Abstract
Vaccinia virus intracellular mature virus (IMV) binds to glycosaminoglycans (GAGs) on cells via three virion proteins, H3L, A27L, and D8L. In this study, we demonstrated that binding of IMV to BSC40 cells was competitively inhibited by soluble laminin but not by fibronectin or collagen V, suggesting that this cell surface extracellular matrix (ECM) protein may play a role in vaccinia virus entry. Moreover, IMV infection of GAG− sog9 cells was also inhibited by laminin, demonstrating that virion binding to laminin does not involve a prior interaction with GAGs. Furthermore, comparative envelope protein analyses of wild-type vaccinia virus strain Western Reserve, which binds to laminin, and of a mutant virus, IA27L, which does not, showed that the A26L open reading frame (ORF), encoding an envelope protein, was mutated in IA27L, resulting in A26L being absent from the IMV. Expression of the wild-type A26L ORF in IA27L resulted in laminin binding activity. Moreover, recombinant A26L protein bound to laminin in vitro with a high affinity, providing direct evidence that A26L is the laminin binding protein on IMV. In summary, these results reveal a novel role for the vaccinia viral envelope protein A26L in binding to the ECM protein laminin, an association that is proposed to facilitate IMV entry.
Vaccinia virus, the prototype of the Orthopoxvirus genus of the family Poxviridae, infects many cell lines and animals (16) and produces several forms of infectious particles, namely, intracellular mature virus (IMV), intracellular enveloped virus, cell-associated enveloped virus, and extracellular enveloped virus (EEV) (30). The membrane structure of IMV is different from that of intracellular enveloped virus, which again is different from that of cell-associated enveloped virus and EEV (47). Although IMV and EEV were originally thought to enter cells through different mechanisms (28, 39, 46), recent studies have revealed that a conserved viral fusion complex is required for the penetration of both (5, 24, 32, 33, 36-38, 42, 43).
The molecular mechanism of vaccinia virus entry remains largely unknown, partly due to the complexity of the virion, since the IMV contains more than 70 viral proteins, as shown by proteomic analysis (9, 34, 50). In addition, vaccinia virus is promiscuous in its infectivity, which may indicate redundant pathways of cell entry. Most studies have shown that viral cores are delivered into cells by fusion of the IMV envelope with the plasma membrane (2, 7, 14, 28, 46), but endocytosis was also recently reported (44). Due to the wide infectivity of the virus, we are interested in understanding how vaccinia virus IMV recognizes, and binds to, cells. We previously showed that the IMV contains three envelope proteins which bind to cell surface glycosaminoglycans (GAGs) during virus entry. Vaccinia viral H3L and A27L bind to heparan sulfates, and D8L binds to chondroitin sulfates (10, 20, 21, 27). Mutant viruses in which H3L or D8L gene expression is prevented (H3L− or D8L−, respectively), although less virulent in animals, remain infectious (27, 31). In addition, sog9 cells, which are defective in GAG expression, remained susceptible to IMV infections, implying that additional cellular molecules mediate vaccinia virus entry.
Cell surface extracellular matrix (ECM) proteins are ubiquitously expressed and are involved in a variety of cellular processes, including cell adhesion, migration, and survival (15, 41). Here, we report that we identified a viral envelope protein mediating virus and ECM protein laminin (LN) interaction.
MATERIALS AND METHODS
Cells, viruses, and reagents.
BSC40 cells, obtained from S. Pennathur, and sog9 cells, obtained from F. Tufaro (3), were cultured in Dulbecco's modified Eagle's medium supplemented with 10% calf serum (both from Biological Industries, Israel). The vaccinia virus Western Reserve (WR) strain was used in this study. A mutant virus, WR32-7/Ind14K, derived from the Western Reserve strain, was obtained from G. Smith (35); in this study, for simplicity, it will be referred to as IA27L. IA27L contains an A27L open reading frame (ORF) under IPTG (isopropyl-β-d-thiogalactopyranoside) regulation, so it was propagated in culture medium containing 5 mM IPTG (35). The virus vMJ360, expressing lacZ from a viral early promoter, was obtained from B. Moss (13). All viruses were grown on BSC40 cells, and IMVs were purified by two successive centrifugations through 36% and 25 to 40% sucrose gradient layers as described previously (25). Human laminin, fibronectin (FN), and collagen V (CN) were purchased from Sigma.
Preparation of recombinant GST-A26L(127-500) fusion protein and rabbit immunization.
Full-length A26L has 500 amino acids but was found to form insoluble inclusion bodies upon expression as a glutathione (GSH) S-transferase (GST) fusion protein. This problem was overcome using a construct consisting of residues 127 to 500 of A26L fused to GST [GST-A26L(127-500)], and the purified protein was used as an antigen to generate antibodies in a New Zealand White rabbit. The A26L ORF, encoding amino acids 127 to 500, was amplified by PCR using the following two primers (restriction sites are underlined): 5′ primer, 5′-CCCGGATCCTTTTGTATATCTGGT-3′, and 3′ primer, 5′-GCGGTCGACTTATAAAATCGTAGATCTCCC-3′. The PCR product was digested with BamHI and SalI and cloned into pGEX-4T-1b, resulting in plasmid pGEX-A26, which was transformed into Escherichia coli BL21(DE3) and grown overnight. The culture was then diluted 1:100, and growth continued until the optical density at 600 nm reached 0.6 to 0.8, before induction with 0.2 mM IPTG for 30 min at 37°C. The bacteria were then harvested and lysed by incubation for 5 min at 4°C in lysis buffer (1% Triton X-100, 2 mM EDTA, 0.1% β-mercaptoethanol, 0.2 mM phenylmethylsulfonyl fluoride, and 0.5 mM benzamidine), and the recombinant protein was bound to glutathione-Sepharose 4B beads (Amersham Biosciences), eluted with 10 mM glutathione in 50 mM Tris-HCl buffer, pH 8.0, and dialyzed against phosphate-buffered saline (PBS).
For antiserum production, 500 μg of dialyzed GST-A26L(127-500) was mixed with complete adjuvant and injected intramuscularly into a New Zealand White rabbit. The rabbit was boosted with 250 μg of GST-A26L(127-500) five times at 2-week intervals and bled 1 week after the fifth boost. The anti-A26L antiserum (named B10) recognized A26L on immunoblots up to a dilution of 1:1,000.
Membrane protein extraction from IMV and mass spectrum analysis of the protein of interest.
Vaccinia virus IMV particles were extracted with 1% NP-40 containing 50 mM dithiothreitol (DTT) to separate the membrane and core fractions, as described previously (8). In brief, purified IMV (108 PFU) was incubated for 1 h at 37°C in 1% NP-40, 50 mM Tris-HCl, pH 7.5, and 50 mM dithiothreitol, and the insoluble and soluble fractions were separated by centrifugation at 14,000 × g for 30 min at 4°C. Proteins in the pellet and supernatant were separated on 7% sodium dodecyl sulfate (SDS) gels and detected by silver staining or immunoblotting. After silver staining, the protein band was excised and subjected to tryptic digestion and mass spectrum analysis performed by EverNew Biotech Inc. The 11 tryptic peptides obtained were used to search the data bank, and the protein identity was determined.
Construction of IA27L-A26WR virus. (i) Construction of plasmids expressing the A26L ORF from the WR strain for insertion into IA27L.
The IA27L virus, generated from the wild-type Western Reserve strain, was previously described as WR32-7/Ind14K (35). In the IA27L virus, the A27L locus is inactivated by Ecogpt insertion and the insertion of another copy of the A27L gene under IPTG regulation into the tk locus (35). To insert the A26L (WR) ORF into IA27L, the 5′- and 3′-flanking sequences generated in the PCRs were derived, respectively, from the A25L ORF and A28L ORF. The 5′-flanking sequence was generated by PCR using IA27L vaccinia viral genomic DNA as template and the following two primers (XhoI and KpnI restriction sites are underlined): 5′-GGGCTCGAGTTACCCGATTGTAGTTAA-3′ and 5′-GGGGGTACCTAGATAATGATTAATGTT-3′. The PCR product was digested with XhoI and KpnI and cloned into the pBlueScript KS(−) vector (Stratagene), resulting in plasmid pA25-KS(−). The full-length A26L ORF was generated by PCR using vaccinia virus Western Reserve genome as the template and the following two primers (XhoI and SalI restriction sites are underlined): 5′-GGGCTCGAGTTATAAAATCGTAGATCT-3′ and 5′-GGGGTCGACGAATTTCATTTTGTTTTTTTCTATGCTATAAATGGCGAACATTATAAAT-3′. The A26L (WR) PCR DNA fragment was cloned into plasmid pA25-KS(−) to produce plasmid pA25/A26L(WR)-KS(−). The 3-kb p11k-lacZ gene expression cassette was isolated from pSC11-5t by SalI and PstI digestion as described previously (8) and ligated with the A25/A26L(WR)-KS(−) plasmid to create plasmid A25/A26L(WR)/lacZ-KS(−). The 3′-flanking fragment was generated by PCR using IA27L viral genomic DNA as template and the following two primers (NotI and SmaI restriction sites are underlined): 5′-CCCGCGGCCGCGAATGTTGTTTGTTAATG-3′ and 5′-AAACCCGGGTCATGTCTGGATCCGTCG-3′. The resulting 3′-flanking fragment was inserted into pA25/A26L(WR)/lacZ-KS(−) to create plasmid pA25/A26L(WR)/lacZ/A28-KS(−). The sequences of the PCR fragments were confirmed by DNA sequencing.
(ii) Generation of recombinant IA27L-A26WR virus.
293T cells (106) were infected with IA27L virus and subsequently transfected with 2 μg of pA25/A26L(WR)/lacZ/A28-KS(−). The cells were cultured in normal medium supplemented with 5 mM IPTG and harvested 2 days postinfection (p.i.). Recombinant IA27L-A26WR virus was isolated by three rounds of plaque purification in agar containing 150 μg/ml of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as described previously (8).
One-step growth curve analysis.
BSC40 cells were infected with each vaccinia virus at a multiplicity of infection (MOI) of 5 PFU per cell, cultures were harvested at different times p.i., and the IMV titers in the lysates were determined on BSC40 cells as described previously (8). To determine the EEV titers, medium was collected at 24 h p.i. and, after a brief centrifugation (200 × g, 5 min) to remove cell debris, a monoclonal antibody, 2D5, against IMV L1R protein (1:500) (23) was added to the medium and the mixture was rotated for 30 min at 4°C to block IMV contamination in the medium, as described previously (23). The mixture was then added to BSC40 cells for plaque determination under an agar overlay.
Immunoblot analysis.
Viral proteins from purified IMVs or extracts (1% NP-40 and 50 mM DTT) from virus-infected cells were fractionated by electrophoresis on 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes for immunoblot analysis with anti-A26L (1:1,000 dilution), anti-D8L (1:1,000 dilution), or anti-H3L (1:1,000 dilution) antibodies as described previously (8).
Blocking assay using soluble ECM proteins.
To test whether soluble ECM proteins blocked the binding of vaccinia virus to cells, we measured IMV binding to cells using plaque assays (21) or confocal immunofluorescence microscopy (48). Depending on the assay sensitivity we used different amount of viruses for binding assays. The plaque assays are very sensitive, so we used 150 PFU in each experiment. For immunofluorescence microscopy, we used an MOI of 10 or 20 PFU per cell for each experiment. The results from the two assays were consistent. In brief, vaccinia virus was incubated for 30 min at 4°C with various amounts of the soluble ECM protein and then the mixtures were added to BSC40 cells and incubated at 4°C for 30 min. The inoculum was then removed, and the cells were washed and processed for plaque assays under agar overlay or, alternatively, were fixed with 4% paraformaldehyde and bound virions were visualized by staining with anti-L1R antibody and confocal microscopy as described previously (11, 48). When viral early gene expression was monitored, BSC40 cells were infected at an MOI of 5 PFU per cell with vMJ360 (expressing lacZ from a viral early promoter) and harvested at 2 h p.i. and β-galactosidase (β-Gal) activity was measured using ONPG (o-nitrophenyl-β-d-galactopyranoside) (18) or X-Gal (10, 13).
Analysis of in vitro binding of soluble ECM protein by SPR.
Recombinant GST-A26L(127-500) protein, induced and purified as described above, was complexed with biotinylated glutathione as described below. Biotinylated GSH was prepared by adding 5 mM GSH (pH 7.0 in PBS) to 10 mM Ez-Link maleimide PEO2-biotin (Pierce Chemical Co.) in PBS, the mixture was gently shaken overnight at room temperature, and then 2-mercaptoethanol was added to a final concentration of 5 mM and the sample was incubated at room temperature for 1 h to deactivate excess 10 mM Ez-Link maleimide PEO2-biotin. GST-A26L(127-500) (5 μM) and the biotinylated GSH were incubated for 1 h at room temperature, and then the mixture was dialyzed against PBS for 24 h at room temperature to remove nonbound biotinylated GSH and used for BIAcore analysis. Real-time surface plasmon resonance (SPR) experiments were performed on a BIAcore Biosensor 3000 system (BIAcore, Uppsala, Sweden) using 10 mM HEPES, pH 7.0, 0.15 M NaCl, 0.005% (wt/vol) Polysorbate 20 as running buffer at a flow rate of 10 μl/minute. The biotinylated GSH/GST-A26L(127-500) complex was immobilized on the surface of sensor chips coated with streptavidin (SA) purchased from BIAcore by injection at a flow rate of 10 μl/min. Each ECM protein (1 μM) or laminin at four different concentrations (0.125, 0.25, 0.5, and 1 μM) was then injected for 180 s for binding, followed by injection of running buffer for 300 s for dissociation. Parallel injections of laminin onto a biotinylated GSH-GST complex immobilized on an SA sensor chip using the same procedure were performed to measure background binding. The ligand-coated surface was regenerated by two successive injections of 120 μl of 1 M NaCl.
RESULTS
Laminin binds to wild-type vaccinia virus IMV and blocks virion binding to BSC40 cells.
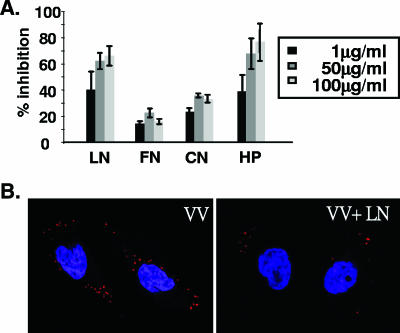
Purified vaccinia virus IMV virions were incubated for 60 min at 4°C with various soluble ECM proteins, and then the mixture was incubated with BSC40 cells and the amount of IMV bound was determined. As shown in Fig. 1A, at concentrations of 1, 50, and 100 μg/ml, soluble LN caused a significant block of the binding of IMVs to cells (∼70% of the maximum) in the plaque assays, whereas FN and CN did not. Heparin (HP), used as a positive control (10), blocked up to 80% of IMV binding in a dose-dependent manner. Figure 1B shows confocal microcopy results, demonstrating inhibition of IMV binding by soluble laminin. These results showed that IMV contains a laminin binding protein and suggested a possibility that IMVs bind to cell surface laminin during virus infection.
FIG. 1.
Laminin blocks IMV binding to BSC40 cells. (A) Different concentrations of soluble LN, FN, CN, or HP were mixed with purified IMV (150 PFU) for 30 min at 4°C and then added to cells for 30 min at 4°C, and the percent inhibition of virus binding to cells was determined by plaque assay as described in Materials and Methods. (B) Confocal immunofluorescence microscopy showing that soluble laminin reduces IMV binding to BSC40 cells. Cells were infected in the absence (VV) or presence (VV+LN) of 100 μg/ml of laminin at an MOI of 20 PFU per cell for 30 min at 4°C, and cell-bound virions were visualized by staining with anti-L1R antibody (red stain) and confocal microscopy as previously described (11, 48). The nucleus is stained blue with DAPI (4′,6′-diamidino-2-phenylindole).
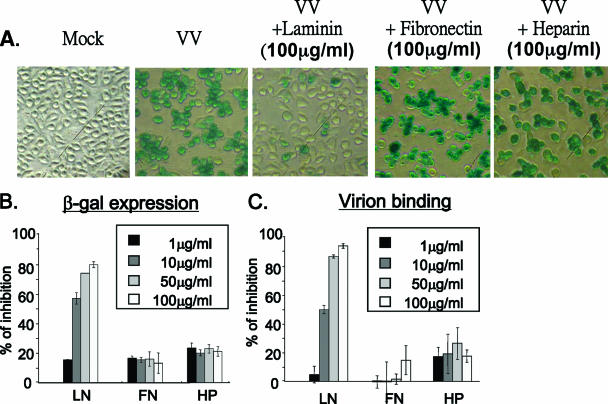
To determine whether IMV binding to laminin involved prior binding to GAGs, we used the sog9 cell line, which does not express any GAGs on the cell surface (3). Figures 2A and B show that viral infection of sog9 cells, as measured by expression of the early marker gene lacZ, was not blocked by heparin (negative control) but was significantly reduced by incubation of the IMVs with soluble laminin. Direct virion binding assays also showed that the amount of IMVs bound to cells was reduced by soluble laminin (Fig. 2C), consistent with the results of the early gene assays (Fig. 2B). The results suggested that vaccinia virus IMV binds to the cell surface ECM protein laminin during virus entry and that this interaction can occur independently of the binding of IMV to GAGs.
FIG. 2.
Soluble laminin inhibits vaccinia virus infection of sog9 cells. (A) sog9 cells were infected with vMJ360 (expressing lacZ from a viral early promoter) at an MOI of 5 PFU per cell in the presence (100 μg/ml) or absence of soluble laminin, fibronectin, or heparin as described in the legend to Fig. 1. The infected cells were fixed at 3 h p.i. in 0.5% paraformaldehyde and analyzed for β-Gal activity by X-Gal staining. (B) Experiments were performed as for panel A except that different concentrations (0, 1, 10, 50, and 100 μg/ml) of soluble laminin, fibronectin, or heparin were used and β-Gal activity was quantified by an ONPG assay as described previously (18). (C) Experiments were performed as for panel B except that the cells were harvested and cell-bound IMV particles were determined by confocal immunofluorescence analysis as described in Materials and Methods.
The putative viral laminin binding protein is not one of the previously identified GAG binding proteins and is absent in IA27L virus.
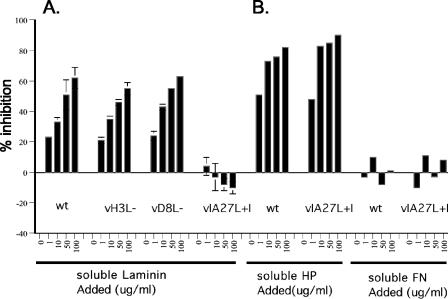
To identify the viral envelope protein that binds to laminin, we tested two previously described mutant viruses, H3L− and D8L−, in which the GAG binding proteins, H3L and D8L, respectively, are not expressed (21, 27). Figure 3A shows that H3L− and D8L− infections of cells were similarly inhibited by soluble laminin, showing that neither H3L nor D8L is responsible for the binding of IMV to laminin. In contrast, infection with WR32-7/Ind14K virus (designated in this article as IA27L virus), a previously described mutant virus that expresses A27L under IPTG regulation (35), was not blocked by soluble laminin, showing that IA27L IMV does not contain the viral protein that binds to laminin. This inability to bind to laminin appears to be a unique difference between IA27L virus and wild-type virus, since the binding of both viruses was affected identically by preincubation with heparin or fibronectin (Fig. 3B). These results indicate that the putative envelope protein that mediates IMV binding to laminin is not H3L or D8L and is absent in IA27L IMVs.
FIG. 3.
Laminin blocks the binding of wild-type vaccinia virus IMV to BSC40 cells, but not that of IA27L. Purified IMVs from wild-type vaccinia virus (wt), H3L−, D8L−, or IA27L were mixed for 30 min at 4°C with different concentrations (0, 1, 10, 50, and 100 μg/ml) of soluble laminin (A) or HP or FN (B) and then were used to infect BSC40 cells, and the percent inhibition of virus binding to cells was determined as described in Materials and Methods.
IA27L virus expresses a truncated A26L envelope protein that is not incorporated into IMV particles.
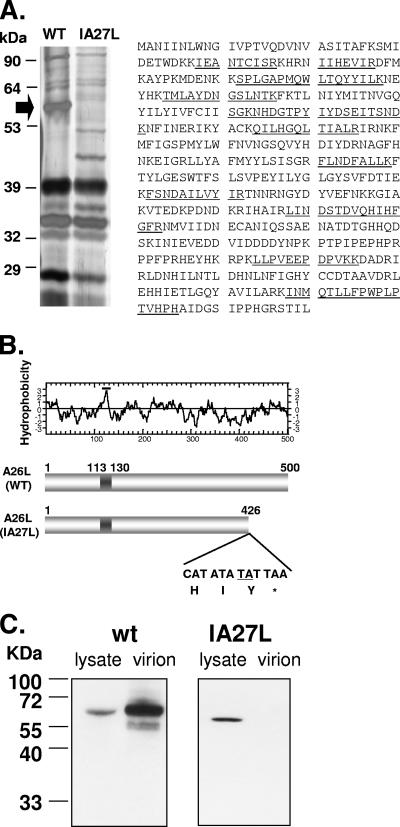
IA27L was derived from vaccinia virus Western Reserve strain mutant virus 48-7, which is reported to have some deletions in the left end of the viral genome (35). Since IA27L has not been sequenced, it was not possible to perform viral genome alignment to locate the ORFs that are altered in IA27L. To identify the envelope protein that binds to laminin, we extracted viral envelope proteins from both wild-type virus and IA27L and compared their profiles by 7% SDS-polyacrylamide gel electrophoresis. As shown in Fig. 4A, an abundant protein with a molecular mass of about 60 kDa was detected in wild-type virus but not in IA27L. The band was excised and subjected to mass spectroscopy analyses, and the protein was identified as vaccinia virus envelope P4c protein (also known as 4c) (29, 45). Vaccinia virus P4c from the Western Reserve strain, an orthologue of Copenhagen strain A26L, is a 500-amino-acid protein encoded by WR149 ORF (http://www.poxvirus.org/), with a possible hydrophobic region at residues 113 to 130 (Fig. 4B). For simplicity, we will refer to WR149 as A26L throughout the text. To determine why A26L was absent in IA27L IMV particles, we sequenced the A26L ORF in IA27L virus and found a TA insertion in the coding region (Fig. 4B), which resulted in an in-frame termination downstream of the insertion site, producing a truncated A26L polypeptide of 426 amino acids. We then prepared anti-A26L antibodies by injecting a New Zealand White rabbit with a recombinant GST protein consisting of residues 127 to 500 of A26L fused to GST, GST-A26L(127-500); used these in immunoblot analyses of cell lysates and purified IMVs from wild-type virus and IA27L; and found that a full-length 60-kDa protein was detected in cells infected with wild-type vaccinia virus and a smaller truncated form was detected in cells infected with IA27L (Fig. 4C). Most importantly, the wild-type A26L was detected in purified wild-type IMVs, but the truncated protein was not detected in IA27L IMVs, demonstrating that the truncation abolished the incorporation of A26L into IMV particles. Incorporation of other envelope proteins, such as H3L and D8L, into the virions was not affected (data not shown).
FIG. 4.
A26L is absent in envelope proteins extracted from IA27L. (A) Silver staining of a 7% SDS-polyacrylamide gel showing IMV membrane proteins extracted from wild-type virus (WT) and IA27L using 1% NP-40 and 50 mM DTT (left panel). The arrow indicates the 58-kDa protein excised for protein identification by mass spectroscopy. Right panel: amino acid sequence of the A26L/WR149 protein taken from http://www.poxvirus.org/; the 11 tryptic peptides detected by mass spectrometry are underlined. (B) Upper panel: hydrophobicity plot of A26L from vaccinia virus Western Reserve strain. Region 113-130 contains hydrophobic residues and is a potential transmembrane region. Lower panel: insertion of nucleotides TA (underlined) in IA27L and the resulting premature termination codon TAA. WT, wild type. (C) Immunoblot analysis of A26L in lysates and purified IMVs from BSC40 cells infected with wild-type (wt) virus or IA27L using anti-A26L antibody.
Construction of IA27L-A26WR virus expressing wild-type A26L.
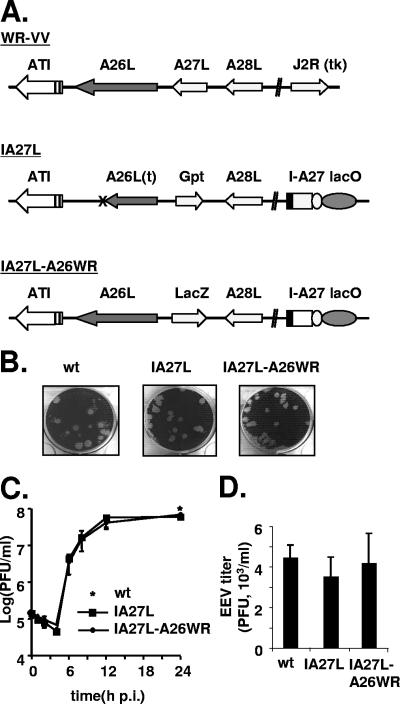
To directly demonstrate that A26L was responsible for the binding of IMVs to laminin, we replaced the truncated A26L ORF in IA27L virus with the wild-type gene by homologous recombination (Fig. 5A). The resulting virus, IA27L-A26WR, expressed a wild-type A26L ORF from a viral late promoter and a lacZ marker gene for virus purification. IA27L-A26WR was purified, and the plaque morphology was found to be similar to that of the parental IA27L virus (Fig. 5B). One-step growth analyses of IA27L-A26WR showed that the yield of IMVs (Fig. 5C) and EEVs (Fig. 5D) from the infected cells was not affected by A26L expression.
FIG. 5.
Generation of recombinant IA27L-A26WR. (A) Schematic diagrams of wild-type vaccinia virus Western Reserve strain (WR-VV), IA27L, and IA27L expressing wild-type A26L (IA27L-A26WR). The A26L and J2R (tk) loci in the virus genomes are shown. In IA27L, the A27L locus is inactivated by Gpt insertion, and a gene cassette with an IPTG-inducible A27L ORF (I-A27) and the lacI repressor gene (lacO) is inserted into the J2R locus (35). The arrows indicate the direction of transcription. Expression of lacZ is driven by a viral late promoter, p11k. The X marks the TA insertion site in the A26L ORF in IA27L virus. (B) Plaque morphology of wild-type virus (wt), IA27L, and IA27L-A26WR after crystal violet staining. (C) IMV yield in one-step growth analyses of IA27L and IA27L-A26WR in BSC40 cells. BSC40 cells were infected with each virus at an MOI of 5 PFU per cell, incubated in culture medium with 5 mM IPTG, and harvested at the indicated time for plaque assays. The star shows the yield of wild-type virus (wt) at 24 h p.i. (D) EEV yield in one-step growth analyses of wild-type virus (wt), IA27L, and IA27L-A26WR. BSC40 cells were infected as described for panel C, the culture medium was collected at 24 h p.i. and mixed with anti-L1R antibody to block contaminating IMV and then was used to infect BSC40 cells, and the EEV titer was determined.
IA27L-A26WR virus expressing full-length A26L in IMVs binds to laminin during virus infection.
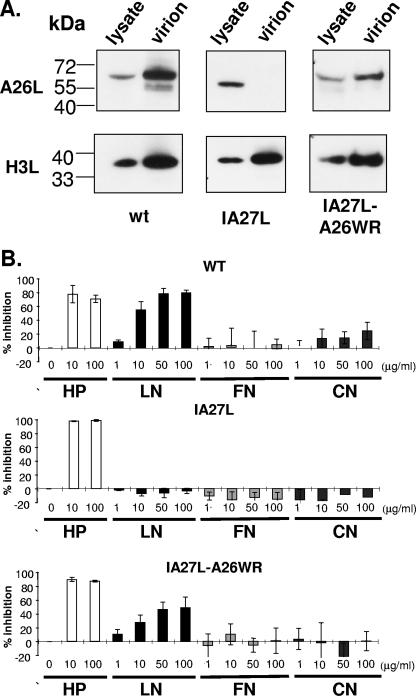
We then performed immunoblot analyses and showed that the full-length 60-kDa A26L was detected in cells infected with IA27L-A26WR (Fig. 6A). More importantly, as with the wild-type virus, full-length A26L was incorporated into IA27L-A26WR IMV particles, demonstrating that the C-terminal 75 amino acids are essential for A26L assembly into IMV particles. Levels of the control H3L envelope protein were similar in all three viruses.
FIG. 6.
Expression of full-length A26L in IA27L-A26WR virus allows IMV binding to laminin. (A) Immunoblot analyses of A26L expression in cell lysates (24 h p.i.) and in purified virions from cells infected with wild-type virus (wt), IA27L, or IA27L-A26WR using anti-A26L or anti-H3L antibodies. (B) Soluble laminin blocks the binding of IA27L-A26WR to BSC40 cells. Soluble HP, LN, FN, and CN at the indicated concentrations were tested for their abilities to block the binding of wild-type (WT), IA27L, and IA27L-A26WR IMV virions to BSC40 cells, as described in the legend to Fig. 1.
To directly demonstrate that the full-length A26L-WR in IMVs is responsible for laminin binding, we purified IMVs from cells infected with wild-type virus, IA27L, or IA27L-A26WR, and performed soluble laminin protein blocking assays as described above. As shown in Fig. 6B, in contrast to IA27L, the binding of IA27L-A26WR was blocked by soluble laminin to an extent similar to that of wild-type virus, confirming an essential role of A26L in laminin recognition. Consistent with all previous experiments, the insertion of wild-type A26L into IA27L to produce IA27L-A26WR did not alter virus binding to other agents, such as heparin, fibronectin, and collagen V (Fig. 6B). We therefore conclude that A26L is the laminin binding envelope protein on vaccinia virus IMV.
The region containing amino acids 127 to 500 of A26L binds to laminin in vitro.
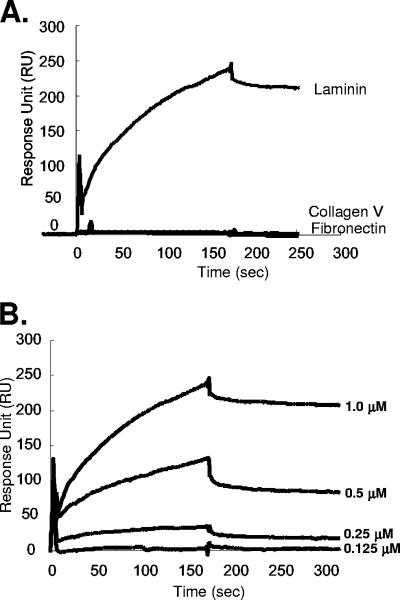
In an attempt to provide further direct evidence that A26L binds to laminin, we cloned and expressed a GST fusion protein containing the full-length A26L protein; however, this was not soluble and aggregated in inclusion bodies during preparation (data not shown). Another GST fusion construct containing amino acids 127 to 500 of A26L, GST-A26L(127-500), was soluble and was purified and immobilized on chips for SPR studies of the in vitro binding of laminin; measurements using soluble ECM proteins and the control protein, GST, were performed in parallel to determine the background signal. As shown in Fig. 7A, laminin, but not collagen V or fibronectin, bound to the immobilized GST-A26L protein. Soluble ECM bound to GST weakly (data not shown). Binding of laminin at four different concentrations to GST-A26L was measured (Fig. 7B), and after removal of the background binding to control GST, a binding affinity constant, KA, of 7.54 × 107 M−1 was obtained from the association and dissociation curves. This binding affinity constant is comparable to that previously reported for laminin binding to a single binding site antibody (22).
FIG. 7.
(A) SPR analysis of the binding of A26L(127-500) to laminin, collagen V, or fibronectin. Biotinylated GST-A26L(127-500) was immobilized on an SA sensor flow chip and tested for the binding of 1 μM laminin, collagen V, or fibronectin as described in Materials and Methods; the first 180 s represent association, and the next 300 s represent dissociation. (B) SPR sensorgrams of laminin binding after removal of the background binding of laminin to biotinylated GST using four different concentrations of laminin (0.125, 0.25, 0.5, and 1.0 μM) with a flow rate of 10 μl/minute to extract the association/dissociation kinetics parameters. As a result, a kon of 1.5 × 104 M−1 s−1, a koff of 2.0 × 10−4 s−1, and a KA (=kon/koff) of 7.5 × 107 M−1 were determined for the A26L and laminin interaction (χ2, 9.6). The binding rate constants were derived from a 1:1 Langmuir binding model (1). The association/dissociation kinetics were measured and recorded by SPR as described in Materials and Methods.
DISCUSSION
We investigated the binding of vaccinia virus IMV to ECM proteins during virus entry for several reasons. First of all, vaccinia virus infects many, if not all, mammalian cell lines, so it is expected that viral envelope proteins will bind to diverse cell surface components during cell entry. Second, although highly charged GAGs are ubiquitously expressed on the cell surface and are known to help virion recruitment to cells, this type of interaction is considered to be low affinity (19). Third, genetic analyses using mutant viruses lacking individual GAGs, such as H3L− and D8L− mutants, or GAG− sog9 cells have suggested the existence of other molecules involved in virion binding (21, 27). Fourth, virus infection induces cell morphological changes, including cell detachment and rounding up, implying that virus entry affects cell adhesion to the ECM. Although ECM proteins are not transmembrane receptor proteins per se, they are secreted as very abundant components on the cell surface and are intimately associated with diverse receptors on the cell surface. Thus, viral binding to ECM proteins may provide advantages, such as an increased virion binding affinity, recruitment of potential cellular receptor molecules, and viral control of cellular signaling transduction. Recently, studies of several pathogens, Treponema pallidum, Leptospira interrogans, and papillomavirus, revealed that they all bind to laminin, suggesting a conserved role of ECM in pathogen invasion (4, 6, 12).
The present study showed that wild-type vaccinia virus IMVs contained an envelope protein, A26L, that binds to laminin on cells. Soluble laminin blocked IMV binding to BSC40 cells, suggesting that A26L binding to laminin is important for virus entry. However, we cannot exclude the possibility that, because laminin is a 900-kDa protein, binding to A26L on the virus surface could obscure other viral proteins and have steric effects that prevent entry. The mutated A26L ORF in IA27L virus was found to result in the production of a truncated form of A26L protein of only 426 amino acids, which was not packaged into IMVs, and the resulting IMV particles did not bind to laminin. When the wild-type A26L ORF was expressed in IA27L, A26L was packaged into IMVs and laminin binding activity was restored, confirming that it is responsible for the binding of IMV to laminin. Furthermore, direct binding of A26L to laminin was demonstrated in vitro.
The fact that IA27L is infectious indicates that A26L is not essential for vaccinia virus entry. This is not totally unexpected for the following reasons. First of all, A26L ORF is not well conserved, being intact in some, and mutated in other, poxviruses (http://www.poxvirus.org); orthologues expressing an intact protein are found in vaccinia virus, cowpox virus, monkeypox virus, and variola virus. Second, binding to GAGs and ECM proteins could provide functional redundancy for IMV attachment. One indication of this comes from previous studies of D8L gene inactivation. Inactivation of the D8L gene in wild-type vaccinia virus has no effect on virus infection of cell cultures (31), whereas its inactivation in IA27L virus reduces IMV infectivity 10-fold (21), suggesting that IMV infectivity loss due to D8L inactivation is rescued by A26L expressed in wild-type virus but not in IA27L. Furthermore, it is conceivable that a complex virus, such as vaccinia virus, recognizes diverse cellular molecules in addition to laminin on permissive cells. For example, IA27L infection of sog9 cells was not blocked by heparin and laminin, suggesting that IMV binding to sog9 cells can occur by an as-yet-unidentified mechanism.
A26L (previously known as P4c or 4c) was one of the first structural proteins identified in IMV particles in the Western Reserve strain of vaccinia virus (26). Stern and Dales (40) suggested that the surface tubules of IMVs contains a major 58-kDa protein and that a rabbit antiserum prepared against surface tubules neutralized virus infectivity and suppressed cell fusion. However, a subsequent study showed that, although A26L is present in surface tubules, it is not the major component and, ironically, A26L appears not to be immunogenic when tested with anti-vaccinia virus sera from human and rabbits (49). A26L was later shown to mediate IMV progeny trafficking in cells (29, 45). Ulaeto et al. (45) showed that A26L is detected in IMVs, but not in EEVs, and proposed a role for A26L in the differentiation switch determining the final fate of virion progeny as IMVs rather than EEVs. Moreover, A26L was shown to interact with A-type inclusion protein to direct IMV progeny into A-type inclusion bodies in infected cells (29). Our present study provides evidence for a new role of A26L in the vaccinia virus life cycle.
Compared to binding to GAGs, the binding of A26L to laminin may provide an additional advantage for IMV recruitment to cells. A26L binds to laminin with a higher affinity than that of the protein-GAG interaction, suggesting a more efficient interaction. In addition, A26L binding to laminin during virus entry may affect cellular stress responses. It is well documented that cell adhesion via the ECM is important for cell survival and that disruption of the cell surface receptor-ECM interaction results in anoikis, a term describing apoptosis induced by cell detachment (17). Immediately after infection, cells infected with IA27L-A26WR exhibited a slower cell rounding-up phenotype than those infected with IA27L (data not shown), suggesting that cell-associated A26L helps maintain cell adhesion. Whether any specific cellular signaling is regulated by A26L will be investigated in the future.
Acknowledgments
This work was supported by grants from the Academia Sinica (AS95IMB1) and National Science Council of R.O.C. (NSC95-2627-M-001-004).
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Adamson, A. W. 1967. Physical chemistry of surfaces, p. 397-401. Interscience, New York, NY.
- 2.Armstrong, J. A., D. H. Metz, and M. R. Young. 1973. The mode of entry of vaccinia virus into L cells. J. Gen. Virol. 21:533-537. [DOI] [PubMed] [Google Scholar]
- 3.Banfield, B. W., Y. Leduc, L. Esford, K. Schubert, and F. Tufaro. 1995. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J. Virol. 69:3290-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbosa, A. S., P. A. Abreu, F. O. Neves, M. V. Atzingen, M. M. Watanabe, M. L. Vieira, Z. M. Morais, S. A. Vasconcellos, and A. L. Nascimento. 2006. A newly identified leptospiral adhesin mediates attachment to laminin. Infect. Immun. 74:6356-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, E., T. G. Senkevich, and B. Moss. 2006. Vaccinia virus F9 virion membrane protein is required for entry but not virus assembly, in contrast to the related L1 protein. J. Virol. 80:9455-9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cameron, C. E., N. L. Brouwer, L. M. Tisch, and J. M. Kuroiwa. 2005. Defining the interaction of the Treponema pallidum adhesin Tp0751 with laminin. Infect. Immun. 73:7485-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, A., and D. H. Metz. 1976. Further investigations on the mode of entry of vaccinia virus into cells. J. Gen. Virol. 32:275-282. [DOI] [PubMed] [Google Scholar]
- 8.Chiu, W. L., and W. Chang. 2002. Vaccinia virus J1R protein: a viral membrane protein that is essential for virion morphogenesis. J. Virol. 76:9575-9587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chung, C. S., C. H. Chen, M. Y. Ho, C. Y. Huang, C. L. Liao, and W. Chang. 2006. Vaccinia virus proteome: identification of proteins in vaccinia virus intracellular mature virion particles. J. Virol. 80:2127-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung, C. S., J. C. Hsiao, Y. S. Chang, and W. Chang. 1998. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J. Virol. 72:1577-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung, C. S., C. Y. Huang, and W. Chang. 2005. Vaccinia virus penetration requires cholesterol and results in specific viral envelope proteins associated with lipid rafts. J. Virol. 79:1623-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Culp, T. D., L. R. Budgeon, M. P. Marinkovich, G. Meneguzzi, and N. D. Christensen. 2006. Keratinocyte-secreted laminin 5 can function as a transient receptor for human papillomaviruses by binding virions and transferring them to adjacent cells. J. Virol. 80:8940-8950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davison, A. J., and B. Moss. 1989. Structure of vaccinia virus early promoters. J. Mol. Biol. 210:749-769. [DOI] [PubMed] [Google Scholar]
- 14.Doms, R. W., R. Blumenthal, and B. Moss. 1990. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J. Virol. 64:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekblom, M., M. Falk, K. Salmivirta, M. Durbeej, and P. Ekblom. 1998. Laminin isoforms and epithelial development. Ann. N. Y. Acad. Sci. 857:194-211. [DOI] [PubMed] [Google Scholar]
- 16.Fenner, F. 1990. Poxviruses, p. 2113-2133. In B. Fields and D. M. Knipe (ed.), Fields virology. Raven Press, New York, NY.
- 17.Frisch, S. M., and H. Francis. 1994. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 124:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guarente, L., and M. Ptashne. 1981. Fusion of Escherichia coli lacZ to the cytochrome c gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 78:2199-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hileman, R. E., J. R. Fromm, J. M. Weiler, and R. J. Linhardt. 1998. Glycosaminoglycan-protein interactions: definition of consensus sites in glycosaminoglycan binding proteins. Bioessays 20:156-167. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao, J. C., C. S. Chung, and W. Chang. 1998. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 72:8374-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsiao, J. C., C. S. Chung, and W. Chang. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J. Virol. 73:8750-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, B. C., S. Davern, and S. J. Kennel. 2006. Mono and bivalent binding of a scFv and covalent diabody to murine laminin-1 using radioiodinated proteins and SPR measurements: effects on tissue retention in vivo. J. Immunol. Methods 313:149-160. [DOI] [PubMed] [Google Scholar]
- 23.Ichihashi, Y., and M. Oie. 1996. Neutralizing epitope on penetration protein of vaccinia virus. Virology 220:491-494. [DOI] [PubMed] [Google Scholar]
- 24.Izmailyan, R. A., C. Y. Huang, S. Mohammad, S. N. Isaacs, and W. Chang. 2006. The envelope G3L protein is essential for entry of vaccinia virus into host cells. J. Virol. 80:8402-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katz, E., and B. Moss. 1970. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc. Natl. Acad. Sci. USA 66:677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin, C. L., C. S. Chung, H. G. Heine, and W. Chang. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Locker, J. K., A. Kuehn, S. Schleich, G. Rutter, H. Hohenberg, R. Wepf, and G. Griffiths. 2000. Entry of the two infectious forms of vaccinia virus at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell 11:2497-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKelvey, T. A., S. C. Andrews, S. E. Miller, C. A. Ray, and D. J. Pickup. 2002. Identification of the orthopoxvirus p4c gene, which encodes a structural protein that directs intracellular mature virus particles into A-type inclusions. J. Virol. 76:11216-11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 31.Niles, E. G., and J. Seto. 1988. Vaccinia virus gene D8 encodes a virion transmembrane protein. J. Virol. 62:3772-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ojeda, S., A. Domi, and B. Moss. 2006. Vaccinia virus G9 protein is an essential component of the poxvirus entry-fusion complex. J. Virol. 80:9822-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ojeda, S., T. G. Senkevich, and B. Moss. 2006. Entry of vaccinia virus and cell-cell fusion require a highly conserved cysteine-rich membrane protein encoded by the A16L gene. J. Virol. 80:51-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resch, W., K. K. Hixson, R. J. Moore, M. S. Lipton, and B. Moss. 25 September 2006, posting date. Protein composition of the vaccinia virus mature virion. Virology doi: 10.1016/j.virol.2008.08.025. [DOI] [PubMed]
- 35.Rodriguez, J. F., and G. L. Smith. 1990. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 18:5347-5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Senkevich, T. G., and B. Moss. 2005. Vaccinia virus H2 protein is an essential component of a complex involved in virus entry and cell-cell fusion. J. Virol. 79:4744-4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Senkevich, T. G., S. Ojeda, A. Townsley, G. E. Nelson, and B. Moss. 2005. Poxvirus multiprotein entry-fusion complex. Proc. Natl. Acad. Sci. USA 102:18572-18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senkevich, T. G., B. M. Ward, and B. Moss. 2004. Vaccinia virus entry into cells is dependent on a virion surface protein encoded by the A28L gene. J. Virol. 78:2357-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, G. L., and A. Vanderplasschen. 1998. Extracellular enveloped vaccinia virus. Entry, egress, and evasion. Adv. Exp. Med. Biol. 440:395-414. [PubMed] [Google Scholar]
- 40.Stern, W., and S. Dales. 1976. Biogenesis of vaccinia: isolation and characterization of a surface component that elicits antibody suppressing infectivity and cell-cell fusion. Virology 75:232-241. [DOI] [PubMed] [Google Scholar]
- 41.Timpl, R., D. Tisi, J. F. Talts, Z. Andac, T. Sasaki, and E. Hohenester. 2000. Structure and function of laminin LG modules. Matrix Biol. 19:309-317. [DOI] [PubMed] [Google Scholar]
- 42.Townsley, A. C., T. G. Senkevich, and B. Moss. 2005. The product of the vaccinia virus L5R gene is a fourth membrane protein encoded by all poxviruses that is required for cell entry and cell-cell fusion. J. Virol. 79:10988-10998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Townsley, A. C., T. G. Senkevich, and B. Moss. 2005. Vaccinia virus A21 virion membrane protein is required for cell entry and fusion. J. Virol. 79:9458-9469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Townsley, A. C., A. S. Weisberg, T. R. Wagenaar, and B. Moss. 2006. Vaccinia virus entry into cells via a low-pH-dependent endosomal pathway. J. Virol. 80:8899-8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ulaeto, D., D. Grosenbach, and D. E. Hruby. 1996. The vaccinia virus 4c and A-type inclusion proteins are specific markers for the intracellular mature virus particle. J. Virol. 70:3372-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanderplasschen, A., M. Hollinshead, and G. L. Smith. 1998. Intracellular and extracellular vaccinia virions enter cells by different mechanisms. J. Gen. Virol. 79:877-887. [DOI] [PubMed] [Google Scholar]
- 47.Vanderplasschen, A., E. Mathew, M. Hollinshead, R. B. Sim, and G. L. Smith. 1998. Extracellular enveloped vaccinia virus is resistant to complement because of incorporation of host complement control proteins into its envelope. Proc. Natl. Acad. Sci. USA 95:7544-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanderplasschen, A., and G. L. Smith. 1997. A novel virus binding assay using confocal microscopy: demonstration that the intracellular and extracellular vaccinia virions bind to different cellular receptors. J. Virol. 71:4032-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilton, S., A. R. Mohandas, and S. Dales. 1995. Organization of the vaccinia envelope and relationship to the structure of intracellular mature virions. Virology 214:503-511. [DOI] [PubMed] [Google Scholar]
- 50.Yoder, J. D., T. S. Chen, C. R. Gagnier, S. Vemulapalli, C. S. Maier, and D. E. Hruby. 2006. Pox proteomics: mass spectrometry analysis and identification of vaccinia virion proteins. Virol. J. 3:10. [DOI] [PMC free article] [PubMed] [Google Scholar]