Abstract
Genotypic population-based methods could be faster and less expensive than phenotypic recombinant assays for determining human immunodeficiency virus type 1 (HIV-1) coreceptor usage in patient samples, but their clinical use requires good genotype-phenotype correlation and concordance with clonal analyses. We have assessed these requirements by clonal analysis of the V1 to V3 env PCR products of 26 patients infected with subtype B HIV-1. We used the resulting set of molecular clones, all sequenced and characterized using a single-cycle recombinant virus phenotypic entry assay, to reevaluate genotype-phenotype correlations. Combining the previously described 11/25 and net charge rules for the V3 genotype improved the prediction of HIV-1 coreceptor usage. We also evaluated the concordance of population-based and clonal analyses for predicting the coreceptor usage of HIV-1 quasispecies. Our population-based recombinant phenotypic assay and direct sequencing of V3 were similarly sensitive for detecting the presence of minor species in the virus population, and both correlated well with clonal analysis. The improved genotype-phenotype correlation obtained by combining two simple genotypic rules and the good concordance with clonal analyses suggest that direct sequencing of V3 is a valuable alternative to population-based recombinant phenotypic assays.
The chemokine receptors CCR5 and CXCR4 are the main coreceptors for human immunodeficiency virus type 1 (HIV-1) entry into target cells (2, 12, 13), and virus strains can be classified as R5, R5X4, and X4 variants depending on their use of one or both coreceptors (3).
Coreceptor usage determines the tropism of the virus for target cells and is thus critical for HIV-1 pathogenesis both throughout the natural course of infection (4, 28) and during antiretroviral therapy (10, 11, 19). Precise characterization of HIV-1 coreceptor usage is thus clinically relevant and of increasing importance because of the future therapeutic use of inhibitors of HIV-1 entry specific for CCR5 and CXCR4 coreceptors. However, the high genetic variability of HIV-1 and the complex constitution of the resulting virus quasispecies hamper the determination of virus coreceptor usage in a given HIV-infected individual. Hence, population-based assessment of HIV-1 tropism, as it is usually performed, could lead to a biased evaluation of the coreceptor usage of minor species in the virus population. It is also still unclear whether HIV-1 isolates that use both CCR5 and CXCR4 entry coreceptors are mainly a mixture of pure R5 and X4 monotropic variants or contain truly R5X4 dualtropic virus clones.
Phenotypic and genotypic methods have been developed to assess HIV-1 coreceptor usage. The major genotypic determinants for HIV-1 coreceptor usage lie in the V1-V2 and V3 variable loops of the gp120 envelope glycoprotein (6, 20, 29). Minimal changes in the V3 amino acid sequence are sufficient to switch coreceptor usage from CCR5 to CXCR4, and key mutations for CXCR4 usage have been identified, notably substitutions with basic residues at V3 positions 11 and/or 25 (8, 9, 14, 15, 26). An increased net charge of V3 is also associated with the use of CXCR4 by HIV-1 (1, 5, 15, 24). Bioinformatic tools have been developed to predict HIV-1 coreceptor usage from the amino acid sequence of V3, taking into account the key amino acids at positions 11 and 25, plus other sites in V3 that differ between CCR5- and CXCR4-using strains (5, 21, 26). Although the V3 amino acid sequence critically influences HIV-1 coreceptor usage, additional variations in the V1-V2 sequence could also influence HIV-1 coreceptor usage (6, 7, 16, 17, 23), but relatively few sets of genotype-phenotype data are available for regions other than the V3 region. The standard phenotypic assays used to identify HIV-1 coreceptor usage required 10 to 20 days of culture to detect virus replication and cytopathic effects on indicator cell lines bearing CD4 and CCR5 or CXCR4. By contrast, sensitive recombinant virus assays can detect coreceptor-restricted virus entry in a single-cycle assay (30). The standard assays could have led to a significant proportion of misdetection of CXCR4 coreceptor usage and hence misinterpretations of the data used for genotype-phenotype correlations.
We have investigated the suitability of direct sequencing of V3 as an alternative to population-based phenotypic assays for determining HIV-1 coreceptor usage in patient samples. Such a genotypic approach requires both good genotype-phenotype correlations and sufficient sensitivity to detect minor species in the virus population before it can be used clinically. We precisely described the constitution of virus quasispecies by using clonal analysis of V1-V3 env PCR products from the peripheral blood mononuclear cells (PBMCs) of 26 patients infected with subtype B HIV-1. The resulting set of molecular clones, all sequenced and characterized using a single-cycle recombinant virus phenotypic entry assay, were then used to reevaluate the genotype-phenotype correlations in these clones. Lastly, we evaluated the concordance between population-based and clonal analyses for predicting the coreceptor usage of HIV-1 quasispecies.
MATERIALS AND METHODS
Study subjects and samples.
We studied 26 patients infected with subtype B HIV-1 who were enrolled in the ANRS EP32 protocol at the Department of Infectious Diseases of Toulouse University Hospital, France. Samples of peripheral whole blood were obtained from each patient, and the Ficoll-separated PBMCs were stored at −80°C. This research was approved by the institutional review boards of Toulouse University Hospital and the Pasteur Institute, Paris, France.
PCR amplification of the V1-V3 region of HIV-1 env.
A nested PCR was used to amplify from PBMCs a sequence of the HIV-1 env gene spanning the V1-V2 and V3 regions. Both primary and nested PCRs were performed with a high-fidelity proofreading polymerase (Expand High Fidelity Plus PCR system; Roche Diagnostics), with the following PCR conditions: 2 min of initial denaturation at 94°C, 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C for 35 cycles. The outer primer pair used in the PCRs was 5′-CATGCCTGTGTACCCACAGA-3′ and 5′-CAGTAGAAAAATTCCCCTCCACA-3′, and the inner pair was 5′-ACCCCAACCCACAAGAAGTA-3′ and 5′-CCCCTCCACAATTAAAACTGT-3′. Samples were treated separately, and negative controls were systematically included. The primers used were located within conserved regions outside known regions linked to HIV-1 tropism, without any preferential matches among known sequences of R5, R5X4, and X4 viruses (data not shown).
Direct sequencing of V3 from bulk PCR products.
The V3 region was directly sequenced from bulk V1-V3 PCR products in both directions by the dideoxy chain termination method (BigDye Terminator v.3.1; Applied Biosystems) on an ABI 3130 DNA sequencer. The primers used for V3 sequencing were 5′-ACAATGYACACATGGAATTARGCCA-3′ and 5′-CCCCTCCACAATTAAAACTGT-3′. Direct sequencing was performed blinded to the clonal analyses. Minority species were detected when the automated sequencer electrophoregram showed a second base peak.
Cloning of V1-V3 PCR products.
The V1-V3 PCR products from the 26 patients were subjected to clonal analysis using a TOPO-TA cloning kit (Invitrogen). Plasmid DNAs containing V1-V3 inserts were purified using a QIAprep spin miniprep kit (QIAGEN) and sequenced in both directions.
Analysis of sequence data.
Multiple alignments were done with CLUSTALW 1.83, and sequence alignments were edited manually using BioEdit software. Detailed sequence alignments for each patient are given in Fig. S2 in the supplemental material of reference 31. Phylogenetic analysis showed distinct clusters of sequences for each patient that excluded any possibility of sample contamination or mix-up (data not shown). N-linked glycosylation sites in V1-V3 amino acid sequences were analyzed using the N-Glycosite tool at http://www.hiv.lanl.gov/content/hiv-db/GLYCOSITE/glycosite.html (31).
Phenotypic characterization of HIV-1 coreceptor usage.
The phenotype of HIV-1 coreceptor usage was determined using a recombinant virus assay (30). Briefly, subconfluent 293 T cells in 25-cm2 flasks were transfected using the calcium phosphate precipitation method with 8 μg of NheI-linearized 43-ΔV vector DNA (V1-V3-deleted pNL4-3) previously mixed in the same tube with 800 ng of PCR-amplified DNA from the patient sample. The V1-V3 PCR products spanned the V1-V3 region deleted from the 43-ΔV vector, with approximately 150-bp extensions on each side to allow homologous recombination during transfection. The recombinant virus released into the transfected 293 T-cell supernatant was used to infect indicator cells bearing CD4 and CCR5 (373-CCR5 cells) or CXCR4 (373-CXCR4 cells) coreceptors (22). These indicator cell lines carried an inducible LTR-LacZ cassette, which allows colorimetric assessment of virus infection by HIV-1 Tat-induced β-galactosidase expression 48 h after infection.
Statistical analysis.
The Kruskal-Wallis test was used to compare the numbers of N-glycosylation sites on R5, R5X4, and X4 viruses. Cuzick's test was used to assess a trend in V1 and V2 lengths across R5, R5X4, and X4 viruses. All tests were two sided, and P values of <0.05 were considered statistically significant. Statistical analysis was performed with Stata 8.2.
Nucleotide sequence accession numbers.
The sequences of the V1-V3 env molecular clones were given GenBank accession numbers DQ136796 to DQ137123.
RESULTS
Genotypic and phenotypic characterization of a set of V1-V3 env molecular clones.
We generated a set of V1-V3 env molecular clones, all sequenced and characterized using a single-cycle recombinant virus phenotypic entry assay, to evaluate genotype-phenotype correlations. The V1-V3 nucleotide sequences of a total of 436 molecular clones obtained from the 26 patients studied were analyzable. Of these, 368 had an open reading frame. Two patients (patients 11 and 12) harbored high frequencies of defective viruses with stop codons because of a high rate of G-to-A hypermutation (data not shown). When multiple copies of a single virus clone were found in a given virus population (n = 102 total multiple copies), virus clones were selected only once for determining HIV-1 coreceptor usage, giving a total of 266 different V1-V3 molecular clones. The phenotype of the coreceptor usage of V1-V3 recombinant virus was successfully determined for 218 of these 266 clones; it could not be determined for the remaining 48 clones, as no signals of virus infection were detected using 373-CCR5 or 373-CXCR4 indicator cell lines. This could be due to a defect in homologous recombination between the V1-V3 sequence and the 43-ΔV vector or to a defect in the infectivity of the recombinant viruses. We thus used a final sequence data set of 218 nonredundant V1-V3 molecular clones obtained from the 26 patients to evaluate genotype-phenotype correlations.
V1-V2 changes alone are not sufficient to cause CCR5-to-CXCR4 coreceptor switching independent of V3.
We found differences in the V1-V2 sequences of R5, R5X4, and X4 viruses, with a trend toward increased V1 and V2 length from R5 to R5X4 viruses and from R5X4 to X4 viruses (P < 0.001) (data not shown), but there was no statistically significant difference in the N-linked glycosylation patterns (P = 0.51) (data not shown). However, it is unclear whether changes in V1-V2 alone can cause coreceptor switching independent of V3. We identified the molecular clones in each virus population that were redundant for V3 but unique for V1-V2 (n = 100 total) to assess the impact of V1-V2 changes on coreceptor usage independent of V3. We found only seven clones from two patients (patients 1 and 21) in which changes in V1-V2 alone could account for their use or not of the CCR5 coreceptor in addition to CXCR4 (R5X4 versus X4 phenotype). However, we found no clone in which changes in V1-V2 alone were sufficient to explain a switch from an R5 to an R5X4 or X4 phenotype independent of changes in V3.
The combination of the 11/25 and net charge rules for the V3 genotype improves the prediction of HIV-1 coreceptor usage.
We reassessed the sensitivity, specificity, and positive and negative predictive values of two simple genotypic rules for the V3 genotype that have previously been proposed to predict HIV-1 coreceptor usage by using the 118 molecular clones that were nonredundant for both the V1-V2 and V3 regions (i.e., excluding the 100 clones with redundant V3 sequences). The 11/25 rule proposes that a basic amino acid (R or K) at either the 11 or 25 position of the V3 region is associated with CXCR4 coreceptor usage, whereas acidic or neutral amino acids at these positions are associated with CCR5 coreceptor usage (8, 9, 14, 15, 26). The net charge rule proposes that a net charge of ≥+5 for the V3 loop (calculated by subtracting the number of negatively charged amino acids [D and E] from the number of positively charged ones [K and R]) is associated with CXCR4 coreceptor usage, whereas a net charge of <+5 is associated with CCR5 coreceptor usage (1, 5, 15, 24). However, pure X4 monotropic and R5X4 dualtropic viruses cannot be identified by these genotypic rules; they both were classified as CXCR4-using viruses (R5X4/X4 phenotype in Tables 1 and 2).
TABLE 1.
Phenotypes of HIV-1 molecular clones based on V3 11/25 amino acids and net charge
| V3 genotype | Net charge | No. of clones with indicated phenotype
|
|
|---|---|---|---|
| R5 | R5X4/X4 | ||
| R at position 11 | <+5 | 0 | 0 |
| ≥+5 | 0 | 14 | |
| K at position 25 | <+5 | 0 | 0 |
| ≥+5 | 0 | 9 | |
| R at position 25 | <+5 | 6 | 0 |
| ≥+5 | 0 | 7 | |
| No R or K at position 11 or 25 | <+5 | 74 | 0 |
| ≥+5 | 6 | 2 | |
TABLE 2.
Performance of the 11/25 and/or net charge rule for genotypic prediction of CXCR4 coreceptor usage on HIV-1 molecular clonesa
| Genotypic rule(s) | Expected phenotype | No. of clones with indicated phenotype
|
Sens (%) | Spe (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|---|
| R5 | R5X4/X4 | ||||||
| One basic residue at position 11 or 25 | R5X4/X4 | 6 | 30 | 93.8 | 93 | 83.3 | 97.6 |
| No basic residue at position 11 or 25 | R5 | 80 | 2 | ||||
| Net charge of ≥+5 | R5X4/X4 | 6 | 32 | 100 | 93 | 84.2 | 100 |
| Net charge of <+5 | R5 | 80 | 0 | ||||
| One basic residue at position 11 or 25 and net charge of ≥+5 | R5X4/X4 | 0 | 30 | 93.8 | 100 | 100 | 97.6 |
| No concomitant basic residue at position 11 or 25 and net charge of ≥+5 | R5 | 86 | 2 | ||||
Basic residues refer to amino acids R and K. Sens, sensitivity; Spe, specificity; PPV, positive predictive value; NPV, negative predictive value.
An R amino acid residue was found at V3 position 11 (11R genotype) in 14 of our molecular clones, and a K amino acid was found at V3 position 25 (25K genotype) in 9 clones (Table 1). All of these clones with an 11R or a 25K genotype had a concordant X4 or R5X4 phenotype, as predicted by the 11/25 rule. They all also had a net charge of ≥+5, and both rules were thus in agreement for predicting CXCR4 coreceptor usage for these clones. Thirteen clones had an R amino acid at V3 position 25 (Table 1). However, only 7 of these 13 clones had a concordant X4 or R5X4 phenotype, whereas the 6 remaining clones had a discordant R5 phenotype. The 11/25 rule thus seems to be less reliable for predicting CXCR4 coreceptor usage for the 25R genotype than for the 11R and 25K genotypes, as already reported (18). The seven clones with a 25R genotype and a concordant X4 or R5X4 phenotype also had a net charge of ≥+5, whereas the remaining six clones with a 25R genotype but a discordant R5 phenotype had a net charge of <+5. By combining the requirements of the 11/25 and net charge rules for predicting coreceptor usage, all of the 13 clones harboring a 25R genotype could thus be correctly classified as CXCR4- or CCR5-using viruses. Similarly, the combination of the two rules also reduces the number of clones that would be misclassified as CXCR4-using viruses solely on the basis of a net charge of ≥+5. Hence, six of the eight clones with a net charge of ≥+5 but no concomitant key mutation at V3 position 11 or 25 were then correctly reclassified as CCR5-using viruses by combining the two rules (Table 1).
The combination of the 11/25 and net charge rules thus improves the sensitivity (93.8%) and specificity (100%) and positive (100%) and negative (96.7%) predictive values for CXCR4 coreceptor usage prediction in our set of molecular clones compared to the use of the 11/25 rule and the net charge rule separately (Table 2).
Population-based phenotypic characterization of HIV-1 coreceptor usage: concordance with clonal analysis.
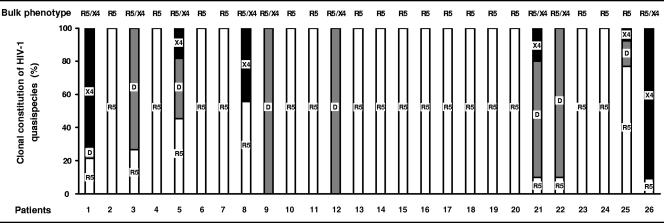
Clonal analysis of PCR products provides a randomly sampled image of the virus population present in a patient's sample, but such an approach is not suitable for routine analysis. However, the assessment of HIV-1 tropism in a population, as it is usually performed, could lead to a biased evaluation of the coreceptor usage of minor species in the virus population. We tested the sensitivity of phenotypic analysis of a bulk HIV-1 population by using our recombinant virus phenotypic entry assay for detecting CXCR4-using clones in our 26 patients. Phenotypic analysis of uncloned V1-V3 PCR products revealed virus populations with a pure R5 phenotype in 17 patients and a mixed R5 and X4 phenotype in 9 patients. However, it is still unclear whether such mixed R5 and X4 phenotypes in populations are due to a mixture of pure R5 and X4 monotropic virus clones or to truly R5X4 dualtropic virus clones. Clonal analysis of the virus populations of these nine patients showed that two patients (patients 8 and 26) harbored a mixture of pure R5 and X4 monotropic virus clones, while the other seven harbored some truly R5X4 dualtropic virus clones. The phenotypes of the bulk virus population and the proportion of R5, R5X4, and X4 virus molecular clones making up the quasispecies of each patient are summarized in Fig. 1. Phenotypic analysis of the bulk virus population was concordant with the clonal constitution of HIV-1 quasispecies in all but one patient. This patient (patient 23) harbored minority X4 and R5X4 clones (accounting for about 20% of the clonal population) that were not detected by bulk phenotypic analysis.
FIG. 1.
Phenotypic characterization of HIV-1 coreceptor usage by bulk and clonal analyses of virus quasispecies. The bulk phenotype obtained with our recombinant virus phenotypic entry assay of uncloned env PCR products for the 26 patients is indicated at the top of the figure. The results obtained by bulk and clonal analyses were compared using the proportions of R5 (white), R5X4 dualtropic D (gray), and X4 (black) molecular clones making up the quasispecies for each patient. Genetically unique (n = 218) and redundant (n = 102) molecular clones are included in the descriptions of the quasispecies since all contributed to the bulk signal obtained when the phenotype of the entire virus population was tested.
Performance of direct sequencing of the V3 region of env to predict the coreceptor usage of HIV-1 quasispecies.
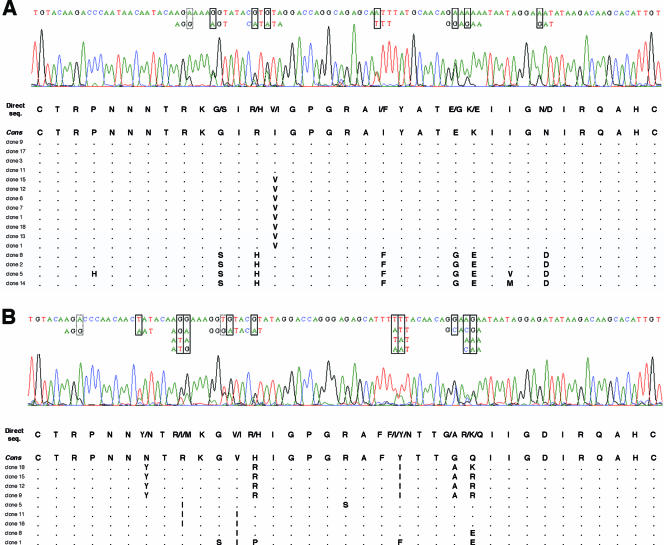
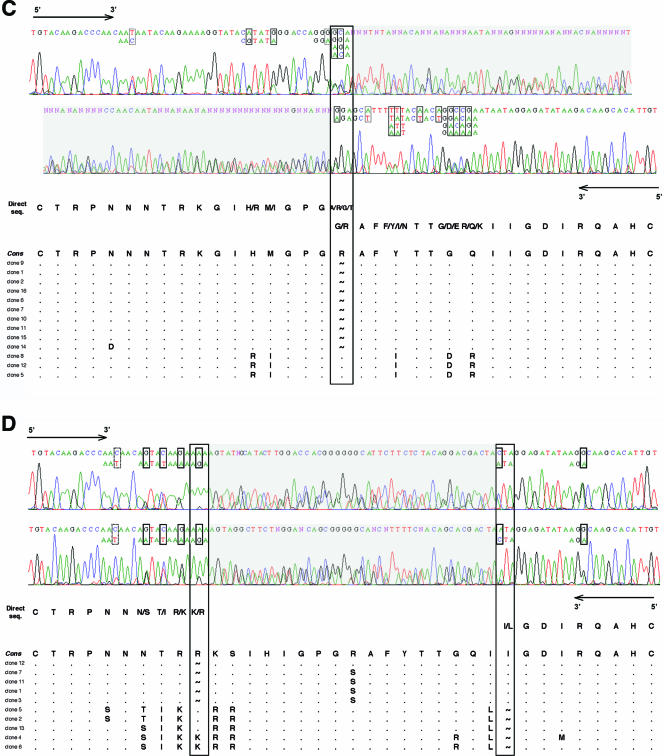
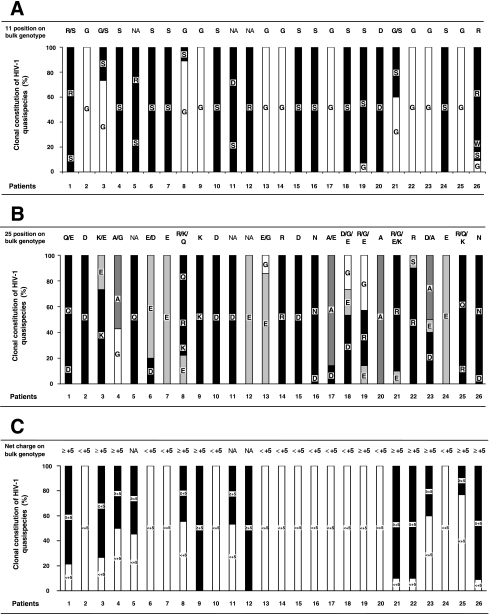
Given the good correlation between the V3 genotype and the entry phenotype obtained by combining the 11/25 and net charge rules, we examined the feasibility of using direct sequencing of V3 from bulk PCR products to assess the coreceptor usage of a given HIV-1 population rather than bulk phenotypic analysis. Direct sequencing of bulk PCR products, performed blinded to their clone content, was used to determine the V3 genotype of the majority virus population and to detect minor variants when the automated sequencer electrophoregram showed a second base peak (Fig. 2). We determined the V3 genotype in 23 of 26 patients. A stop codon in the V3 sequence from two patients (patients 11 and 12) harboring a high frequency of defective viruses prevented the determination of their V3 genotype. The full V3 genotype of a third patient (patient 5) was not determined by direct sequencing because of multiple-length polymorphism in V3 (Fig. 2D). The presence of CXCR4-using viruses in HIV-1 quasispecies was suspected when an R or K amino acid at V3 position 11 or 25 was detected in major or minor species on the electrophoregram, concomitant with a maximum net charge of ≥+5 (when considering the combination of codons that resulted in the highest net charge). Our direct sequencing detected minor species accounting for more than about 20% of the virus population, a result similar to that obtained with bulk phenotypic analysis. However, it can be difficult to determine the V3 genotype by directly sequencing bulk PCR products in some individuals harboring a great variety of virus quasispecies. The length polymorphism of V3, although it is rarer than that of V1-V2, can result in a frameshift and hence overlapping peaks on the electrophoregram (Fig. 2C and D). The genotype of a virus population harboring clones with a codon insertion or deletion at a single position in V3 can still be determined by sequencing the PCR products in the sense direction from the start to the insertion/deletion site and in the antisense direction from the end back to the insertion/deletion site (patient 25 [Fig. 2C] and patient 1 [data not shown] virus populations). However, it was impossible to determine the entire V3 genotype when insertions or deletions occurred at multiple positions of V3 (patient 5 virus population) (Fig. 2D). Nevertheless, the 23 of 26 V3 genotypes of bulk PCR products that were analyzable globally correlated well with the clonal analyses in terms of V3 positions 11 (Fig. 3A) and 25 (Fig. 3B) and the net charge (Fig. 3C). Combining the 11/25 and net charge rules ensured that the V3 genotype obtained by direct sequencing accurately predicted the coreceptor usage in all but one patient (patient 19).
FIG. 2.
Electrophoregrams of V3 obtained by direct sequencing: concordance with clonal genotypic analysis. Minor species were detected when the automated sequencer electrophoregram showed a second base peak. Various nucleotides are boxed (solid, nonsynonymous changes; dashed, synonymous changes), and all possible combinations are shown when contiguous nucleotide polymorphisms are present in a given codon. The V3 genotype is shown with all possible amino acids deduced from direct sequencing (seq.) and compared to aligned amino acid sequences obtained by clonal analysis. Genetically unique (n = 218) and redundant (n = 102) molecular clones have been included in the description of the quasispecies since all contributed to the bulk sequence obtained when the entire virus population was sequenced. (A and B) Successful genotyping of V3 by direct sequencing. Examples of patient 3 and 8 virus populations. Automated sequencer electrophoregrams obtained from bulk PCR products of patient 3 (A) and patient 8 (B) virus populations. The PCR products were sequenced in both directions, but only the 5′-to-3′ strand is shown. (C and D) Difficulties in genotyping of V3 because of length polymorphisms. Examples of patient 25 and 5 virus populations. Automated sequencer electrophoregrams for bulk PCR products of patient 25 (C) and patient 5 (D) virus populations. Both the forward 5′-to-3′ and reverse 3′-to-5′ strands are shown. The single (C) and multiple (D) insertion/deletion sites responsible for a downstream frameshift in certain virus clones are boxed. A tilde represents a gap inserted in the amino acid sequence of a clone to maintain the alignment. Electrophoregrams that could not be analyzed because of overlapping peaks are shaded gray. The V3 genotype of the patient 25 virus population was determined from the start to the insertion/deletion site in the sense direction and from the end back to the insertion/deletion site in the antisense direction (C). Insertion/deletion sites at two different positions within V3 prevented the determination of the full genotype of the patient 5 virus population (D). Cons, consensus sequence.
FIG. 3.
Genotypic characterization of V3 positions 11 and 25 and net charge by bulk and clonal analyses of virus quasispecies. The bulk genotype obtained by direct sequencing of uncloned env PCR products for the 26 patients is indicated at the top of the figure. NA (not analyzable) is indicated if the bulk genotype could not be determined by direct sequencing. The results obtained by bulk and clonal analyses were compared using the proportions of molecular clones making up the quasispecies for each patient. Genetically unique (n = 218) and redundant (n = 102) molecular clones are included in the description of the quasispecies since all contributed to the bulk sequence obtained when the entire virus population was sequenced. (A) Bulk and clonal genotypic analyses of V3 position 11. (B) Bulk and clonal genotypic analyses of V3 position 25. (C) Bulk and clonal genotypic analyses of the V3 net charge.
DISCUSSION
The therapeutic use of HIV-1 entry inhibitors will require efficient affordable methods for determining how circulating viruses in HIV-infected patients use coreceptors. Genotyping of V3 env could be an easier way to predict HIV-1 coreceptor usage than recombinant phenotypic assays. It implies that there is a good correlation between the genotype and the phenotype, but the V3 env sequences previously used to study this correlation were based on relatively insensitive phenotypic assays. In addition, the precise genotypic or phenotypic determination of virus coreceptor usage in HIV-infected patients is hampered by the great diversity of virus quasispecies.
The data for V1-V3 molecular clones from our 26 patients were obtained using a single-cycle recombinant virus phenotypic entry assay and used to study the correlation between the V1-V3 sequences of clones and their associated phenotypes. We found that changes in V1-V2 are not sufficient to cause a switch from the CCR5 to the CXCR4 coreceptor if there is no change in V3. We found only a few virus clones in which V1-V2 changes alone could account for their use or not of the CCR5 coreceptor in addition to CXCR4 (R5X4 versus X4 phenotype). Changes in V1-V2 may be important for maintaining the virus replicative capacity by compensating for deleterious V3 mutations during coreceptor switching (25, 27).
We have studied the correlation between the V3 genotype and phenotype in our molecular clones, as V3 is most important for determining HIV-1 coreceptor usage. We reassessed the performance of published genotypic rules for predicting CXCR4 usage (8, 9, 14, 15). The best predictions of CXCR4 usage were obtained when both the 11/25 and net charge rules were applied, giving excellent sensitivity and specificity. This combination significantly improved the prediction of CXCR4 usage by reducing the number of cases in which the genotype and the phenotype did not agree. The presence of an R at V3 position 25 is the least reliable of the 11/25 basic residues for predicting CXCR4 usage, and it has been proposed that only 25K be considered (18). However, we find that viruses harboring 25R but displaying a discordant R5 phenotype all have a net charge of <+5. A combination of the two rules thus correctly predicts the coreceptor usage of 25R viruses without losing any sensitivity. Sequences with a net charge of ≥+5 but no associated R or K amino acid at V3 position 11 or 25 also account for some discordant genotype-phenotype results, and again, this bias is greatly reduced by combining the two rules. We calculated the V3 net charge by subtracting the number of negatively charged amino acids, D and E, from the number of positively charged ones, K and R. However, it is unclear whether an H amino acid should also be included in the number of positively charged amino acids to calculate the V3 net charge; both methods have been used in previous studies (15, 18, 21). We tested both methods on the data for our molecular clones. The (K + R) − (D + E) rule was better at discriminating between R5 and R5X4 or X4 viruses than the (K + R + H) − (D + E) rule, regardless of the net charge cutoff chosen (data not shown).
Somewhat surprisingly, the sensitivity for detecting CXCR4-using viruses was much better than that reported previously, even when the 11/25 and net charge rules were assessed separately (21, 26). This may be because standard phenotypic assays significantly misdetected the CXCR4 coreceptor usage in the viruses previously used to correlate genotype and phenotype. GHOST cells have commonly been used in phenotypic assays, but these cells express low levels of CXCR4, which could have given false-negative results. In contrast, both U87 and U373 indicator cells stably express high levels of CD4 and chemokine coreceptors on their surfaces. Nevertheless, the sensitivities of standard phenotypic assays using U87 indicator cells are also likely to be suboptimal since these assays require 10 to 20 days of culture to detect virus replication and cytopathic effects. We have investigated the correlation between genotype and phenotype by using a large set of virus molecular clones that were all characterized with a single-cycle recombinant virus phenotypic entry assay. Hence, the good correlation between V3 genotype and virus phenotype we found could be due to more-accurate detection of CXCR4-using viruses. Previous studies also reported that the 11/25 and net charge rules were better at predicting the syncytium-inducing (SI) phenotype than at predicting CXCR4 coreceptor usage (18, 21, 26). This may be because there is no absolute concordance between SI and X4 genotype determinants and/or because of differences in the sensitivity and standardization of phenotypic techniques for detecting SI and CXCR4-using viruses.
Bioinformatic tools have been developed for predicting HIV-1 coreceptor usage from the V3 genotype, taking into account the key residues 11 and 25 plus additional sites in V3 that differ between CCR5- and CXCR4-using strains (21, 26). We find a general agreement between the results for our molecular clones obtained using the combined 11/25 and net charge rules and those obtained with a position-specific scoring matrix (PSSM) (21). The two X4 virus clones missed when using the combined 11/25 and net charge rules were correctly predicted by the PSSM, but five virus clones that were assessed correctly by the combined 11/25 and net charge rules were wrongly predicted by the PSSM test (data not shown). Our findings thus confirm that the coreceptor usage of HIV-1 molecular clones can reliably be predicted using the V3 genotype.
However, clonal analysis is not suitable for routine clinical practice; efficient population-based methods are needed. These may be genotype or phenotype analyses. Direct sequencing of V3 could be easier and less expensive than recombinant phenotypic assays for predicting HIV-1 coreceptor usage in routine practice. Our results indicate that the V3 bulk sequence was generally in agreement with clonal analysis for most of our patients. We determined the V3 genotype of the virus in most of our patients and accurately predicted HIV-1 coreceptor usage in all but one of them, although a great diversity of virus quasispecies or length polymorphisms can prevent the determination of the V3 genotype by direct sequencing in some patients.
Our detailed analysis of the coreceptor usage of HIV-1 quasispecies shows that population-based approaches are generally in good agreement with clonal analyses, even if the complex clonal constitution of virus populations can make it difficult to determine the coreceptor usage of HIV-1 quasispecies. The V3 genotype is a sensitive and specific indicator of CXCR4 coreceptor usage, so direct V3 sequencing could be a valuable alternative to phenotypic assays. These results could have important implications for monitoring HIV-1 coreceptor usage in a clinical setting, as HIV-1 entry inhibitors specific for CCR5 and CXCR4 coreceptors are currently being developed.
Acknowledgments
We thank F. Mammano for providing the 43-ΔV vector and M. Alizon for providing the U373 indicator cell lines. We also thank L. Cuzin, F. Balsarin, M. Barone, and C. Delourme for assistance in monitoring the ANRS EP32 study. The English text was checked by Owen Parkes.
P. Delobel received a doctoral fellowship from the College des Universitaires de Maladies Infectieuses et Tropicales and from the Pasteur Institute (CMIT-IP program). This study was supported by the Pasteur Institute and the French National Agency for AIDS Research (ANRS EP32 research grant).
This work is dedicated to the memory of Nicole Israël.
Footnotes
Published ahead of print on 28 February 2007.
REFERENCES
- 1.Albert, J., P. Stalhandske, S. Marquina, J. Karis, R. A. Fouchier, E. Norrby, and F. Chiodi. 1996. Biological phenotype of HIV type 2 isolates correlates with V3 genotype. AIDS Res. Hum. Retrovir. 12:821-828. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib, G., C. Combadiere, C. C. Broder, Y. Feng, P. E. Kennedy, P. M. Murphy, and E. A. Berger. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955-1958. [DOI] [PubMed] [Google Scholar]
- 3.Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 4.Berger, E. A., P. M. Murphy, and J. M. Farber. 1999. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17:657-700. [DOI] [PubMed] [Google Scholar]
- 5.Briggs, D. R., D. L. Tuttle, J. W. Sleasman, and M. M. Goodenow. 2000. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages). AIDS 14:2937-2939. [DOI] [PubMed] [Google Scholar]
- 6.Carrillo, A., and L. Ratner. 1996. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J. Virol. 70:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro, B., K. Wehrly, J. Nishio, and S. Perryman. 1996. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J. Virol. 70:9055-9059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Jong, J.-J., A. De Ronde, W. Keulen, M. Tersmette, and J. Goudsmit. 1992. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. J. Virol. 66:6777-6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Jong, J.-J., J. Goudsmit, W. Keulen, B. Klaver, W. Krone, M. Tersmette, and A. de Ronde. 1992. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication capacity. J. Virol. 66:757-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delobel, P., M. T. Nugeyre, M. Cazabat, K. Sandres-Saune, C. Pasquier, L. Cuzin, B. Marchou, P. Massip, R. Cheynier, F. Barre-Sinoussi, J. Izopet, and N. Israel. 2006. Naive T-cell depletion related to infection by X4 human immunodeficiency virus type 1 in poor immunological responders to highly active antiretroviral therapy. J. Virol. 80:10229-10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delobel, P., K. Sandres-Saune, M. Cazabat, C. Pasquier, B. Marchou, P. Massip, and J. Izopet. 2005. R5 to X4 switch of the predominant HIV-1 population in cellular reservoirs during effective highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 38:382-392. [DOI] [PubMed] [Google Scholar]
- 12.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 13.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 14.Fouchier, R. A., M. Brouwer, S. M. Broersen, and H. Schuitemaker. 1995. Simple determination of human immunodeficiency virus type 1 syncytium-inducing V3 genotype by PCR. J. Clin. Microbiol. 33:906-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fouchier, R. A., M. Groenink, N. A. Kootstra, M. Tersmette, H. G. Huisman, F. Miedema, and H. Schuitemaker. 1992. Phenotype-associated sequence variation in the third variable domain of the human immunodeficiency virus type 1 gp120 molecule. J. Virol. 66:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groenink, M., A. C. Andeweg, R. A. M. Fouchier, S. Broersen, R. C. M. van der Jagt, H. Schuitemaker, R. E. Y. de Goede, M. L. Bosch, H. G. Huisman, and M. Tersmette. 1992. Phenotype-associated env gene variation among eight related human immunodeficiency virus type 1 clones: evidence for in vivo recombination and determinants of cytotropism outside the V3 domain. J. Virol. 66:6175-6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groenink, M., R. A. Fouchier, S. Broersen, C. H. Baker, M. Koot, A. B. van't Wout, H. G. Huisman, F. Miedema, M. Tersmette, and H. Schuitemaker. 1993. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science 260:1513-1516. [DOI] [PubMed] [Google Scholar]
- 18.Hoffman, N. G., F. Seillier-Moiseiwitsch, J. Ahn, J. M. Walker, and R. Swanstrom. 2002. Variability in the human immunodeficiency virus type 1 gp120 Env protein linked to phenotype-associated changes in the V3 loop. J. Virol. 76:3852-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunt, P. W., P. R. Harrigan, W. Huang, M. Bates, D. W. Williamson, J. M. McCune, R. W. Price, S. S. Spudich, H. Lampiris, R. Hoh, T. Leigler, J. N. Martin, and S. G. Deeks. 2006. Prevalence of CXCR4 tropism among antiretroviral-treated HIV-1-infected patients with detectable viremia. J. Infect. Dis. 194:926-930. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253:71-74. [DOI] [PubMed] [Google Scholar]
- 21.Jensen, M. A., F.-S. Li, A. B. van 't Wout, D. C. Nickle, D. Shriner, H.-X. He, S. McLaughlin, R. Shankarappa, J. B. Margolick, and J. I. Mullins. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376-13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Labrosse, B., A. Brelot, N. Heveker, N. Sol, D. Schols, E. De Clercq, and M. Alizon. 1998. Determinants for sensitivity of human immunodeficiency virus coreceptor CXCR4 to the bicyclam AMD3100. J. Virol. 72:6381-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masciotra, S., S. M. Owen, D. Rudolph, C. Yang, B. Wang, N. Saksena, T. Spira, S. Dhawan, and R. B. Lal. 2002. Temporal relationship between V1V2 variation, macrophage replication, and coreceptor adaptation during HIV-1 disease progression. AIDS 16:1887-1898. [DOI] [PubMed] [Google Scholar]
- 24.Michael, N. L., G. Chang, P. K. Ehrenberg, M. T. Vahey, and R. R. Redfield. 1993. HIV-1 proviral genotypes from the peripheral blood mononuclear cells of an infected patient are differentially represented in expressed sequences. J. Acquir. Immune Defic. Syndr. 6:1073-1085. [PubMed] [Google Scholar]
- 25.Pastore, C., R. Nedellec, A. Ramos, S. Pontow, L. Ratner, and D. E. Mosier. 2006. Human immunodeficiency virus type 1 coreceptor switching: V1/V2 gain-of-fitness mutations compensate for V3 loss-of-fitness mutations. J. Virol. 80:750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Resch, W., N. Hoffman, and R. Swanstrom. 2001. Improved success of phenotype prediction of the human immunodeficiency virus type 1 from envelope variable loop 3 sequence using neural networks. Virology 288:51-62. [DOI] [PubMed] [Google Scholar]
- 27.Schuitemaker, H., R. A. Fouchier, S. Broersen, M. Groenink, M. Koot, A. B. van 't Wout, H. G. Huisman, M. Tersmette, and F. Miedema. 1995. Envelope V2 configuration and HIV-1 phenotype: clarification. Science 268:115. [DOI] [PubMed] [Google Scholar]
- 28.Schuitemaker, H., M. Koot, N. A. Kootstra, M. W. Dercksen, R. E. de Goede, R. P. van Steenwijk, J. M. Lange, J. K. Schattenkerk, F. Miedema, and M. Tersmette. 1992. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus population. J. Virol. 66:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1991. Macrophage and T cell-line tropisms of HIV-1 are determined by specific regions of the envelope gp120 gene. Nature 349:167-169. [DOI] [PubMed] [Google Scholar]
- 30.Trouplin, V., F. Salvatori, F. Cappello, V. Obry, A. Brelot, N. Heveker, M. Alizon, G. Scarlatti, F. Clavel, and F. Mammano. 2001. Determination of coreceptor usage of human immunodeficiency virus type 1 from patient plasma samples by using a recombinant phenotypic assay. J. Virol. 75:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, M., B. Gaschen, W. Blay, B. Foley, N. Haigwood, C. Kuiken, and B. Korber. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14:1229-1246. [DOI] [PubMed] [Google Scholar]