ability of the Beijing strains within the different Beijing sublineages to spread and cause disease.
MATERIALS AND METHODS
Urban setting.
As part of an ongoing molecular epidemiological study, an attempt was made to collect sputum samples at diagnosis from all new and retreatment tuberculosis cases who attended primary health care clinics and who were resident in an epidemiological field site in Cape Town, Western Cape, South Africa (3), during the period January 1993 to December 2004. Sputum smear microscopy (fluorescent staining) and/or culture (BACTEC 460, MGIT 960, or Löwenstein-Jensen medium) was performed by the National Health Laboratory Service or the Stellenbosch University laboratories to confirm the presence of M. tuberculosis. Clinical and demographic data including previous history of tuberculosis, gender and age, smear positivity, and drug susceptibility test results (if requested) were recorded in a database. Chest radiography was not done routinely and therefore was not included in this study. A case was considered as smear positive if any sputum sample, collected within the first 2 months after diagnosis, was found to be smear positive. Human immunodeficiency virus (HIV) testing was not routinely done in the initial years of this study although a recent survey of 366 new adult smear-positive tuberculosis cases (2000 to 2002) in this epidemiological field site showed that 10% were HIV positive.
Rural setting.
Sputum isolates were routinely collected at diagnosis from all high-risk tuberculosis patients (patients whose sputum failed to convert after the intensive phase of therapy or patients who had previously been treated with antituberculosis drugs) who were attending primary health care clinics and who were resident in the Boland, Overberg, Karoo, and Southern Cape region of the Western Cape, South Africa, during the period February 2003 to August 2004. This region is located 150 to 500 km northeast of the epidemiological field site in Cape Town. Sputum smear microscopy (fluorescent staining) and/or culture (BACTEC 460, MGIT 960, or Löwenstein-Jensen medium) was performed by the National Health Laboratory Service or the Stellenbosch University laboratories to confirm the presence of M. tuberculosis. Clinical and demographic data including previous history of tuberculosis, gender and age, smear positivity, and drug susceptibility test results were recorded in a database. A survey of 384 tuberculosis retreatment patients (2003 to 2004) in this study setting showed that 13% were HIV coinfected. This study setting was included to enable a comparison of the M. tuberculosis strain population structures in two independent communities.
This study was approved by the Ethics Committee (Institutional Review Board) of Stellenbosch University.
Drug susceptibility testing.
Drug susceptibility testing was done by the National Health Laboratory Service, using the indirect proportion method on Middlebrook medium containing critical concentrations of 0.2 μg/ml isoniazid and 30 μg/ml rifampin. In this study drug resistance was defined as resistance to either isoniazid or rifampin or a combination of isoniazid and rifampin (multidrug-resistant [MDR] tuberculosis). Drug susceptibility testing for other resistance markers was not routinely done.
Genotypic classification.
Sputum isolates from patients resident in the urban epidemiological field site were subcultured on Löwenstein-Jensen medium and genotyped by IS6110 DNA fingerprinting (22) and spoligotyping according to internationally standardized methods (11). Sputum isolates from patients resident in the rural setting were cultured on BACTEC 12B medium, and a boiled aliquot was genotyped by spoligotyping according to internationally standardized methods (11). Subcultures of the isolates from the rural setting were not available for IS6110 DNA fingerprinting.
Isolates were classified as members of the Beijing genotype if they had the characteristic Beijing spoligotype (10). Only the first M. tuberculosis isolate from each case with a Beijing strain was included for subsequent genetic analysis.
Genomic comparison.
Each Beijing strain from each patient was subjected to PCR amplification in a reaction mixture containing 0.2 μg DNA template, 5 μl Q buffer, 2.5 μl 10× buffer, 2 μl 25 mM MgCl2, 4 μl 10 mM deoxynucleoside triphosphates, 1 μl of each primer (50 pmol/μl) (Table 1), and 0.125 μl HotStarTaq DNA polymerase (QIAGEN, Germany) and made up to 25 μl with H2O. Primers 1 to 22 (Table 1) were used in combination with the universal forward primer and the internal control primers to determine the presence or absence of IS6110 elements at specific chromosomal loci (2). Primer set 23 to 26 (Table 1) was used to determine the presence or absence of SNPs in the mutT2 (codon 58), mutT4 (codon 48), and ogt (codons 12 and 37) genes (15) by the amplification refractory mutation system method. Primer set 27 to 30 (Table 1) was used to determine whether RDs (RD105, RD181, RD150, and RD142) (19) were present or absent. Primer set 31 to 40 (Table 1) was used to determine the presence or absence of sSNPs at chromosomal positions 797736, 909166, 1477596, 1548149, 1692069, 1892017, 2376135, 2532616, 2825581, and 4137829 (9) relative to the H37Rv whole-genome sequence (6) by the amplification refractory mutation system method. Amplification was initiated by incubation at 95°C for 15 min, followed by 35 to 45 cycles at 94°C for 1 min, annealing temperature (Table 1) for 1 min, and 72°C for 1 min. After the last cycle, the samples were incubated at 72°C for 10 min. PCR amplification products were electrophoretically fractionated in 3.0% agarose in 1× Tris-buffered EDTA, pH 8.3, at 6 V/cm for 4 h and visualized by staining with ethidium bromide. The existence of a mutational event was determined by the presence or absence and the size of the respective PCR product.
TABLE 1.
Primer sequences
| Primer set | Primer namee | Sequence (5′ to 3′) | Tmf (°C) |
|---|---|---|---|
| Internal control | Internal control | TCC CAG TGA CGT TGC CTT C | 62 |
| Internal control | GAG CAG CAG TGG AAT TTC GC | 62 | |
| Universal forward | Universal forward | TTC AAC CAT CGC CGC CTC TAC | 62 |
| 1 | IS21-1 (1262963a) | GTC GCC GGA GTT GAA GAA GCT GAA CC | 62 |
| 2 | IS21-2 (absenta) | CGA TCA ATG TTC CGC CTA ATT GAA CC | 62 |
| 3 | IS21-3 (3797825a) | AAT GCA GAC GAC GCG ACG ATT GAA CC | 62 |
| 4 | IS21-4 (1592a) | CAG CGA CAC TCA CAG CCA ATT GAA CC | 62 |
| 5 | IS21-5 (1543972a) | CTG CAC CGC GCG CAA CGA GGT GAA CC | 62 |
| 6 | IS21-6 (2263627a) | TCC AGG CAC CAG CAT CAA GGT GAA CC | 62 |
| 7 | IS21-7 (2038898a) | CGT TTG TGG GTG TCC GGT ATT GAA CC | 62 |
| 8 | IS21-8 (2367677)a | TGT CGC CAG TTA CGC ACG AGT GAA CC | 62 |
| 9 | IS21-9 (3549199a) | CGC GGA GCC GTC GGC CGC GGT GAA CC | 62 |
| 10 | IS21-10 (888990a) | TGA GGG GGT GTT GAG GTT GGT GAA CC | 62 |
| 11 | IS21-11 (3379025a) | GGG TTT TAA AAA AGT CGC TGT GAA CC | 62 |
| 12 | IS21-12 (3127932a) | GAT GGC ACG GCC GAC CTG AAT GAA CC | 62 |
| 13 | IS21-13 (3547342a) | GAC CGC GGT TGG GTG GAC ATT GAA CC | 62 |
| 14 | IS21-14 (3378553a) | CCC GCG CCT GCG CAA TTG GCT GAA CC | 62 |
| 15 | IS21-15 (3844681a) | GCG ATG TGG ATT GTG TCG GGT GAA CC | 62 |
| 16 | IS21-16 (1986638a) | ATC TGT TCA TCT CCG ACC TGT GAA CC | 62 |
| 17 | IS21-17 (absenta) | CGC GCG GTT CGA ATA CGG TGT GAA CCC | 62 |
| 18 | IS21-18 (2163649a) | TAC CAG CGA CGT TAA ACG GAT GAA CC | 62 |
| 19 | IS21-19 (3493910a) | GAA CCC CGG CTC CGC CTC GAT GAA CC | 62 |
| 20 | IS21-20 (1657015a) | GAG GCT CCT TTC GAC CGC GTT GAA CC | 62 |
| 21 | IS21-21 (2366892a) | GCG TGA TGT GCA CCA TAG TGT GAA CC | 62 |
| 22 | IS21-22 (2634022a) | GGT GCG GGG TCG GGG CCG TTT GAA CC | 62 |
| 23 | mutT2 (codon 58)b wt F | AGA GCT CGC CGA AGA ACC GG | 70 |
| mutT2 (codon 58b) mut F | AGA GCT CGC CGA AGA ACC GC | 70 | |
| mutT2 (codon 58b) R | AAG CAG ATG CAC GCG ATA GG | 70 | |
| 24 | mutT4 (codon 48b) F | AGC CGA GAA TCA CAT GGA CG | 68 |
| mutT4 (codon 48b) wt R | CGA GGT GAG CGG GAT CG | 68 | |
| mutT4 (codon 48b) mut R | CGA GGT GAG CGG GAT CC | 68 | |
| 25 | ogt (codon 12b) F | CCG CAG GAG AAG ATC GCA T | 68 |
| ogt (codon 12b) wt R | GCC CGG CCA GGG TTA ATA GC | 68 | |
| ogt (codon 12b) mut R | GCC CGG CCA GGG TTA ATA GT | 68 | |
| 26 | ogt (codon 37b) F | CCA TCG GGC CAT TAA CCC T | 66 |
| ogt (codon 37b) wt R | TCG GGT GTC CAG TGT GCG C | 66 | |
| ogt (codon 37b) mut R | TCG GGT GTC CAG TGT GCG A | 66 | |
| 27 | RD105c F | ACA GCG CGG GTC ATA TCA C | 62 |
| RD105c int | GCA ACA CCC GCT TGT CTT TG | 62 | |
| RD105c R | AAC CAG CTC CTC GAC GCT ATC | 62 | |
| 28 | RD142c F | CCG GTG GTA CGG GTA TTT CC | 62 |
| RD142c int | GCT CGA GCA TGA TCA GCA AAG | 62 | |
| RD142c R | TAG CAC CAG TAC CGG ATG TCC | 62 | |
| 29 | RD150c F | AGT GCT GGC AAT AGC GGT TG | 62 |
| RD150c int | CAC CGG CAC TTA CCA TCT CG | 62 | |
| RD150c R | CCA GCA CTT GTT GCA ACT TCG | 62 | |
| 30 | RD181c F | AAA TCC GCC CAT ACC CGT C | 62 |
| RD181c R | AGC TTC GAC TGG CCA TAG GC | 62 | |
| 31 | 797736d wt F | CAG CTC ATC TGT TGA TGT TC | 66 |
| 797736d mut F | CAG CTC ATC TGT TGA TGT TT | 66 | |
| 797736d R | ACG CCA TAA GCA CCT TCA CAC | 66 | |
| 32 | 909166d wt F | CAC GTT TGA CCG GAT CCC GC | 69 |
| 909166d mut F | CAC GTT TGA CCG GAT CCC GT | 69 | |
| 909166d R | CGT CCC AAG GAG CAG TCA AG | 69 | |
| 33 | 1477596d wt F | GCC CGG CCA GGG TTA ATA GC | 69 |
| 1477596d mut F | GCC CGG CCA GGG TTA ATA GT | 69 | |
| 1477596d R | CCG GTA AAC CAA TGG CCA C | 69 | |
| 34 | 1548149d F | CGA CGT GAC ATG GCT GGA TT | 68 |
| 1548149d wt R | CCG ACA ACG TCA GCG ATA CC | 68 | |
| 1548149d mut R | CCG ACA ACG TCA GCG ATA CT | 68 | |
| 35 | 1692069d F | CGA CCC GTG ATC GCA TGT A | 69 |
| 1692069d wt R | CAC CGA GCT CAC CGG CCT TT | 69 | |
| 1692069d mut R | CAC CGA GCT CAC CGG CCT TC | 69 | |
| 36 | 1892017d wt F | CCA CGT TTC TTG ATG CCT AT | 66 |
| 1892017d mut F | CCA CGT TTC TTG ATG CCT AC | 66 | |
| 1892017d R | TAT CGA GGC CGA CGA AAG G | 66 | |
| 37 | 2376135d wt F | GTT GAT GTA TAT CGC GGA CA | 66 |
| 2376135d mut F | GTT GAT GTA TAT CGC GGA CG | 66 | |
| 2376135d R | GCC GCC GAA TTA GAA CAG C | 66 | |
| 38 | 2532616d F | CTG CTT CGA CAC CTT TAA CGC | 72 |
| 2532616d wt R | ATG CCC AAC GCC GCA GTG GGC | 72 | |
| 2532616d mut R | ATG CCC AAC GCC GCA GTG GGT | 72 | |
| 39 | 2825581d wt F | TGA CGG TCG GAT TCT TGG GT | 68 |
| 2825581d mut F | TGA CGG TCG GAT TCT TGG GG | 68 | |
| 2825581d R | AGC CCA CGA GAT ACT GAG CG | 68 | |
| 40 | 4137829d wt F | AGA TGG CCT ACC GGA TCA CC | 69 |
| 4137829d mut F | AGA TGG CCT ACC GGA TCA CT | 69 | |
| 4137829d R | GAC GCA GTC GCA ACA GTT CAC | 69 |
Phylogenetic analysis.
The evolutionary state(s) for the SNPs in the mismatch repair genes and the 11 Beijing ST sSNPs was assigned according to the DNA sequence of H37Rv (6). IS6110 insertion sites were assigned according to the absence or presence of an IS6110 element at a specific chromosomal site (IS6110 insertions at positions 3547342 and 3378553 were excluded from the final analysis as they showed a high degree of homoplasy). The RDs were assigned according to their absence or presence. The complete set of evolutionary states for the different mutational events or subsets thereof was subjected to phylogenetic analysis using the neighbor joining algorithm (PAUP* 4.0; Phylogenetic Analysis Using Parsimony [*Other Methods], version 4b10; Sinauer Associates, Sunderland, MA). Bootstrapping was performed to establish a degree of statistical support for branch nodes (8). A consensus tree rooted to M. bovis was generated using the program Contree (PAUP* 4.0). Only branches which occurred in >50% of the bootstrap trees were included in the final tree. Each branch was defined as a Beijing sublineage.
Statistical analysis.
The z test for proportions was used for testing frequency of occurrence of cases in the respective Beijing sublineages. The Fisher exact test was used to determine the association between Beijing sublineage and gender, new versus retreatment cases, smear positivity within 2 months of diagnosis, drug resistance (for patients resident in either the urban or the rural setting), and IS6110 clustering (urban setting only). In all cases, a P value of 0.05 was used as the cutoff level for significance.
RESULTS
Genotyping showed that 325/1,525 patients (21%) from the urban epidemiological field site in Cape Town and 270/904 patients (30%) from the rural setting in the Boland, Overberg, Karoo, and Southern Cape regions of the Western Cape had tuberculosis with an M. tuberculosis Beijing strain. Drug susceptibility testing showed that of the patients with Beijing strains, 50 (15%) from the urban setting and 63 (23%) from the rural setting had drug-resistant tuberculosis.
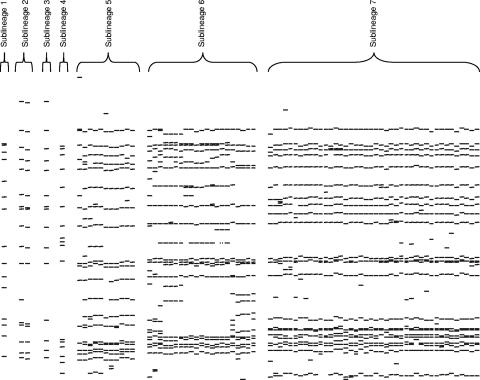
Phylogenetic analysis according to the neighbor joining algorithm grouped the Beijing strains from the two settings into four STs, ST11, ST19, ST22, and a novel ST (Beijing sublineages 6 and 7) (Fig. 1, left). The evolutionary history of these STs was further resolved according to SNPs in the mismatch repair genes, mutations induced by IS6110 insertion, and the structure of the RDs (Fig. 1, right). This phylogenetic tree was statistically robust, and the predicted evolutionary order was congruent with the tree based only on sSNPs (compare left and right sides of Fig. 1). This phylogeny demonstrates that the Beijing genotype is a monophyletic clade, defined by the RD105 deletion; IS6110 insertions at chromosome positions 1592, 1986638, 3127932, 3549199, and 3844681; and an sSNP at position 1548149 (Fig. 1, right). Subsequent evolution divided the Beijing strains into seven Beijing sublineages (Fig. 1, right) according to RDs (19), sSNPs (9), SNPs in the mismatch repair genes (15), and IS6110 insertions (2). The evolutionary order suggests that the RD181 deletion and the mutT4 (codon 48) SNP occurred before ST19, while the SNPs in the mutT2 and ogt mismatch repair genes occurred after ST19 and before ST10. Beijing sublineages 3 to 6 were distinguished by a combination of specific IS6110 insertions and/or sSNPs, while Beijing sublineage 7 was characterized by the RD150 deletion. The overall topology of the tree suggests divergent evolution, and this is supported by the increasing complexity in the IS6110 banding patterns between Beijing sublineage 1 (closest to the ancestor) and Beijing sublineage 7 (most distant from the ancestor) isolates from the urban setting (Fig. 2).
FIG. 1.
The evolutionary history of M. tuberculosis Beijing strains from an urban and a rural setting in the Western Cape, South Africa. (Left) Phylogenetic reconstruction based on sSNPs (9) using the bootstrapping and neighbor joining algorithms (PAUP* 4.0). The tree was rooted to M. bovis. Bootstrap values are given at internal nodes. The chromosomal positions of the predominant sSNPs are given at the nodes where they occur. All branches with a zero length were collapsed. (Right) Phylogenetic reconstruction based on sSNPs (9), SNPs in mismatch repair genes (15), IS6110 insertion points (2), and RDs (19) using the bootstrapping and neighbor joining algorithms (PAUP* 4.0). The tree was rooted to M. bovis. Bootstrap values are given at internal nodes. The respective evolutionary events are indicated at nodes where they occur (the order in which these events occurred is unknown). All branches with a zero length were collapsed. The scale indicates the number of steps per unit length.
FIG. 2.
IS6110 DNA fingerprints of M. tuberculosis Beijing strains from the urban setting. M. tuberculosis isolates were classified by IS6110 DNA fingerprinting according to the internationally standardized method (22). The DNA fingerprints from representative isolates are grouped according to their Beijing sublineage. Only a single representative of each IS6110 DNA fingerprint is shown.
Beijing strains with STs ST03, ST08, ST10, ST17, ST50, ST25, ST26, and ST27; strains with the RD142 deletion; and strains with IS6110 insertions at positions 2038898, 2163649, and 2367677 were absent from this study setting although Beijing strains with these evolutionary events have been described elsewhere (2, 9, 15, 19).
In order to determine whether the evolutionary process had altered the ability of drug-sensitive Beijing strains to spread and cause disease, the frequency of occurrence of strains from the more recently evolved Beijing sublineage 7 was compared to that of strains belonging to the more distantly evolved Beijing sublineages 2 to 6. This analysis showed an overabundance of Beijing sublineage 7 strains in the urban population (Table 2) (P < 0.001 from the z test for the hypothesis that proportion of sublineage 7 cases = 0.14) as well as in the rural population (Table 3) (P < 0.001 from the z test for the hypothesis that proportion of sublineage 7 cases = 0.14). In addition, strain clustering according to IS6110 DNA fingerprinting in the urban community was strongly associated with the Beijing sublineage 7 (Table 2) (odds ratio, 5.82; 95% confidence interval, 3.13 to 10.82 [P < 0.001]). (A similar analysis could not be done for isolates from the rural setting as cultures were not available for DNA fingerprinting.) Together, this suggests that strains from sublineage 7 have evolved a phenotype which allows them to transmit and cause disease more frequently than strains from sublineages 2 to 6.
TABLE 2.
Clinical, demographic, and molecular epidemiological data from patients resident in the urban epidemiological field site in Cape Town, South Africa
| Drug sensitivity and Beijing sublineage | Total no. of patients | No. (%) male | Avg age (yrs) | No. (%) of cases
|
No. of strains | No. of unique strains | No. of clusters | No. (%) of cases in clusters | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| New | Smear result
|
Monoresistant to:
|
MDR | |||||||||||
| Negative | Positive | Unknown | Isoniazid | Rifampin | ||||||||||
| Drug sensitive | ||||||||||||||
| 1 | 0 | |||||||||||||
| 2 | 4 | 2 (50) | 38.5 | 2 (50) | 3 (75) | 1 (25) | 2 | 2 | 4 (100) | |||||
| 3 | 2 | 2 (100) | 35.3 | 1 (50) | 1 (50) | 1 (50) | 1 | 1 | 2 (100) | |||||
| 4 | 1 | 22 | 1 (100) | 1 (100) | 1 | 1 | ||||||||
| 5 | 7 | 4 (57) | 36.9 | 3 (43) | 2 (29) | 4 (57) | 1 (14) | 6 | 5 | 1 | 2 (29) | |||
| 6 | 34 | 22 (65) | 34.5 | 23 (68) | 7 (20) | 24 (71) | 3 (9) | 18 | 12 | 6 | 22 (65) | |||
| 7 | 227 | 144 (63) | 35 | 136 (60) | 56 (24) | 156 (70) | 15 (6) | 44 | 31 | 13 | 196 (86) | |||
| Drug resistant | ||||||||||||||
| 1 | 1 | 1 (100) | 25.7 | 1 (100) | 1 (100) | 1 | 1 | |||||||
| 2 | 0 | |||||||||||||
| 3 | 0 | |||||||||||||
| 4 | 0 | |||||||||||||
| 5 | 33 | 15 (45) | 36.3 | 15 (45) | 9 (27) | 22 (67) | 2 (6) | 33 (100) | 6 | 4 | 2 | 29 (88) | ||
| 6 | 8 | 6 (75) | 36.7 | 2 (25) | 3 (38) | 5 (62) | 7 (87) | 1 (13) | 5 | 4 | 1 | 4 (50) | ||
| 7 | 8 | 3 (37) | 35.9 | 7 (87) | 3 (38) | 5 (62) | 2 (25) | 6 (75) | 5 | 4 | 1 | 4 (50) | ||
TABLE 3.
Clinical and demographic data from patients resident in the rural setting of the Boland, Overberg, Karoo, and Southern Cape region of the Western Cape, South Africa
| Drug sensitivity and Beijing sublineage | Total no. of patients | No. (%) male | Avg age (yrs) | No. (%) of cases
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| New | Smear result
|
Monoresistant to:
|
MDR | |||||||
| Negative | Positive | Unknown | Isoniazid | Rifampin | ||||||
| Drug sensitive | ||||||||||
| 1 | 0 | |||||||||
| 2 | 0 | |||||||||
| 3 | 0 | |||||||||
| 4 | 1 | 29 | 1 (100) | 1 (100) | ||||||
| 5 | 7 | 4 (57) | 36.3 | 2 (29) | 5 (71) | |||||
| 6 | 68 | 42 (62) | 27.7 | 22 (32) | 37 (55) | 24 (35) | 7 (10) | |||
| 7 | 131 | 72 (55) | 34.5 | 45 (34) | 58 (44) | 54 (41) | 19 (15) | |||
| Drug resistant | ||||||||||
| 1 | 3 | 2 (67) | 29.7 | NAa | 2 (67) | 1 (33) | 3 (100) | |||
| 2 | 2 | 23.5 | NA | 1 (50) | 1 (50) | 2 (100) | ||||
| 3 | 3 | 1 (33) | 31 | NA | 1 (33) | 1 (33) | 1 (33) | 1 (33) | 1 (33) | 1 (33) |
| 4 | 1 | 1 (100) | 39 | NA | 1 (100) | |||||
| 5 | 5 | 3 (60) | 34.4 | NA | 2 (40) | 1 (20) | 2 (40) | 5 (100) | ||
| 6 | 30 | 20 (67) | 32.1 | NA | 13 (43) | 12 (40) | 5 (17) | 9 (30) | 1 (3) | 20 (67) |
| 7 | 19 | 14 (74) | 29.3 | NA | 9 (47) | 8 (42) | 2 (11) | 6 (32) | 1 (5) | 12 (63) |
NA, not available.
To determine whether the frequency of occurrence of drug-susceptible strains could be explained by either a demographic or a clinical phenomenon, the demographic and clinical data from patients (in both settings) were compared according to their strains' Beijing sublineage. No association could be shown between Beijing sublineage and age, gender, previous history of tuberculosis, and smear positivity within the first 2 months following diagnosis.
To determine whether evolution has altered the ability of the Beijing sublineages to acquire drug resistance, phenotypic drug susceptibility data were compared according to the respective strains' Beijing sublineage. The results from the 50 patients resident in the urban setting showed no association between the Beijing sublineage and the number of strains with drug resistance (Table 2). However, a high level of transmission of resistant strains was seen in Beijing sublineage 5, which was associated with MDR tuberculosis (Table 2). Similarly, the high number of drug-resistant tuberculosis cases seen with Beijing sublineages 6 and 7 in the rural setting was associated with MDR tuberculosis (Table 3).
DISCUSSION
In this study we have used genetic information to predict evolutionary relationships between strains within the Beijing evolutionary lineage in order to test the hypothesis that the evolutionary process has altered the ability of these strains to spread and cause disease. According to the phylogenetic prediction we demonstrate that the Beijing strain family is a monophyletic clade which could be divided into seven sublineages. The predicted evolutionary order was congruent with the “gold standard” tree based on sSNPs (9). The observation that many of the evolutionary events have also been described for Beijing isolates from different geographical regions suggests that these evolutionary events occurred in the distant past and that the resulting progeny (including both distantly and more recently evolved clones) were subsequently imported into South Africa (13).
In this study, cases with a sublineage 7 strain were overrepresented, suggesting that the evolutionary process has had a positive influence on the strain's ability to spread and cause disease. This concept was further supported by an association between sublineage 7 and transmissibility. Similarly, in a previous study, a significant association was observed between extrathoracic tuberculosis and a subset of Beijing strains characterized by either RD142 or RD150 deletion (12). Together these data support the hypothesis that phenotypic differences may exist within the Beijing strain family (7, 19). Molecular epidemiological studies also support the notion that the more recently evolved strains (termed typical Beijing strains) are adapted to spread and cause disease, given their frequency of occurrence, in comparison to distantly evolved strains (termed atypical Beijing strains) (14, 18). However, factors including social behavior which could lead to the spread of sublineage 7 strains were not investigated in this study. An alternative explanation for the high frequency of occurrence of the Beijing sublineage 7 strains in South Africa could be a strong founder effect. However, comparison between Beijing genotypes from South Africa and East Asia showed that at least nine different Beijing founder strains were introduced into South Africa (data not shown). The spectrum of founder strains is likely to reflect that in East Asia at the time of importation into South Africa since the East Asia immigrants were derived from many different locations throughout the region. Thus, it is probable that the subsequent spread of sublineage 7 in South Africa represents evolutionary selection of an individual sublineage rather than a founder effect.
The observation of a changing epidemiological picture concurrent with the accumulation of evolutionary events cannot be linked to the sSNPs analyzed as these events are thought to be largely neutral (9). Conversely, mutation in the mismatch repair genes (15), IS6110 integration (2), and chromosomal deletions (19) may influence the strains' ability to spread and cause disease by altering or eliminating gene function. Mutation in the mismatch repair genes has been hypothesized to enhance the ability of the Beijing strains to acquire drug resistance (15). IS6110 insertion is thought to influence the characteristics of the Beijing strains by altering the function of genes by either causing knockouts or disrupting or enhancing promoter activity (2). The accumulation of chromosomal deletions has been suggested to contribute to an altered virulence in the Beijing strain family (19), and in a previous study, the analysis of 100 M. tuberculosis strains suggested an association between deletion and an altered phenotype (20). In our study the most pathogenic strains, as measured by their frequency of occurrence, were associated with the accumulation of deletions, including RD150. The true extent of chromosomal evolution within the Beijing strain family almost certainly extends beyond the evolutionary events tested in this study, and the influence of such events on the phenotype of different strains remains to be determined.
Our study design differs from previous studies as we analyzed intrastrain family phenotypic-genotypic correlations compared to interstrain family phenotypic-genotypic correlations (5, 7, 16, 17, 21). In our study, the analysis of patient demographic and clinical data failed to demonstrate an association with any of the Beijing strain sublineages. This implies that the appearance of secondary cases occurs independently of the proportion of infectious (smear-positive) cases in the different sublineages, thereby suggesting that the characteristics of the strains from the different sublineages could be the key factors influencing the ability to spread and cause disease.
We noted the presence of drug resistance in all of the Beijing sublineages and noted that the frequency of occurrence of the drug resistance phenotype was not associated with the most recently evolved Beijing sublineage. The absence of spread of drug resistance in the most recently evolved Beijing sublineage could be explained by the fitness cost as a result of the acquisition of mutations in the genes conferring drug resistance (1). The high number of resistant cases in sublineage 5 (urban setting) and sublineages 6 and 7 (rural settings) was associated with MDR tuberculosis and clearly indicates that MDR tuberculosis can spread within communities if not managed and treated appropriately.
In summary, we suggest that the pathogenic characteristics of M. tuberculosis strains are not conserved within defined strain families but rather that individual lineages within strain families have evolved unique pathogenic characteristics.
Acknowledgments
TB in the 21st Century Consortium, NRF (2054201), and the Harry Crossley Foundation and the European Commission 6th Framework Program on Research Technological Development Demonstration (project no. 037919) are thanked for financially supporting this project.
We thank the nurses and data analysts for the collection and processing of clinical data. We thank I. Toms, Department of Health, City of Cape Town, and are indebted to the residents of the epidemiological field site.
Footnotes
Published ahead of print on 14 March 2007.
REFERENCES
- 1.Andersson, D. I. 2006. The biological cost of mutational antibiotic resistance: any practical conclusions? Curr. Opin. Microbiol. 9:461-465. [DOI] [PubMed] [Google Scholar]
- 2.Beggs, M. L., K. D. Eisenach, and M. D. Cave. 2000. Mapping of IS6110 insertion sites in two epidemic strains of Mycobacterium tuberculosis. J. Clin. Microbiol. 38:2923-2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyers, N., R. P. Gie, H. L. Zietsman, M. Kunneke, J. Hauman, M. Tatley, and P. R. Donald. 1996. The use of a geographical information system (GIS) to evaluate the distribution of tuberculosis in a high-incidence community. S. Afr. Med. J. 86:40-41, 44. [PubMed] [Google Scholar]
- 4.Bifani, P. J., B. Mathema, N. E. Kurepina, and B. N. Kreiswirth. 2002. Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol. 10:45-52. [DOI] [PubMed] [Google Scholar]
- 5.Borgdorff, M. W., H. van Deutekom, P. E. de Haas, K. Kremer, and D. van Soolingen. 2004. Mycobacterium tuberculosis, Beijing genotype strains not associated with radiological presentation of pulmonary tuberculosis. Tuberculosis (Edinburgh) 84:337-340. [DOI] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Drobniewski, F., Y. Balabanova, V. Nikolayevsky, M. Ruddy, S. Kuznetzov, S. Zakharova, A. Melentyev, and I. Fedorin. 2005. Drug-resistant tuberculosis, clinical virulence, and the dominance of the Beijing strain family in Russia. JAMA 293:2726-2731. [DOI] [PubMed] [Google Scholar]
- 8.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783-793. [DOI] [PubMed] [Google Scholar]
- 9.Filliol, I., A. S. Motiwala, M. Cavatore, W. Qi, M. H. Hazbón, M. Bobadilla del Valle, J. Fyfe, L. García-García, N. Rastogi, C. Sola, T. Zozio, M. I. Guerrero, C. I. León, J. Crabtree, S. Angiuoli, K. D. Eisenach, R. Durmaz, M. L. Joloba, A. Rendón, J. Sifuentes-Osornio, A. Ponce de León, M. D. Cave, R. Fleischmann, T. S. Whittam, and D. Alland. 2006. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 188:759-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glynn, J. R., J. Whiteley, P. J. Bifani, K. Kremer, and D. van Soolingen. 2002. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerg. Infect. Dis. 8:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. Van Embden. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong, Y., M. D. Cave, L. Zhang, B. Foxman, C. F. Marrs, J. H. Bates, and Z. H. Yang. 2006. Population-based study of deletions in five different genomic regions of Mycobacterium tuberculosis and possible clinical relevance of the deletions. J. Clin. Microbiol. 44:3940-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mokrousov, I., H. M. Ly, T. Otten, N. N. Lan, B. Vyshnevskyi, S. Hoffner, and O. Narvskaya. 2005. Origin and primary dispersal of the Mycobacterium tuberculosis Beijing genotype: clues from human phylogeography. Genome Res. 15:1357-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mokrousov, I., O. Narvskaya, T. Otten, A. Vyazovaya, E. Limeschenko, L. Steklova, and B. Vyshnevskyi. 2002. Phylogenetic reconstruction within Mycobacterium tuberculosis Beijing genotype in northwestern Russia. Res. Microbiol. 153:629-637. [DOI] [PubMed] [Google Scholar]
- 15.Rad, M. E., P. Bifani, C. Martin, K. Kremer, S. Samper, J. Rauzier, B. Kreiswirth, J. Blazquez, M. Jouan, D. van Soolingen, and B. Gicquel. 2003. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerg. Infect. Dis. 9:838-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun, Y. J., A. S. Lee, S. Y. Wong, and N. I. Paton. 2006. Association of Mycobacterium tuberculosis Beijing genotype with tuberculosis relapse in Singapore. Epidemiol. Infect. 134:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun, Y. J., T. K. Lim, A. K. Ong, B. C. Ho, G. T. Seah, and N. I. Paton. 2006. Tuberculosis associated with Mycobacterium tuberculosis Beijing and non-Beijing genotypes: a clinical and immunological comparison. BMC Infect. Dis. 6:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toungoussova, O. S., P. Sandven, A. O. Mariandyshev, N. I. Nizovtseva, G. Bjune, and D. A. Caugant. 2002. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. J. Clin. Microbiol. 40:1930-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsolaki, A. G., S. Gagneux, A. S. Pym, Y.-O. L. Goguet de la Salmoniere, B. N. Kreiswirth, D. Van Soolingen, and P. M. Small. 2005. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J. Clin. Microbiol. 43:3185-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. USA 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Crevel, R., R. H. Nelwan, W. de Lenne, Y. Veeraragu, A. G. van der Zanden, Z. Amin, J. W. van der Meer, and D. van Soolingen. 2001. Mycobacterium tuberculosis Beijing genotype strains associated with febrile response to treatment. Emerg. Infect. Dis. 7:880-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Soolingen, D., L. Qian, P. E. de Haas, J. T. Douglas, H. Traore, F. Portaels, H. Z. Qing, D. Enkhsaikan, P. Nymadawa, and J. D. van Embden. 1995. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J. Clin. Microbiol. 33:3234-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]