Abstract
Staphylococcus saprophyticus is a common cause of urinary tract infections (UTIs) in women. Little is known about the molecular epidemiology of S. saprophyticus UTIs. In the current study, we compared 76 isolates of S. saprophyticus prospectively isolated from women with uncomplicated UTI participating in a randomized placebo-controlled treatment trial performed in northern Sweden from 1995 to 1997 with 50 strains obtained in 2006 from five different locations in northern Europe with pulsed-field gel electrophoresis (PFGE). The aim was to elucidate the molecular epidemiology of this uropathogenic species and to investigate whether specific clones are associated with UTI in women. A total of 47 different PFGE profiles were detected among the 126 analyzed isolates. Ten clusters consisting of 5 to 12 isolates each showing PFGE DNA similarity of >85% were identified. Several clusters of genetically highly related isolates were detected in the original trial as well as among isolates obtained during 2006 from different locations. In the original trial, clonal persistence was found among 16 of 21 (76%) patients examined in the placebo group at follow-up 8 to 10 days after inclusion, indicating a low spontaneous short-time bacteriological cure rate. We conclude that multiple clones of S. saprophyticus were causing lower UTIs in women. The result suggests that some human-pathogenic clones of S. saprophyticus are spread over large geographical distances and that such clones may persist over long periods of time.
Staphylococcus saprophyticus is a coagulase-negative staphylococcus associated primarily with community-acquired lower urinary tract infection (UTI) in young and middle-aged women (9). In this category of patients, S. saprophyticus is second only to Escherichia coli as the causative agent of UTIs (5, 19, 22). Complications of S. saprophyticus infection such as recurrent infection, acute pyelonephritis, nephrolithiasis, septicemia, and endocarditis have been documented but are all rare (2, 17). Compared with other uropathogens, S. saprophyticus differs in seasonal variation and geographic distribution. UTI has been documented mostly in women in the northern hemisphere, where the prevalence of colonization and infection is more frequent during late summer and autumn (3, 5, 13, 18, 22).
However, little is known about the molecular epidemiology of S. saprophyticus UTIs, and it is unknown if specific strains or clones are related to human infections.
In the present study, we compared 76 isolates of S. saprophyticus prospectively isolated from women with uncomplicated UTI participating in a clinical trial from 1995 to 1997 with 50 strains obtained in 2006 from five different locations in northern Europe with pulsed-field gel electrophoresis (PFGE). The aims of the present study were to elucidate the molecular epidemiology of this species and to investigate whether specific clones are associated with UTI in women.
MATERIALS AND METHODS
Origin of isolates.
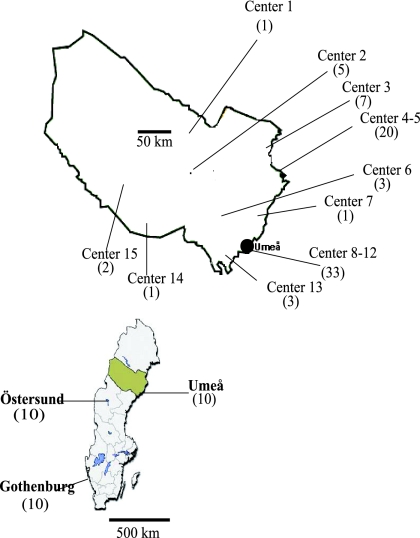
The strains of S. saprophyticus examined in this study originated from a subset of women included in a prospective, multicenter, randomized, double-blind, placebo-controlled trial comparing different dosing regimens of pivmecillinam for community-acquired lower UTI (6, 7). The trial was performed at 18 primary health care centers in the county of Västerbotten in northern Sweden between April 1995 and December 1997 (Fig. 1). A total of 1,143 patients were randomized to treatment with pivmecillinam or placebo. Subjects were evaluated clinically, and urine samples were obtained at inclusion and at two follow-up visits (Fig. 2). The Ethics Committee of Umeå University, Sweden, approved the trial, and informed written consent was obtained from each included patient. Details concerning the design and the general outcome of the trial have previously been described (4, 6, 7). These strains were compared with an additional 50 clinical isolates of S. saprophyticus obtained from five different locations in northern Europe. Ten strains each were obtained from the Departments of Clinical Bacteriology in Umeå and Östersund (both located in the northern part of Sweden) and the Department of Clinical Microbiology, Sahlgrenska University Hospital, Gothenburg (located in southern Sweden), as well as from the National Center for Antimicrobials and Infection Control, Statens Serum Institute, Copenhagen, Denmark, and the Institute for Hygiene and Microbiology, Ruhr University, Bochum, Germany. All these strains were isolated in 2005 and 2006, except for five strains from Copenhagen, which were collected during 1995.
FIG. 1.
Map of Sweden depicting the county of Västerbotten, which covers an area 55.401 square km and has a population of 258,000 inhabitants. The location of the 15 centers that contributed isolates of S. saprophyticus in the original trial and the numbers of isolates obtained from each center are shown in the enlargement of the county.
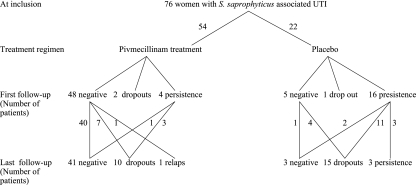
FIG. 2.
Flow chart of patients with S. saprophyticus UTI randomized to treatment with pivmecillinam or placebo in the original trial and bacteriological outcome at follow-up 8 to 10 days and 35 to 49 days after inclusion, respectively.
Bacteriological methods.
Urine samples in the original trial were analyzed within 24 h at the Department of Clinical Bacteriology, University Hospital of Umeå. Significant or nonsignificant uropathogens were quantified (CFU/ml) as previously described (6, 7). For the present study, all coagulase-negative staphylococcal isolates were reanalyzed according to a previously described species identification system (15). This reevaluation identified 76 isolates of S. saprophyticus and 10 isolates of other coagulase-negative staphylococcus species.
The additional isolates from 2005 to 2006 were identified by routine methods including analysis on chromogenic agar (CM0949, Chromogenic UTI medium; Oxoid Ltd., Basingstoke, United Kingdom) (1). All S. saprophyticus isolates demonstrated a zone diameter breakpoint of <13 mm using a 5-μg disk of novobiocin (ISA medium; Oxoid Ltd.) (15).
PFGE and calculation of genetic similarity.
DNA was prepared from a 3-ml bacterial culture in Todd-Hewitt broth (Difco Laboratories, Detroit, MI) as previously described (14). In summary, the DNA was digested using SmaI (MBI Fermentas, Villinius, Latvia), and the DNA fragments were separated by PFGE in a GenePath apparatus (Bio-Rad Laboratories, Hercules, CA) using the program for Staphylococcus aureus (program 14) (50-500 kb) for 19.7 h according to the manufacturer's instructions (Bio-Rad). Gels were stained in 1 mg/liter ethidium bromide, distained, and photographed under UV illumination. Genetic similarity between isolates was calculated using GelCompar II 4.0 (Applied Maths, Sint-Martens-Latem, Belgium) using the Dice coefficient and the unweighted-pair group method with arithmetic mean with 1.0% tolerance and 0.5% optimization settings. S. aureus NCTC 8325 was included as a reference in every sixth to seventh lane to allow normalization of the electrophoretic pattern. Bands sizes below 36 kb were not analyzed (16).
The discriminatory power of the PFGE typing method was calculated by using the Simpson index of diversity (D) (10).
PFGE interpretation and definitions.
Bacterial isolates were defined as being genetically “indistinguishable” if their PFGE profile was 100% similar and as being “closely related” if they showed PFGE profiles with at least 85% similarity, corresponding to a two- to three-band difference, which is consistent with one single genetic event (21). Isolates with PFGE profiles showing less than 85% similarity were defined as being genetically unrelated strains. A similarity threshold of >85% defined clusters. Bacterial persistence was defined as the identification of an isolate with an identical PFGE profile at inclusion and in follow-up cultures with no negative culture in between. Relapse was defined as the identification of an isolate with an identical PFGE profile compared with the primary infecting isolate with a negative culture in between. Reinfection was defined as the identification of an isolate with an unrelated PFGE profile occurring after a negative culture or a culture with a different species.
RESULTS
PFGE.
A total of 100 isolates of S. saprophyticus were identified in the original trial: 76 at inclusion and 20 and 4 at first and last follow-ups, respectively (Fig. 2). PFGE revealed 8 to 12 distinct bands for each isolate, and several of the DNA restriction fragments were shared by the majority of the strains (Fig. 3). Analysis of the 76 isolates identified at inclusion in the original trail showed 37 different profiles, 24 of which were singletons. The population was thus rather heterogeneous and composed of multiple genetic clusters containing small groups of related isolates. These groups consisted of three to eight isolates each (Fig. 3) and were identified from women attending different primary health care centers as well as during the entire study period. No obvious association in the PFGE banding pattern was detected with regard to the year of isolation or the origin of the strains.
FIG. 3.
Analyses of genetic similarity of 126 S. saprophyticus isolates using PFGE and GelCompar II software and their origins. The horizontal upper bar represents genetic similarity (percent). The dotted lines in the middle represent digitalized transformations of the PFGE DNA pattern. Numbers in the right column represent different centers participating in the original trial, and the names of cities describe the origins of the strains obtained in 2006. The arrows at the far right mark 10 clusters with at least 5 to 12 isolates each.
A similar heterogeneous picture was found when the original 76 isolates were compared with the 45 and 5 S. saprophyticus isolates collected in 2005 to 2006 and 1995, respectively (Fig. 3). A total of 47 PFGE profiles were found among these 126 isolates, 21 of which consisted of only one single isolate. Ten clusters each consisting of at least 5 to 12 isolates were detected. Some isolates with indistinguishable PFGE profiles were identified from various geographic locations. Specifically, identical PFGE profiles were detected when isolates in the original trial were compared with isolates from Umeå in 2006, Gothenburg in 2006, Copenhagen in 2005, and Bochum in 2006. Moreover, four isolates from Östersund, two isolates from Copenhagen in 1995, and two isolates from Copenhagen in 2005 had identical PFGE patterns (Fig. 3).
Calculation of D at the 85% similarity level for all 126 S. saprophyticus strains resulted in a D value of 0.971 (0.963 to 0.980).
Bacteriological outcome in the original trial.
The clinical outcome in the two treatment arms is shown in Fig. 2. A persistent infection with a strain showing a PFGE profile identical to that of the primary infecting strain was found among 16/21 (76%) and 3/6 (50%) examined patients in the placebo group at the first and last follow-ups, respectively. Eleven of the 16 patients with persistent infection in the placebo group were classified as “clinical treatment failures” and were given appropriate antibiotic treatment, while the remaining 5 were regarded as being “clinically cured,” three of whom persisted at the last follow-up (Fig. 2). Reinfection with an unrelated strain of S. saprophyticus was not detected in any of the patients. The majority of the strains included (49 of 76 isolates) were isolated during the late summer season, with a peak in September (23 isolates).
DISCUSSION
The present study is, to our knowledge, the first analysis of the molecular epidemiology of S. saprophyticus among women with community-acquired UTI. PFGE analyses demonstrated that a rather heterogeneous population with numerous clones of S. saprophyticus was associated with UTI. No specific uropathogenic clone was identified among isolates in the original trial or among the isolates from 2005 to 2006. Several clusters of genetically highly related isolates and some isolates that exhibited indistinguishable PFGE profiles were detected in the original trial as well as among isolates from the year 2006. This suggests that there are human-pathogenic clones of S. saprophyticus spread over large geographical distances and that these clones persisted over the time period from 1995 to 2006. The infectious source of S. saprophyticus causing UTI in women is unknown. However, it has been shown that S. saprophyticus is widely spread in the ecosystem and that a variety of food products are contaminated, leading to the colonization of the human gastrointestinal tract (8). Furthermore, rectal, vaginal, or urethral colonization with S. saprophyticus has been associated with UTI (17). As the origins of food products are both domestic and international, humans in a specific area are most likely exposed to S. saprophyticus strains originating from different geographic regions and perhaps other countries. Domestic and international travel may also facilitate the acquisition of S. saprophyticus strains from other geographic locations. Larger patient populations including strains from various sources and locations should be sampled to test if our results reflect the general epidemiology of S. saprophyticus UTIs and to further explore the possible existence of specific uropathogenic strains. Such studies may also clarify the seasonal variation of S. saprophyticus UTI observed in the current study and in a number of previous publications (5, 13, 22).
PFGE is an established method for molecular epidemiological analyses of many coagulase-negative species of staphylococcus (11). Our results, showing that all patients with bacterial persistence at follow-up carried isolates with PFGE profiles that were identical to that of the primary infecting strain, indicate that PFGE is also a useful tool for epidemiological analyses of S. saprophyticus. Moreover, the high D value calculated using Simpson's diversity index confirms that PFGE has a high discriminatory power for S. saprophyticus typing (20).
In general, the PFGE analyses demonstrated a relatively uniform DNA banding pattern with many shared band sizes among tested isolates of S. saprophyticus. This indicates a stable core genome with little genomic rearrangements among S. saprophyticus strains. This is consistent with a recent finding of low abundance of mobile genetic elements and open reading frames encoding transposition in S. saprophyticus (12). In contrast, it was found that both S. aureus and Staphylococcus epidermidis harbor many genetic elements that may mediate rearrangements of their respective genomes. We hypothesize that the relative homogeneity of PFGE patterns of S. saprophyticus may be related to a low genomic content of elements that mediate recombination. Analysis of S. saprophyticus strains from different geographical locations with other molecular typing methods may confirm this hypothesis.
In the original trial, clonal persistence was common among the placebo-treated patients, and the majority of these patients were classified as clinical treatment failures. Similar results were recently presented when E. coli strains isolated in the original trial were analyzed using PFGE (4). In that study, 110/156 (70%) patients in the placebo group had a persistent E. coli infection at the first follow-up. A subset of 54 strains was analyzed with PFGE, and 94% of these isolates showed PFGE patterns that were identical to that of the primary infecting strain. Those and our results support previous findings showing that the clinical course, short-term persistence rate, and spontaneous cure rate for S. saprophyticus UTI resembles that of E. coli infections (3, 6, 7).
In conclusion, PFGE analysis of S. saprophyticus causing lower UTI in women demonstrated the existence of a rather heterogeneous population consisting of small groups of related isolates. Indistinguishable isolates were identified in different countries 11 years apart, indicating the persistence and geographical spread of some clones of S. saprophyticus. Moreover, persistent infection was common among placebo-treated patients, demonstrating a low spontaneous bacteriological cure rate in UTI caused by S. saprophyticus.
Acknowledgments
We thank Christina Welinder-Olsson, Department of Clinical Microbiology, Sahlgrenska University Hospital Gothenburg; Karin Ejrnaes, National Center for Antimicrobials and Infection Control, Statens Serum Institute, Copenhagen, Denmark; and Florian Szabados, Institute for Hygiene and Microbiology, Ruhr University Bochum, Germany, for kindly providing us with isolates of S. saprophyticus. We also thank Anders Johansson for valuable comments.
This work was supported by grants from the Research and Development Unit, Jämtland County Council, Sweden, and the Medical Faculty of Umeå University, Umeå, Sweden.
Footnotes
Published ahead of print on 7 March 2007.
REFERENCES
- 1.Aspevall, O., B. Osterman, R. Dittmer, L. Sten, E. Lindback, and U. Forsum. 2002. Performance of four chromogenic urine culture media after one or two days of incubation compared with reference media. J. Clin. Microbiol. 40:1500-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi, S. H., J. H. Woo, J. Y. Jeong, N. J. Kim, M. N. Kim, Y. S. Kim, and J. Ryu. 2006. Clinical significance of Staphylococcus saprophyticus identified on blood culture in a tertiary care hospital. Diagn. Microbiol. Infect. Dis. 56:337-339. [DOI] [PubMed] [Google Scholar]
- 3.Colodner, R., S. Ken-Dror, B. Kavenshtock, B. Chazan, and R. Raz. 2006. Epidemiology and clinical characteristics of patients with Staphylococcus saprophyticus bacteriuria in Israel. Infection 34:278-281. [DOI] [PubMed] [Google Scholar]
- 4.Ejrnaes, K., D. Sandvang, B. Lundgren, S. Ferry, S. Holm, T. Monsen, R. Lundholm, and N. Frimodt-Moller. 2006. Pulsed-field gel electrophoresis typing of Escherichia coli strains from samples collected before and after pivmecillinam or placebo treatment of uncomplicated community-acquired urinary tract infection in women. J. Clin. Microbiol. 44:1776-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferry, S., L. G. Burman, and B. Mattsson. 1987. Urinary tract infection in primary health care in northern Sweden. I. Epidemiology. Scand. J. Prim. Health Care 5:123-128. [DOI] [PubMed] [Google Scholar]
- 6.Ferry, S. A., S. E. Holm, H. Stenlund, R. Lundholm, and T. J. Monsen. 2007. Clinical and bacteriological outcome of different doses and duration of pivmecillinam compared with placebo therapy of uncomplicated lower urinary tract infection in women: the LUTIW project. Scand. J. Prim. Health Care 25:49-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferry, S. A., S. E. Holm, H. Stenlund, R. Lundholm, and T. J. Monsen. 2004. The natural course of uncomplicated lower urinary tract infection in women illustrated by a randomized placebo controlled study. Scand. J. Infect. Dis. 36:296-301. [DOI] [PubMed] [Google Scholar]
- 8.Hedman, P., O. Ringertz, B. Eriksson, P. Kvarnfors, M. Andersson, L. Bengtsson, and K. Olsson. 1990. Staphylococcus saprophyticus found to be a common contaminant of food. J. Infect. 21:11-19. [DOI] [PubMed] [Google Scholar]
- 9.Hovelius, B., and P. A. Mardh. 1984. Staphylococcus saprophyticus as a common cause of urinary tract infections. Rev. Infect. Dis. 6:328-337. [DOI] [PubMed] [Google Scholar]
- 10.Hunter, P. R., and M. A. Gaston. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krediet, T. G., E. M. Mascini, E. van Rooij, J. Vlooswijk, A. Paauw, L. J. Gerards, and A. Fleer. 2004. Molecular epidemiology of coagulase-negative staphylococci causing sepsis in a neonatal intensive care unit over an 11-year period. J. Clin. Microbiol. 42:992-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuroda, M., A. Yamashita, H. Hirakawa, M. Kumano, K. Morikawa, M. Higashide, A. Maruyama, Y. Inose, K. Matoba, H. Toh, S. Kuhara, M. Hattori, and T. Ohta. 2005. Whole genome sequence of Staphylococcus saprophyticus reveals the pathogenesis of uncomplicated urinary tract infection. Proc. Natl. Acad. Sci. USA 102:13272-13277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Latham, R. H., K. Running, and W. E. Stamm. 1983. Urinary tract infections in young adult women caused by Staphylococcus saprophyticus. JAMA 250:3063-3066. [PubMed] [Google Scholar]
- 14.Monsen, T., C. Olofsson, M. Ronnmark, and J. Wistrom. 2000. Clonal spread of staphylococci among patients with peritonitis associated with continuous ambulatory peritoneal dialysis. Kidney Int. 57:613-618. [DOI] [PubMed] [Google Scholar]
- 15.Monsen, T., M. Ronnmark, C. Olofsson, and J. Wistrom. 1998. An inexpensive and reliable method for routine identification of staphylococcal species. Eur. J. Clin. Microbiol. Infect. Dis. 17:327-335. [DOI] [PubMed] [Google Scholar]
- 16.Murchan, S., M. E. Kaufmann, A. Deplano, R. de Ryck, M. Struelens, C. E. Zinn, V. Fussing, S. Salmenlinna, J. Vuopio-Varkila, N. El Solh, C. Cuny, W. Witte, P. T. Tassios, N. Legakis, W. van Leeuwen, A. van Belkum, A. Vindel, I. Laconcha, J. Garaizar, S. Haeggman, B. Olsson-Liljequist, U. Ransjo, G. Coombes, and B. Cookson. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raz, R., R. Colodner, and C. M. Kunin. 2005. Who are you—Staphylococcus saprophyticus? Clin. Infect. Dis. 40:896-898. [DOI] [PubMed] [Google Scholar]
- 18.Rupp, M. E., D. E. Soper, and G. L. Archer. 1992. Colonization of the female genital tract with Staphylococcus saprophyticus. J. Clin. Microbiol. 30:2975-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schneider, P. F., and T. V. Riley. 1996. Staphylococcus saprophyticus urinary tract infections: epidemiological data from Western Australia. Eur. J. Epidemiol. 12:51-54. [DOI] [PubMed] [Google Scholar]
- 20.Struelens, M. J. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2-11. [DOI] [PubMed] [Google Scholar]
- 21.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallmark, G., I. Arremark, and B. Telander. 1978. Staphylococcus saprophyticus: a frequent cause of acute urinary tract infection among female outpatients. J. Infect. Dis. 138:791-797. [DOI] [PubMed] [Google Scholar]