Abstract
The function of the alphaherpesvirus UL47 tegument protein has not yet been defined. Nonetheless, previous studies with transfected cells have shown that both the herpes simplex virus type 1 homologue (hUL47, or VP13/14) and the bovine herpesvirus type 1 (BHV-1) homologue (bUL47, or VP8) have the capacity to shuttle between the nucleus and the cytoplasm. Furthermore, hUL47 packaged into the virion has also been shown to bind several individual virus-specific RNA transcripts. Here, we extend these observations and show that hUL47 binds a wide range of RNA species in vitro. It has a high affinity for polyadenylated transcripts but has no apparent selectivity for virus-encoded RNA over cellular RNA. We also show that the virion population of bUL47 binds RNA in vitro. However, while purified recombinant hUL47 retains its RNA binding activity, recombinant bUL47 does not, suggesting that the BHV-1 homologue may require virus-induced modification for its activity. We identify the minimal RNA binding domain in hUL47 as a 26-residue N-terminal peptide containing an arginine-rich motif that is essential but not sufficient for optimal RNA binding, and we demonstrate that this RNA binding domain incorporates the hUL47 minimal nuclear localization signal. In addition, we show that soon after hUL47 is expressed during infection, it colocalizes in the infected cell nucleus with ICP4, the major virus transcriptional activator. Using RNA immunoprecipitations, we demonstrate that hUL47 is also bound in vivo to at least one viral transcript, the ICP0 mRNA. Taken together, these results suggest that hUL47 may play a role in RNA biogenesis in the infected cell.
The protein encoded by the herpes simplex virus type 1 (HSV-1) UL47 gene (hUL47, also known as VP13/14) is a major structural protein of unknown function (29, 56, 58, 59). Early studies of the role of hUL47 revealed that it may play a part in gene expression early in infection, as a virus lacking the hUL47 gene was shown to express reduced levels of immediate-early (IE) proteins (58, 59). In addition, UL47 knockout viruses in pseudorabies virus and Marek's disease virus also replicate more slowly than wild-type viruses (12, 25). The effect on IE gene expression was initially believed to be an indirect one, and it was proposed that hUL47 may in some way modulate the activity of the VP16 tegument protein, the transcriptional activator of IE genes (34, 59). Indeed, as hUL47 is also assembled into the tegument of the virion along with VP16 (29), these two proteins are likely to be delivered to the cell together upon virus entry.
In recent years, it has become clear that hUL47 and its homologues possess a number of intrinsic characteristics that may explain a role in gene expression in addition to or possibly instead of its potential influence on VP16. We have previously characterized UL47 from HSV-1 as a nucleocytoplasmic shuttling protein that exhibits a steady-state nuclear localization when expressed in isolation and during its early stages of expression in virus infection (10, 11). Subsequently, studies from others and ourselves have shown that the bovine herpesvirus type 1 (BHV-1) homologue of UL47 (VP8, or bUL47) is also targeted to the nucleus and has the capacity to shuttle between the nucleus and the cytoplasm (52, 60). Although there are several known functions for such nucleocytoplasmic shuttling proteins, a major role for proteins with this property is in the binding and export of RNA from the nucleus (4), and several virus-encoded RNA binding proteins, such as human immunodeficiency virus type 1 (HIV-1) Rev and HSV-1 ICP27, have been shown to enhance the export of virus-encoded transcripts from the nucleus to the cytoplasm (17, 36, 42). In this respect, it is noteworthy that the HSV-1 virion population of hUL47 binds in vitro to a riboprobe specific for the Us8.5 gene, the major viral transcript to be packaged into virions (44, 45). Reverse transcriptase-PCR (RT-PCR) experiments have also been used to show that hUL47 binds to the α22 mRNA in infected cells (45). Furthermore, we have shown recently that in BHV-1-infected cells, bUL47 shuttling is altered by treatment with the RNA polymerase II inhibitor actinomycin D, a result that is consistent with a potential role for bUL47 in RNA biogenesis (53). Thus, the UL47 proteins share many similarities with HIV-1 Rev and HSV-1 ICP27, both of which also shuttle rapidly between the nuclear and cytoplasmic compartments and have their shuttling inhibited by treatment with actinomycin D (32, 35, 39). In addition, for a number of virus RNA binding proteins, such as HIV-1 Rev, human cytomegalovirus (hCMV) UL69, or Epstein-Barr virus EB2, the RNA binding domains have been characterized as arginine-rich motifs that at least in some examples overlap or are inseparable from their nuclear import signals (19, 28, 50). It is therefore significant that in both UL47 homologues nuclear import has been attributed to arginine-rich motifs present in the N termini of the proteins (11, 52, 60).
Several virus-encoded RNA binding proteins have been shown to bind to specific RNA sequences that often have defined secondary structure. For example, HIV-1 Rev binds to the Rev responsive element (RRE) sequence located in the Env coding region of all long HIV-1 transcripts (5), while HIV-1 Tat binds to the TAR stem-loop sequence present at the 5′ end of all HIV-1 transcripts (8). However, for other RNA binding proteins it has not been possible to define specific target sequences in vitro. In this paper we wished to extend our understanding of UL47 function by addressing the nature of the UL47-RNA interaction. Using Northwestern assays, we show that in vitro hUL47 binds generally to mRNA transcripts of both viral and cellular origin. We further identify a minimal RNA binding motif in hUL47 of 26 residues that is rich in arginines and is inseparable from the nuclear localization signal (NLS) of the protein. We go on to show that hUL47 binds mRNA in vivo and localizes to the major sites of transcription in the infected cell nucleus. Hence, we show that hUL47 shares many similarities with other viral RNA binding proteins and provide further evidence that hUL47 may play a role in RNA biogenesis and/or trafficking during HSV-1 infection.
MATERIALS AND METHODS
Cells.
Vero, BHK, COS-1, and MDBK cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum. Sf9 cells were grown in TC 100× medium (Gibco) supplemented with 10% heat-inactivated fetal bovine serum.
Viruses and plasmids.
HSV-1 was routinely grown in BHK cells or Vero cells and titrated on Vero cells. Extracellular HSV-1 virions were purified on Ficoll gradients from the infected-cell medium of 5 × 108 BHK cells, as described previously (15). Extracellular BHV-1 virions were purified on Ficoll gradients from the infected-cell medium of 5 × 108 MDBK cells, as described previously (15).
The parental HSV-1 strain used in this study was strain 17 (s17). The BHV-1 strain used was P8-2, kindly provided by Vikram Misra, University of Saskatchewan. The recombinant viruses based on s17 expressing yellow fluorescent protein (YFP)-tagged hUL47 (179v), green fluorescent protein (GFP)-VP16 (44v), and GFP-UL49 (166v) have been described previously (10, 15, 26). To generate a recombinant virus expressing both YFP-hUL47 and cyan fluorescent protein (CFP)-ICP4, 179v was used to coinfect cells with a CFP-ICP4-expressing virus also based on s17 that was kindly provided by Roger Everett, Glasgow, United Kingdom (16). Progeny viruses from the coinfection were titrated to obtain individual plaques, and those that exhibited both CFP and YFP fluorescence were plaque purified a further three times. To construct baculoviruses for expression of glutathione S-transferase (GST)-hUL47 and GST-bUL47, the respective UL47 open reading frames were inserted as BamHI fragments into plasmid pAcG2T (Pharmingen), and the resulting plasmids were cotransfected into Sf9 cells with Baculogold DNA (Pharmingen). Recombinant baculovirus was further amplified to produce virus stocks for the purification of GST-hUL47 or GST-bUL47. All smaller fragments of hUL47 were expressed as GST fusion proteins in Escherichia coli from the plasmid pGEX6P-3 (Amersham). Plasmids pMD42, pMD43, and pMD44, expressing residues 1 to 76 (1-76), 1-92, and 1-92ΔR2+3 (all arginine residues in R boxes 2 and 3 mutated to glycines), respectively, as GST fusions, were all generated by transferring the corresponding BamHI fragments from previously constructed GFP expression vectors (11) into the BamHI site of pGEX6P-3. Plasmids pMD45 and pMD46, expressing residues 63-76 and 37-76, respectively, were constructed in the same way following PCR amplification of these regions of hUL47 as BamHI fragments, while pJV87 was constructed by PCR amplification of the region expressing residues 50-76 as a BamHI-EcoRI fragment followed by insertion into BamHI/EcoRI-cut pGEX6P-3. Plasmids pJV82 and pJV83, expressing 1-92ΔR2 and 1-92ΔR3 as GST fusion proteins, were constructed by PCR amplification of the corresponding regions from previously constructed GFP expression vectors (11), generating BamHI-EcoRI fragments that were then inserted into BamHI/EcoRI-cut pGEX6P-3. To construct a plasmid expressing β-galactosidase (β-gal), the β-galactosidase open reading frame was first amplified using primers containing BglII and XbaI sites. This fragment was inserted in place of chicken muscle pyruvate kinase (CMPK) in the plasmid p3PK encoding amino acids 17-476 of CMPK, kindly provided by J. Frangioni (Beth Israel Hospital, Boston, MA), generating plasmid pJV53. Plasmids for the expression of hUL47-β-galactosidase fusion proteins were based on two previously constructed plasmids in which residues 1-76 or 37-76 of hUL47 had been inserted into the plasmid p3PK. In each case, the CMPK open reading frame was replaced by the β-galactosidase open reading frame as described for plasmid pJV53, resulting in plasmids pJV55 and pJV56, respectively. A plasmid expressing residues 63-76 of hUL47 fused to β-galactosidase was constructed by replacing the hUL47 sequences in pJV55 with a DNA fragment for residues 63-76 generated by the annealing of two oligonucleotides. Plasmids pJV84 and pJV85, expressing 1-92ΔR2 and 1-92ΔR3 as β-galactosidase fusion proteins, were constructed by PCR amplification of the corresponding regions as HindIII-BglII fragments from previously constructed GFP expression vectors (11), followed by insertion into pJV53. Plasmids pJV86 and pJV99, expressing residues 50-76 and 50-68, respectively, as β-galactosidase fusion proteins, were constructed by PCR amplification of the relevant regions as HindIII-BglII fragments from pMD10 followed by insertion into pJV53. Plasmids pMD10 and pMD15, expressing wild-type hUL47 or hUL47 with R boxes 2 and 3 mutated to glycines as GFP fusion proteins, have been described previously (11). Plasmids for the in vitro transcription of either genomic ICP0 (pCI-110) or ICP0 (pT7-110) cDNA were kindly provided by Roger Everett, Glasgow.
Antibodies.
Monoclonal anti-GFP for Western blotting and polyclonal anti-GFP for immunoprecipitation were obtained from BD Biosciences. Polyclonal anti-HSV-1 UL49 serum has been described previously (14). The polyclonal anti-HSV-1 UL47 antibody 5283 was raised against full-length GST-UL47. Monoclonal anti-VP16 (LP1) was kindly provided by Helena Browne, University of Cambridge. Monoclonal anti-β-galactosidase antibody was obtained from Promega.
Transfections.
For transfection followed by live-cell fluorescence analysis, COS-1 cells were plated at a density of 105 cells per well of a two-well cover glass chamber (LabTek). For transfection followed by immunofluorescence, COS-1 cells were plated at a density of 2 × 105 cells per well onto 16-mm glass coverslips in individual wells of a six-well plate. Twenty-four hours after plating, the cells were transfected using the calcium phosphate precipitation technique modified with BES [N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid]-buffered saline in place of HEPES-buffered saline.
Microscopy.
Cells infected with virus expressing fluorescent proteins were examined live at the indicated times. Cells transfected with plasmids expressing GFP-tagged fusion proteins were examined live 20 h after transfection. For immunofluorescence analysis of transfected cells, cells on coverslips were fixed 40 h after transfection in 4% paraformaldehyde for 20 min followed by permeabilization for 10 min in 0.5% Triton X-100. The fixed cells were blocked by incubation for 30 min in phosphate-buffered saline (PBS) containing 10% newborn calf serum, and primary anti-β-galactosidase antibody was added at a dilution of 1:200 in the same solution for a further 30 min. Following extensive washing in PBS, fluorescein isothiocyanate-conjugated secondary antibody was added in the same blocking solution, incubated for a further 30 min, and washed extensively in PBS. The coverslips were mounted on glass slides in Vectashield (Vector Labs). All samples were examined using a Zeiss Axiovert S100 TV inverted microscope. Images were acquired with a Photometrics Quantix digital camera and were processed with Metamorph and Adobe Photoshop software.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Protein samples were analyzed by electrophoresis on 10 or 12% polyacrylamide gels, followed by staining with Coomassie blue or transfer to nitrocellulose for analysis by Western or Northwestern blotting. Western blots were processed using horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse secondary antibodies (Bio-Rad) and developed using an enhanced chemiluminescence kit (Pierce).
Northwestern blotting.
Proteins on nitrocellulose membranes were renatured in 10 mM Tris (pH 7.6), 50 mM NaCl, 1 mM EDTA, 0.1% NP-40, 0.02% Ficoll, 0.02% polyvinylpyrrolidone, and 0.02% bovine serum albumin overnight at 4°C and then prehybridized for 1 h at 25°C in the same buffer containing 0.05 mg/ml Saccharomyces cerevisiae tRNA. Hybridization was carried out for 4 h at 25°C in the same buffer containing the relevant 32P-labeled probe. Blots were then washed three times in the same buffer without tRNA and exposed to X-ray film.
Preparation of probes for Northwestern blotting.
For 32P-labeled total RNA probes, cells (infected or uninfected) were first labeled with [32P]orthophosphate prior to harvesting. Confluent monolayers of Vero cells in 10-cm dishes were washed twice with phosphate-free DMEM (ICN) and starved of phosphates by incubation in phosphate-free DMEM supplemented with 10% dialyzed calf serum for 1 h at 37°C. Radioactive labeling was conducted by the addition of 250 μCi of [32P]orthophosphate (Amersham) per dish, and the cells were incubated at 37°C for a further 4 h. After washing off the unincorporated label with PBS, total RNA was prepared using a QIAGEN RNeasy kit. Unlabeled total RNA was prepared in the same way without [32P]orthophosphate labeling. Polyadenylated RNA probes were prepared from total RNA samples (either labeled or unlabeled) by use of Oligotex mRNA spin columns (QIAGEN). RNA binding in Northwestern analyses was competed using yeast tRNA (Sigma), double-stranded sheared salmon sperm DNA (Sigma), and single-stranded sheared salmon sperm DNA, prepared by boiling double-stranded DNA (dsDNA) for 5 min and placing it straight on ice. Radiolabeled single-stranded salmon sperm DNA was prepared by random priming with an Amersham Rediprime kit in the presence of [32P]dCTP. Riboprobes for spliced and unspliced ICP0 RNAs were made by in vitro transcription of linearized plasmids pT7110 and pCI110, respectively, in the presence of [32P]CTP.
Purification of GST-UL47 proteins.
Full-length GST-hUL47 and GST-bUL47 fusion proteins were purified from 108 baculovirus-infected Sf9 cells. Seventy-two hours after infection at a multiplicity of 5, cells were lysed in a Dounce homogenizer with high-salt buffer (500 mM KCl, 20 mM Tris, pH 8, 4 mM MgCl2, 0.4 mM EDTA, 2 mM dithiothreitol, and protease inhibitors). After centrifugation for 15 min at 12 K in the cold, the supernatant was retained. The GST-N-terminal fusion proteins were purified from BL21-CodonPlus-RP strain of E. coli (Stratagene) following induction of transformed cells with 0.6 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were pelleted and resuspended in 2 ml PBS plus protease inhibitors and 1 mM dithiothreitol. Samples were sonicated five times for 10 s each, 1% Triton X-100 was added, and samples were incubated for a further 20 min at 4°C. The supernatant was collected following centrifugation at 12 K for 20 min. GST fusion proteins were purified by the addition of glutathione Sepharose beads to the supernatant followed by 30 min of incubation at room temperature with rocking. The beads were washed three times with PBS, and bound protein was eluted in three 200-μl volumes of 20 mM glutathione in 10 mM Tris-HCl (pH 8).
RNA immunoprecipitations.
One confluent 10-cm dish of Vero cells was infected with the relevant GFP-tagged virus at a multiplicity of 10. Eight hours later, the cells were washed in PBS, harvested in 1 ml of lysis buffer (50 mM Tris, pH 7.6, 150 mM NaCl, 2.5 mM MgCl2, 0.1% NP-40, and protease inhibitors), and left on ice for 15 to 30 min. After a brief vortex, cells were centrifuged in a cold microfuge at 12 K for 15 min, the supernatant was transferred to a fresh tube, and RNasin was added at 1 U/μl. The extract was precleared by incubation with 100 μl of a 50% suspension of protein A Sepharose beads for 5 h at 4°C. Immunoprecipitations were carried out overnight at 4°C on the precleared supernatant, using 5 μl of a polyclonal anti-GFP antibody (BD Biosciences) for 400 μl of extract. Antibody-protein complexes were subsequently purified by a further incubation at 4°C with 50 μl of a 50% suspension of protein A Sepharose beads for 4 h, followed by five 1-ml washes with lysis buffer. After the final wash, the protein A Sepharose beads were resuspended in 300 μl of lysis buffer and extracted once with 300 μl phenol and once with 300 μl phenol-chloroform (pH 4.6). The extracted nucleic acid was ethanol precipitated and resuspended in 100 μl 50 mM Tris (pH 7.6) and 10 mM MgCl2 and treated with 0.5 U/μl RNase-free DNase I (Roche) for 30 min at 30°C. Following extractions with equal volumes of phenol-chloroform (pH 4.6), the samples were ethanol precipitated and resuspended in 25 μl RNase-free water.
RT-PCRs.
One-step RT-PCRs were carried out using a QIAGEN one-step kit. One-fifth of each sample purified from the RNA immunoprecipitations was used to determine the presence of ICP0 mRNA in these samples by amplification across the second intron in the ICP0 gene using a forward primer of sequence 5′ GGACTTTATCTGGACGGGC 3′ and a reverse primer of sequence 5′ TGGTGTTGGTGTTACTGCTG 3′. After 30 cycles of PCR, one-fifth of each sample was analyzed on a 1.2% agarose gel containing ethidium bromide. Control reactions, in which the reverse transcription step was absent, were carried out in parallel to confirm that any product was amplified specifically from RNA.
RESULTS
RNA binding by HSV-1 UL47 and UL49 proteins.
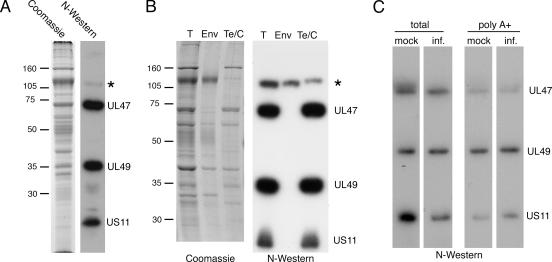
To investigate the nature of RNA binding by hUL47, we first reproduced the results described by Sciortino and coworkers (45), but rather than utilizing a riboprobe specific for a single HSV-1 transcript we used a probe that represented the entire cellular population of RNA molecules synthesized during HSV-1 infection. Vero cells were infected with HSV-1 strain 17 at a multiplicity of 10 and then labeled in vivo with [32P]orthophosphate from 8 to 12 h postinfection. Total 32P-labeled cellular RNA was prepared and used as a probe in a Northwestern blot analysis of HSV-1 virions that had been separated electrophoretically and transferred to nitrocellulose. The results indicate that the RNA probe bound efficiently to three species present in the HSV-1 virions (Fig. 1A). Comparison with the Coomassie blue-stained profile of the virions showed that the two higher-molecular-weight radioactive bands aligned with the UL47 and UL49 (VP22) proteins, as had been shown previously by Sciortino et al. (45) (Fig. 1A). By inference then, and by virtue of its lower molecular mass of around 20 kDa, the lower band was likely to be that of the known RNA binding protein Us11, the third species identified in the previous study (Fig. 1A).
FIG. 1.
HSV-1 virions contain four RNA binding proteins. (A) Purified extracellular virions from HSV-1 s17-infected BHK cells were analyzed by SDS-PAGE followed by Coomassie blue staining or transfer to nitrocellulose. After transfer, the proteins were renatured and subjected to Northwestern (N-Western) blotting using an RNA probe made by purifying total RNA from infected cells labeled for 4 h with [32P]orthophosphate. The asterisk represents an unidentified protein. (B) Extracellular virions from HSV-1 s17 were subjected to fractionation into envelope (Env) and tegument/capsid (Te/C) fractions by detergent treatment of intact virus particles. Following separation of detergent-soluble (envelope) and detergent-insoluble (tegument/capsid) fractions, the samples were analyzed by SDS-PAGE followed by either Coomassie blue staining or transfer to nitrocellulose. Northwestern blotting was carried out with a total RNA probe as described for panel A. T, total. Molecular mass markers (in kilodaltons) are noted at the left of blots. (C) Purified extracellular virions from HSV-1 s17-infected BHK cells were analyzed by SDS-PAGE followed by Northwestern blotting using [32P]orthophosphate-labeled total RNA made from uninfected (mock) or infected (inf.) cells or the polyadenylated fraction of RNA [poly(A+)] purified from [32P]orthophosphate-labeled total RNA made from uninfected or infected cells.
In the Northwestern assay shown in Fig. 1A, a fourth virion protein of around 110 kDa was also observed to bind RNA but with a much lower affinity than UL47, UL49, or Us11 (Fig. 1A). To determine if this additional RNA binding protein had the characteristics of a tegument protein, a similar Northwestern blot procedure was carried out using a total RNA probe on virions that had been detergent stripped to separate envelope (Env) and tegument/capsid fractions (Fig. 1B). Unlike the other RNA binding proteins, which fractionated entirely in the tegument/capsid fraction (Fig. 1B, Te/C lane), our novel RNA binding species fractionated predominantly in the envelope fraction, suggesting it had the characteristics of either an envelope protein or a very loosely associated tegument protein (Fig. 1B). As yet, we have not identified this protein conclusively, but because of its size, potential candidates are the glycoproteins gB, gC, and gH or the tegument protein UL37.
The above-described Northwestern blot analyses were all carried out with a highly heterogeneous probe rather than a single species as has been described previously (45). This may imply that hUL47 has the capacity to bind to a wide range of RNA molecules. To first determine if hUL47 binds only to RNA transcripts synthesized in infected cells, we prepared total RNA probes from both infected and uninfected Vero cells and carried out Northwestern blot analyses on virion samples as described above. Interestingly, cellular RNA from both infected and uninfected cells bound to all three major virion RNA binding proteins, suggesting that none of them exhibits a requirement for RNA transcribed from the viral genome (Fig. 1C, total). We next asked whether hUL47 could bind purely to mRNA transcripts by purifying polyadenylated [poly(A+)] transcripts from the total RNA probes and using these poly(A+) fractions in a Northwestern assay. Although the amount of labeled RNA added in this assay was around 3% of that added for the total RNA probe, all three virion proteins again bound to poly(A+) RNA from either infected or uninfected cells (Fig. 1C).
hUL47 binds with high affinity to RNA.
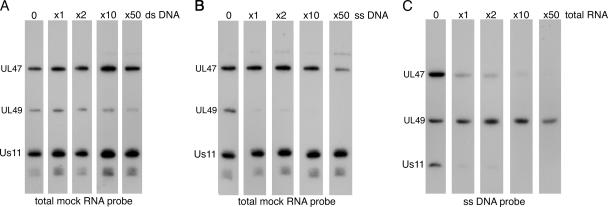
Because UL47 and UL49 from HSV-1 behaved in similar manners in our Northwestern assays and because both have been shown previously to exhibit DNA binding activity (33, 38), we next wished to ensure that the probe binding we were detecting was specific to RNA and was not indicative of a general nonspecific binding of these two tegument proteins to any nucleic acid. As the third protein, Us11, is a well-characterized RNA binding protein (1, 21, 41), it provided us with a control for the assays. To address relative binding affinities of hUL47 and UL49 for RNA, a number of Northwestern assays were carried out using a fixed amount of virions and a fixed amount of probe. Ability to compete for probe binding was assessed by adding increasing amounts of an unlabeled nucleic acid to each Northwestern hybridization. We first asked whether either of the tegument proteins showed any affinity for dsDNA by competing a total RNA probe prepared from uninfected cells with increasing amounts of unlabeled sheared dsDNA (Fig. 2A). The RNA binding of all three proteins was unaffected by up to 50-fold excess of the dsDNA, suggesting that neither UL47 nor UL49 had an affinity for dsDNA. By contrast, when we carried out a similar competition with single-stranded DNA (ssDNA), we observed that while RNA binding by hUL47 and Us11 was unaffected by the presence of excess ssDNA, RNA binding by UL49 was efficiently competed (Fig. 2B). This suggests that UL49 has a higher affinity for ssDNA than RNA. This result was further confirmed by using an ssDNA probe in the Northwestern assay followed by competition with excess unlabeled total RNA (Fig. 2C). In this case, while all three proteins bound the ssDNA probe in the absence of RNA competitor, both hUL47 and Us11 binding were efficiently competed by the addition of equal amounts of competitor, while UL49 binding remained unaffected in the presence of up to 50-fold excess RNA. These results confirm that, like Us11, hUL47 is a potent RNA binding protein. However, they also suggest that although the UL49 protein has the ability to bind RNA in a Northwestern assay, it actually has a greater affinity for ssDNA.
FIG. 2.
Relative binding of HSV-1 virion RNA binding proteins to RNA and DNA. (A and B) Five equal amounts of virions were subjected to SDS-PAGE followed by transfer to nitrocellulose membrane. Each sample was incubated for a Northwestern blot analysis using equal amounts of a total mock RNA probe with increasing amounts of either excess random double-stranded DNA fragments (A) or excess random single-stranded DNA fragments (B). (C) Five equal amounts of virions were subjected to SDS-PAGE followed by transfer to nitrocellulose membrane. Each sample was incubated for a Northwestern blot analysis using equal amounts of a single-stranded DNA probe with increasing amounts of excess total RNA.
hUL47 binds cellular and viral mRNA with equal affinities.
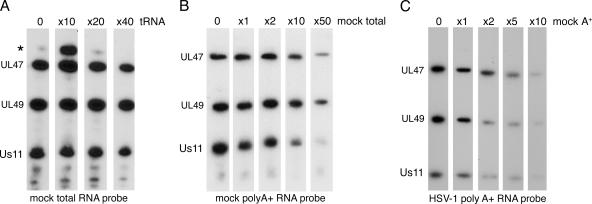
To examine the relative affinities of hUL47 for various classes of RNA, we carried out further competition assays. To first establish if hUL47 preferred regions of dsRNA or ssRNA, a competition was carried out between a total RNA probe purified from uninfected cells and tRNA (Fig. 3A). This demonstrated that the predominantly double-stranded tRNA had little ability to compete for the binding of other RNA molecules to hUL47. To next determine if there was any specificity for poly(A+) RNA over other RNA molecules present in a total RNA probe, we carried out a competition between an uninfected-cell poly(A+) RNA probe and increasing amounts of an unlabeled uninfected-cell total RNA sample (Fig. 3B). hUL47 binding of poly(A+) RNA was not competed by total RNA until it was around 50-fold in excess, suggesting that there is likely to be specificity of hUL47 for polyadenylated mRNA. Binding to Us11 was competed more efficiently than hUL47, presumably due to its known affinity for rRNA (40), a species that would be present in high amounts in a total RNA preparation. Finally, we addressed whether hUL47 exhibits selectivity of viral mRNA molecules over cellular mRNA molecules by carrying out a competition between an infected-cell poly(A+) RNA probe and increasing amounts of an unlabeled uninfected-cell poly(A+) RNA sample (Fig. 3C). In this case, the uninfected-cell poly(A+) RNA competed efficiently for infected-cell poly(A+) RNA binding to hUL47, with probe binding almost entirely depleted by fivefold excess competitor (Fig. 3C). This suggests that, at least in vitro, hUL47 does not exhibit any selectivity for viral mRNA transcripts over cellular mRNA transcripts.
FIG. 3.
Relative binding of hUL47 to different classes of RNA. Five equal amounts of virions were subjected to SDS-PAGE followed by transfer to nitrocellulose membrane. Each sample was incubated for a Northwestern blot analysis using (A) equal amounts of mock total RNA probe with increasing amounts of excess unlabeled tRNA, (B) equal amounts of mock poly(A+) RNA probe with increasing amounts of mock total RNA, and (C) equal amounts of infected-cell poly(A+) RNA probe with increasing amounts of excess mock poly(A+).
RNA binding characteristics of the BHV-1 homologue of UL47 (bUL47).
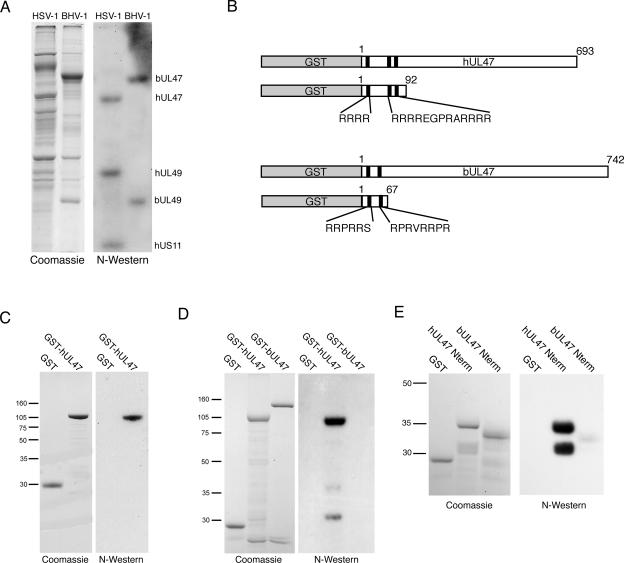
Like HSV-1 UL47, the BHV-1 homologue of UL47 has been shown to be a nucleocytoplasmic shuttling protein (53, 60). Furthermore, we have recently shown that the dynamics of bUL47 shuttling in BHV-1-infected cells is altered by the inhibition of RNA polymerase II transcription with actinomycin D (53), providing indirect evidence for an involvement of this protein in RNA biogenesis. Therefore, to evaluate if bUL47 also has the ability to bind RNA, we carried out a Northwestern assay on both HSV-1 and BHV-1 virions, using a total RNA probe prepared from uninfected cells (Fig. 4A). The blot of BHV-1 virions showed two major RNA binding proteins that align with bUL47 and bUL49 (Fig. 4A). Thus, the property of RNA binding seems to be common to at least two of the alphaherpesvirus UL47 homologues.
FIG. 4.
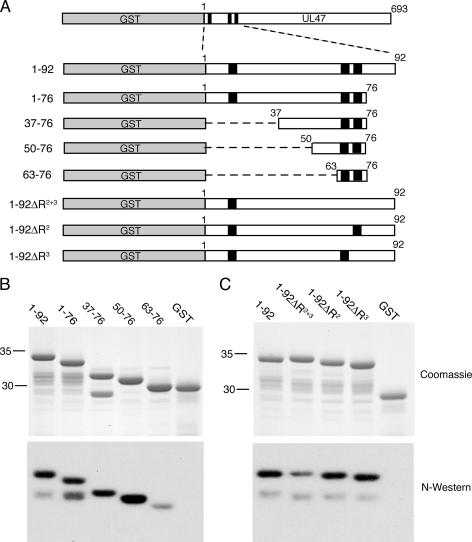
RNA binding by recombinant hUL47 and bUL47. (A) Purified extracellular virions from HSV-1-infected BHK cells or BHV-1-infected MDBK cells were analyzed by SDS-PAGE followed by Coomassie blue staining or transfer to nitrocellulose. After transfer, the proteins were renatured and subjected to Northwestern (N-Western) blotting using an RNA probe made by purifying total RNA from uninfected cells labeled for 4 h with [32P]orthophosphate. (B) Line drawing of the GST-UL47 fusion proteins expressed and purified from baculovirus-infected Sf9 cells (full-length proteins) or E. coli (N-terminal fusion proteins). Black boxes represent arginine-rich motifs located in the N termini of both proteins. (C and D) Equivalent amounts of purified GST and GST-hUL47 (C) or GST, GST-hUL47, and GST-bUL47 (D) were subjected to SDS-PAGE followed by Coomassie blue staining or transfer to nitrocellulose membrane. Northwestern blotting was carried out as described for panel A. (E) Equivalent amounts of purified GST, hUL47 N terminus (Nterm), or bUL47 Nterm were subjected to SDS-PAGE followed by Coomassie blue staining or transfer to nitrocellulose membrane. Northwestern blotting was carried out as described for panel A. Molecular mass markers (in kilodaltons) are noted at the left of blots.
In order to investigate the RNA binding activities of the two UL47 proteins in more detail, we were required to develop an expression system to purify recombinant protein. When hUL47 is expressed in isolation in mammalian cells, one form of the protein (VP14) is synthesized (11). However, in infected cells and extracellular virions hUL47 exists as the doublet VP13/14 (29, 58), implying that a virus-induced modification of hUL47 occurs within the infected cell, resulting in the slightly higher-molecular-weight species VP13. Although the nature of this modification is not yet known, it is plausible that it may be the result of phosphorylation by a virus-encoded kinase, as UL47 has been shown to exist as a phosphoprotein during infection (31). Thus, before attempting to identify the RNA binding domain by using recombinant protein we first needed to determine whether the VP14 species of the hUL47 protein has an intrinsic ability to bind RNA or whether only the virus-induced species VP13 possessed this property. None of our attempts to express full-length hUL47 in E. coli were successful, and therefore to generate purified recombinant hUL47, we constructed a recombinant baculovirus expressing GST-hUL47 by using a Baculogold system (Pharmingen). The GST-hUL47 fusion protein was purified from infected Sf9 cells (Fig. 4B) and analyzed by Northwestern blotting as described above. The results indicate that purified recombinant GST-hUL47 binds efficiently to a total RNA probe, while GST alone exhibits no such binding (Fig. 4C). Hence, hUL47 expressed in the absence of virus infection and therefore unmodified possesses efficient RNA binding activity. However, by contrast, a similar Northwestern blot analysis of purified GST-bUL47 indicated that GST-bUL47 did not retain its ability to bind RNA even though in the same analysis GST-hUL47 RNA binding was evident (Fig. 4D). Taken together, these results suggest that unlike hUL47, bUL47 may require a virus-specific modification to enable its RNA binding activity.
We next wished to identify the RNA binding domain within the hUL47 open reading frame. In this respect, we have previously shown that the N-terminal 92 residues of hUL47 contain three arginine boxes, two of which (R boxes 2 and 3) are required for the nuclear localization of hUL47 (11) (Fig. 4B). Because a well-characterized RNA binding domain is known to be the arginine-rich RNA binding motif, or ARM (55), we examined whether this region of hUL47 was involved in RNA binding. As a comparison, we also used the N-terminal 67 residues of bUL47 fused to GST. We have previously shown that, like the N terminus of hUL47, this region of bUL47 contains two arginine-rich boxes and functions as an NLS (52). Nonetheless, because the full-length recombinant bUL47 did not bind RNA in vitro, the N terminus of the protein provided a useful control for this assay to investigate nonspecific binding of RNA to arginine-rich regions (Fig. 4B). The N-terminal regions of both UL47 proteins were produced and purified as GST fusion proteins, in this case from E. coli (Fig. 4B), and used in a Northwestern assay using a total RNA probe prepared from uninfected cells. While GST alone exhibited no RNA binding activity, residues 1-92 of hUL47 bound efficiently to the RNA probe, indicating that this region contains the intact RNA binding domain of hUL47 (Fig. 4E, compare GST with hUL47 Nterm). By contrast, the arginine-rich N terminus of bUL47 exhibited only residual levels of RNA binding compared to the N terminus of hUL47, demonstrating that a large number of arginine residues is not sufficient to induce RNA binding in this assay (Fig. 4E).
Two arginine boxes in the N terminus of hUL47 are required for RNA binding.
To further refine the RNA binding domain of hUL47, a range of GST fusion proteins, encompassing the N terminus of hUL47, were purified (Fig. 5A) and used in a Northwestern assay with a total RNA probe prepared from uninfected cells. Deletion of all residues C-terminal to the R boxes 2 and 3 (residues 77-92) had little effect on RNA binding of this peptide (Fig. 5B, lane labeled 1-76), suggesting that there was no requirement for the sequence following the R boxes in RNA binding despite the presence of several arginine residues in this region. We then refined the RNA binding domain further by deleting either the first 36 residues (including the first R box) or the first 49 residues from the peptide, resulting in two proteins that bound RNA as efficiently as the longer peptide (Fig. 5B, compare lanes labeled 37-76 and 50-76 with lane labeled 1-76). However, further truncation from the N terminus to produce GST fused to only R boxes 2 and 3 resulted in a peptide that no longer bound efficiently to the RNA probe (Fig. 5B, lane labeled 63-76). Hence, these R boxes are not sufficient for optimal RNA binding, with RNA interaction requiring residues 50-62 in addition to the run of arginines.
FIG. 5.
Defining the RNA binding domain in hUL47. (A) Line drawing of the hUL47 N-terminal GST fusion proteins expressed and purified from E. coli. (B and C) Approximately equivalent amounts of each GST fusion protein, together with GST alone, were subjected to SDS-PAGE followed by Coomassie blue staining or transfer to nitrocellulose membrane. Northwestern (N-Western) blotting was carried out using an RNA probe made by purifying total RNA from uninfected cells labeled for 4 h with [32P]orthophosphate. Molecular mass markers (in kilodaltons) are noted at the left of blots.
To determine the absolute requirement for these arginine residues in RNA binding, we next produced a number of GST fusion proteins incorporating residues 1-92 of hUL47, in which various arginine residues had been mutated to glycine residues. Mutation of all eight arginines in boxes 2 and 3 to glycines in the context of this peptide greatly reduced (but did not abolish) RNA binding by this region of hUL47, confirming that these arginines are important for efficient binding activity (Fig. 5C, lane 1-92ΔR2+3). However, mutation of the individual arginine box 2 or 3 to glycine resulted in peptides that bound RNA as efficiently as the wild-type peptide (Fig. 5C, compare lanes 1-92ΔR2 and 1-92ΔR3 with lane labeled 1-92). Hence, for the property of RNA binding at least, there is redundancy in the positioning of two arginine boxes in this region of the protein.
The hUL47 RNA binding motif is inseparable from its nuclear localization signal.
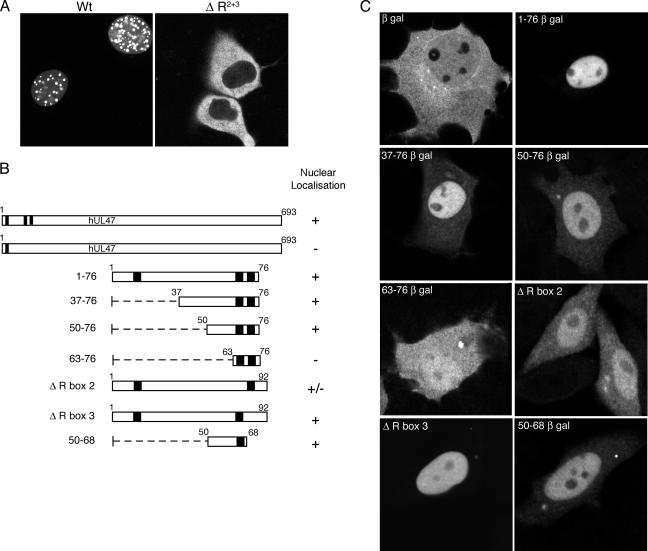
We have shown previously that the arginine boxes involved in RNA binding by hUL47 are also required for nuclear localization of the protein (11) (Fig. 6A) but have not yet defined a minimal transferable NLS for the protein. To determine if it was possible to separate these two properties, we set about defining the minimal region of the hUL47 N terminus that can function as a fully transferable NLS by fusing regions of it to the N terminus of the large, predominantly cytoplasmic protein β-galactosidase (Fig. 6B). Because this reporter protein is too large to diffuse through nuclear pores, it requires active nuclear import via an NLS to accumulate efficiently in the nucleus. The β-gal fusion proteins were expressed in COS-1 cells by transient transfection and analyzed by immunofluorescence 40 h later. While β-gal alone localized in a general cytoplasmic and nuclear pattern (Fig. 6C, β-gal panel), the addition of residues 1-76 of hUL47 to its N terminus resulted in a fusion protein that accumulated entirely in the nucleus (Fig. 6C, 1-76 β-gal panel), confirming that the NLS of hUL47 is contained within this peptide. Removal of the first R box of hUL47 by deletion of the first 36 residues of the protein had little effect on the nuclear targeting of this peptide, while fusion of the RNA binding domain of hUL47 (residues 50-76) to β-gal also resulted in a protein that was predominantly nuclear (Fig. 6C, 50-76 β-gal panel). However, further truncation of this region down to residues 63-76, the region that no longer bound RNA efficiently, completely abrogated nuclear accumulation of this protein (Fig. 6C, 63-76 β-gal panel). Interestingly, while mutation of the arginines in R box 2 to glycines affected the ability of the 1-92 peptide to accumulate in the nucleus, mutation of the arginines in R box 3 to glycines had no effect on nuclear localization (Fig. 6C, compare ΔR box 2 panel with ΔR box 3 panel). Finally, continuing on from this result we fused residues 50 to 68 to β-gal and showed that this region is the minimal region of hUL47 to function as an NLS (Fig. 6C, 50-68 β-gal panel). Thus, both nuclear localization and RNA binding by hUL47 are dictated by the same motif.
FIG. 6.
Defining the NLS in hUL47. (A) COS-1 cells were transfected with plasmids expressing either GFP-hUL47 (wild type [Wt]) or GFP-hUL47 with arginine boxes 2 and 3 mutated to glycines (ΔR2+3). Transfected cells were examined live 20 h after transfection. (B) Line drawing of the N-terminal regions of hUL47 fused to the β-galactosidase reporter. (C) COS-1 cells were transfected with plasmids expressing β-galactosidase alone or the fusions shown in panel B. Forty hours after transfection, the cells were fixed and processed for immunofluorescence by use of an anti-β-galactosidase antibody.
RNA binding by hUL47 in vivo.
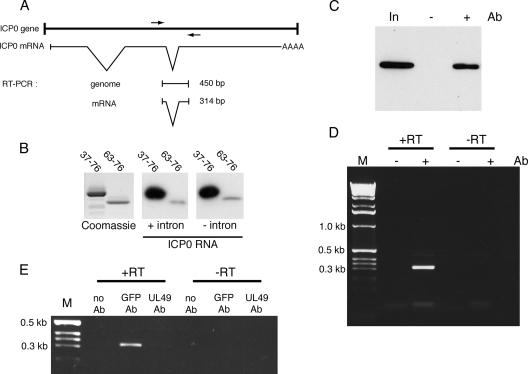
The majority of transcripts synthesized during an HSV-1 infection contain no introns and therefore are not processed through the cellular splicing pathway prior to translation. To determine if the hUL47 RNA binding domain has any preference for RNA containing or lacking an intron, we generated two in vitro-transcribed probes from one of the few HSV-1 genes to contain introns, the IE gene ICP0 (Fig. 7A). In this case, one of the probes was transcribed from the genomic version of the ICP0 gene and therefore retained its intron sequences, and one of them was transcribed from an ICP0 cDNA and therefore lacked its intron sequences. When each of these transcripts was used in a Northwestern assay of the hUL47 minimal RNA binding domain, it was clear that both probes bound efficiently to hUL47 (Fig. 7B). Hence, in this assay at least, hUL47 exhibits no obvious specificity for either spliced or unspliced RNA molecules.
FIG. 7.
RNA binding by hUL47 in vivo. (A) Schematic drawing of the HSV-1 ICP0 gene region, showing the transcribed mRNA with introns spliced out and the RT-PCR products expected from the indicated primers (shown as arrows). (B) Equivalent amounts of GST fused to residues 37-76 or 63-76 of hUL47 were subjected to SDS-PAGE followed by Coomassie blue staining or transfer to nitrocellulose membrane. Northwestern blotting was carried out using an in vitro-transcribed RNA probe representing either genomic ICP0 (+intron) or spliced ICP0 (−intron) mRNA. (C) Vero cells infected with HSV-1 expressing YFP-hUL47 at a multiplicity of 10 were harvested 8 h after infection. Immunoprecipitation was carried out on a soluble extract (In) either in the presence (+) or in the absence (−) of a polyclonal anti-GFP antibody (Ab), and 1/10 of each sample was analyzed by Western blotting using a monoclonal anti-GFP antibody. (D) Nucleic acid was extracted from the immunoprecipitations described for panel C and subjected to RT-PCR using the primers shown in panel A. The reactions were carried out with (+RT) and without (−RT) an initial reverse transcription step. Following RT-PCR, one-fifth of each reaction was analyzed by agarose gel electrophoresis in the presence of ethidium bromide. (E) The RNA immunoprecipitation experiment described for panels C and D was repeated on a soluble extract from cells infected with HSV-1 expressing YFP-hUL47 and precipitated with no antibody, the polyclonal anti-GFP antibody, or a polyclonal anti-UL49 antibody. M, molecular size marker.
We next wished to establish if hUL47 also binds to the ICP0 transcript in vivo. hUL47 was first immunoprecipitated from Vero cells infected with HSV-1 expressing YFP-tagged hUL47, using a polyclonal antibody specific for GFP (Fig. 7C). RNA was then extracted from the immunoprecipitated hUL47 and used in an RT-PCR with primers designed to amplify across the second intron in ICP0, allowing us to distinguish between product synthesized from genomic DNA and that synthesized from spliced RNA (Fig. 7A). The resulting products were then separated by agarose gel electrophoresis. A specific product of the correct size for the ICP0 mRNA was present in the reaction carried out on immunoprecipitated hUL47 but not in the reaction carried out on the control immunoprecipitation where no antibody was included (Fig. 7D, +RT lanes). Furthermore, as no products were synthesized in the absence of the reverse transcription step, the specific product must have been generated from RNA and not from any DNA contaminant (Fig. 7D, −RT lanes). To ensure that the RNA pulldown was specific to the pulldown of UL47 by the GFP antibody, we next used an alternative polyclonal antibody, in this case an anti-UL49 antibody, as a control in an RNA immunoprecipitation assay. Although Western blotting showed that UL49 was efficiently precipitated from the extract with this antibody (data not shown), there was little detectable PCR amplification of the ICP0 RNA in this sample compared to that amplified from the GFP antibody immunoprecipitation, thereby confirming that ICP0 RNA does not bind nonspecifically to a random polyclonal antibody (Fig. 7E, compare GFP Ab with UL49 Ab). In addition, this result also suggests that UL49 is not bound to the ICP0 mRNA in any significant amounts in the infected cell, in spite of its ability to bind RNA by Northwestern blotting in vitro.
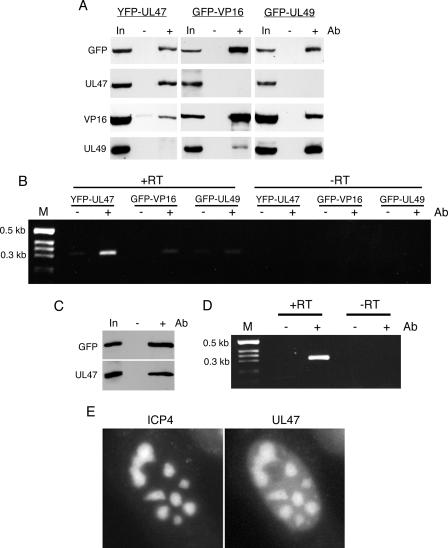
It has recently been shown in a study using a yeast two-hybrid system that UL47 is able to interact with another major tegument protein, VP16 (54), and thus it is possible that ICP0 mRNA present in the UL47 complex had coprecipitated with VP16 or another protein present in the complex rather than directly with UL47. Hence, we next investigated the presence of other tegument proteins in this complex by blotting for VP16 and UL49. At the same time, we blotted similar complexes that had been immunoprecipitated with the GFP antibody from cells infected with HSV-1 expressing either GFP-VP16 or GFP-UL49 (Fig. 8A). The results of these blot analyses confirm that VP16 but not UL49 was coprecipitated in the UL47 complex (Fig. 8A, YFP-UL47). By contrast, UL47 was not coprecipitated in this experiment with VP16 or UL49 (Fig. 8A). However, in agreement with our previous studies (13, 18) VP16 and UL49 coprecipitated with each other (Fig. 8A). Using these same complexes as a starting source of RNA, we next tested for the presence of ICP0 mRNA in the UL47-, VP16-, or UL49-specific immunocomplexes. While the ICP0 RNA was efficiently amplified from precipitated YFP-UL47, there was little evidence of ICP0 RNA in the complex pulled out on the GFP-VP16 or GFP-UL49 proteins (Fig. 8B). This demonstrates that the ICP0 transcript does not precipitate with UL47 by virtue of an interaction with VP16. Furthermore, it also shows that the ICP0 transcript cannot bind nonspecifically in any other GFP immunoprecipitation assay. Interestingly, in one experiment when VP16 was immunoprecipitated at a later time in infection, we were able to demonstrate efficient coimmunoprecipitation of UL47 with VP16 (Fig. 8C). RT-PCR carried out on the same complex indicated that in this case the ICP0 mRNA was present, presumably as a result of binding to the coimmunoprecipitated UL47 (Fig. 8D).
FIG. 8.
ICP0 mRNA binding is specific to complexes containing UL47. (A) Soluble extracts from Vero cells infected with HSV-1 expressing YFP-UL47, GFP-VP16, or GFP-UL49 were immunoprecipitated with the polyclonal anti-GFP antibody (Ab), and the resulting complexes were analyzed by SDS-PAGE followed by Western blotting with antibodies against GFP, UL47, VP16, and UL49. (B) The same complexes from panel A were subjected to RT-PCR using primers specific for ICP0 mRNA. The reactions were carried out with (+RT) and without (−RT) an initial reverse transcription step. One-fifth of each reaction was analyzed by agarose gel electrophoresis in the presence of ethidium bromide. M, molecular size marker. (C) Vero cells infected with HSV-1 expressing GFP-VP16 were harvested late in infection (16 h postinfection), and immunoprecipitation was carried out with polyclonal anti-GFP antibody. The resulting complexes were analyzed by SDS-PAGE followed by Western blotting with antibodies against GFP and UL47. (D) The same complexes from panel C were subjected to RT-PCR with (+RT) and without (−RT) an initial reverse transcription step, using primers specific for ICP0 mRNA. One-fifth of each reaction was analyzed by agarose gel electrophoresis in the presence of ethidium bromide. (E) Vero cells were infected with HSV-1 expressing YFP-hUL47 and CFP-ICP4 at a multiplicity of 5 and examined live 8 h later. In, input extract.
Finally, we addressed the subcellular localization of hUL47 in the infected cell at the time that we had shown it to be bound to viral-specific transcript. It has been shown previously that the major transcriptional activator of HSV-1, the immediate-early protein ICP4, localizes to specific nuclear domains that are considered to be the major sites of RNA transcription in the infected cell nucleus (6, 23, 37). Furthermore, live studies of cells infected with an HSV-1 recombinant expressing CFP-tagged ICP4 have been used to investigate the dynamics of such localization to transcription sites (16). To determine if hUL47 may be targeted to the same sites of viral RNA transcription, we generated an HSV-1 recombinant that expresses both YFP-hUL47 and CFP-ICP4. Although the expression kinetics of these two proteins are quite different, as ICP4 is an immediate-early protein while hUL47 is a true late protein, studies of cells infected with this virus have revealed that at early times of hUL47 expression (up to 8 h), these two proteins colocalize in specific domains within the nucleus (exemplified in Fig. 8E). Taken together, these results indicate that hUL47 can bind RNA in vivo and is targeted to the major nuclear domains where viral RNA transcription occurs. Furthermore, hUL47 may localize to such transcription sites by virtue of its potent RNA binding activity.
DISCUSSION
Although the alphaherpesvirus family of UL47 proteins represents major components of the virus tegument, the role of these proteins in virus infection has to date proved elusive. The main discernible characteristic identified for two members of this family is the ability to shuttle rapidly between the nucleus and the cytoplasm (11, 52, 60). A major role for such proteins is in the export of RNA from the nucleus (4), and a growing number of virus-encoded proteins have now been shown to exhibit nucleocytoplasmic shuttling and to function in viral RNA export pathways (43). Hence, while the exact role of UL47 nucleocytoplasmic shuttling has not yet been defined, it may well be linked to its RNA binding properties that were demonstrated previously by others (45) and were examined here in more detail. Our results have proved conclusively that hUL47 is a true RNA binding protein, exhibiting a strong preference for single-stranded RNA over other nucleic acids in vitro. Furthermore, we have demonstrated RNA binding by hUL47 in vivo. This is in contrast to our findings with VP22, which although previously described as an RNA binding protein (45) would appear to preferentially bind single-stranded DNA in our assays and does not bind to the ICP0 mRNA in our RNA immunoprecipitation studies from infected cells. We were not able to demonstrate any specificity of hUL47 for viral transcripts in vitro, but the protein did appear to bind preferentially to polyadenylated RNA, suggesting an affinity for mRNA of either cellular or viral origin. Such nonspecific RNA binding is not unusual among viral RNA binding proteins, with proteins such as HSV-1 ICP27, influenza virus NP, and hCMV pUL69 also exhibiting little or no sequence specificity (22, 46, 50), unlike the highly sequence-specific binding of HIV-1 Rev and Tat to their respective RRE and TAR RNA elements (5, 9). While it is feasible that this lack of sequence specificity is due to the limitations of the Northwestern assay system, in which proteins may not be properly renatured on the membrane, and that sequence or secondary structure specificity for hUL47 RNA binding may occur within the cell, it is also possible that little or no selectivity is required for hUL47 during infection, because the majority of transcripts being synthesized at that time would be of viral origin. Hence, if hUL47 functions specifically on viral transcripts, these would be the major transcripts that hUL47 would encounter during infection. Nonetheless, in order to determine if any sequence specificity exists in RNA binding by hUL47 it will be necessary to establish the transcripts that it binds in vivo by generating cDNA libraries from coprecipitated RNAs.
There are several reasons why hUL47 may bind and potentially transport mRNA in vivo. First, hUL47 may be involved in circumventing the normal cellular mechanisms of mRNA export from the nucleus. Because the processes of splicing and RNA export are closely linked in the cell (7), the export of viral RNA molecules is problematic for viruses such as HIV-1 and HSV-1 that synthesize unspliced or intronless transcripts, respectively (4, 43, 48, 49). Thus, virus-encoded RNA binding proteins hijack normal cellular pathways involved in protein and RNA export and bypass the need for RNA splicing to occur prior to nuclear export for translation in the cytoplasm (43). Within the cell there are a number of identified nuclear export pathways that different classes of RNA molecules utilize, and several of these have been defined as viral targets (4). In the case of HIV-1 Rev, the karyopherin receptor CRM-1 is utilized to export RRE-containing transcripts, while HSV-1 ICP27 exports intronless mRNAs through the TAP/NXF1 pathway (3, 24, 30, 42, 46). We have demonstrated recently that the BHV-1 UL47 protein contains a novel nuclear export signal that uses an export receptor other than CRM-1 (52). Identification of this potentially novel receptor may enable us to investigate its potential role in viral RNA transport and establish if the same receptor is utilized generally by hUL47 and the other viral homologues.
Another possible reason for hUL47 binding to viral transcripts is to facilitate the packaging of viral mRNAs into assembling virions. It has been shown previously that HSV-1 virions contain a wide range of viral but not cellular mRNAs (44), and the same authors have gone on to show that hUL47 binds at least two viral transcripts packaged into the virion (45). In this situation, hUL47 would be involved in delivering viral mRNAs to the sites of virus and presumably tegument assembly within the cell. Alternatively, hUL47 may be involved in delivering specific transcripts to the correct site in the cytoplasm for translation, so that individual proteins are synthesized at the correct location and/or with their correct binding partners, allowing virus assembly to occur with optimal efficiency.
As mentioned above, HSV-1 encodes the immediate-early protein ICP27, a well-characterized RNA exporting protein (24, 36, 42). However, viruses in which the ICP27 nuclear export signal or RNA binding domain has been mutated are able to replicate (27), suggesting that HSV-1 is still able to process its mRNA transcripts in the absence of these ICP27 activities. Furthermore, ICP27 has been shown to function in RNA transport predominantly at late times in infection (47), and therefore an alternative factor may be involved in RNA transport at earlier times in infection. Interestingly, we have shown that when hUL47 is first expressed in infection it colocalizes in the nucleus with the major viral transcription factor ICP4 and is bound to at least one viral transcript at that time. In addition, we have shown previously that input UL47 from both BHV-1 (53) and HSV-1 (unpublished data) virions is targeted immediately to the nucleus upon virus entry, suggesting that UL47 may play a role there at very early times in infection. However, it remains to be determined if this input population of UL47 has the capacity to bind viral RNA in vivo.
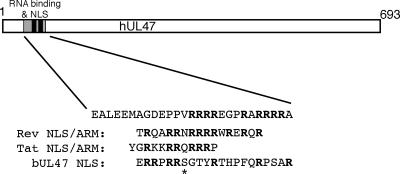
In this paper we have shown that the RNA binding domain within hUL47 is a discrete 26-residue peptide, containing an arginine-rich motif that is essential but not sufficient for RNA binding (Fig. 9). This places hUL47 in a growing group of viral RNA binding proteins, including HIV-1 Rev, HIV Tat, hCMV UL69, and Epstein-Barr virus EB2, that bind through an arginine-rich RNA binding motif, or ARM domain (19, 20, 50, 55, 57). Furthermore, in a scenario reminiscent of the Rev and Tat ARMs and the recently identified ARM in pUL69 (50, 51), we have shown that the domains involved in RNA binding and nuclear localization of hUL47 overlap, and we have not yet been able to separate these activities (Fig. 9). It should be noted that the minimal sequences involved in both of these activities are part of a longer, more complex N-terminal region of hUL47 that contains an additional arginine box (Fig. 4). While this box is not essential, its presence emphasizes that there may be additional redundancy in the sequences involved in nuclear localization and/or RNA binding.
FIG. 9.
Schematic drawing of the hUL47 protein, showing the locations of the RNA binding domain and the NLS. The sequence of the minimal arginine-rich RNA binding motif is shown, together with the HIV-1 Rev and Tat ARMs and the bUL47 NLS. The asterisk denotes a serine residue in bUL47 conserved in several other UL47 homologues.
Our previous studies using GFP as a reporter led us to initially propose that the 13-residue sequence incorporating the arginine-rich motif of R boxes 2 and 3 was sufficient to function as an NLS (11). However, GFP is small enough to diffuse into the nucleus and it is likely that in this situation the small hUL47 arginine motif functioned in nuclear retention of GFP by virtue of its low level of RNA binding, rather than in nuclear import. Here we have shown that this small arginine motif is not able to actively import a large protein into the nucleus but that the NLS of hUL47 requires up to an additional 12 residues upstream of the arginine motif that may be involved in binding to its nuclear import receptor. Although we have yet to identify the nuclear import receptor that hUL47 utilizes, the large number of arginines in the NLS of hUL47 and in that of bUL47 implies that, as for Rev and Tat, the UL47 proteins may use the karyopherin importin β to facilitate transport through the nuclear pores (51, 52). It has been shown previously that importin β binding to the Rev NLS inhibits its RNA binding activity, but only at low levels of RanGTP, as would be the situation in the cytoplasm (51). Thus, as has been suggested previously for Rev and Tat (51), hUL47 may use this arginine-rich domain to switch between import receptor binding in the cytoplasm, where RanGTP is low, and RNA binding in the nucleus, where RanGTP is high, thereby regulating its activity in each cellular compartment.
Our findings in this study have revealed an interesting difference between the activities of the HSV-1 and BHV-1 UL47 proteins. Like hUL47, bUL47 is a nucleocytoplasmic shuttling protein that possesses an arginine-rich NLS sequence at its N terminus (Fig. 9). We have shown here for the first time that, as for virion hUL47, the virion population of bUL47 is a potent RNA binding protein in a Northwestern assay. However, unlike hUL47, which retains its RNA binding when expressed as a recombinant protein, we have been unable to demonstrate RNA binding by recombinant bUL47 expressed as either a GST fusion protein (Fig. 4) or a His-tagged protein (data not shown). Furthermore, the N-terminal region of bUL47, containing its arginine-rich NLS expressed as a GST fusion protein, is also unable to bind RNA. Hence, it would seem that unlike hUL47, bUL47 may require some form of virus-induced modification to make it functional as an RNA binding protein, and by inference this modified bUL47 would be packaged into the virion. A potential candidate for such a modification could be phosphorylation by a virus-encoded kinase, and so it is noteworthy that bUL47 has been shown to be phosphorylated during infection (2). In addition, there are several potential kinase sites within the bUL47 NLS, most notably, the serine residue within the RRPRRS motif (Fig. 9), a sequence that is highly conserved among several other UL47 homologues but not in hUL47 (52). Further studies are required to determine if phosphorylation is a factor involved in RNA binding by bUL47.
In summary, we have shown that the HSV-1 tegument protein hUL47 is a potent RNA binding protein that demonstrates little sequence specificity and binds mRNA through an arginine-rich motif in its N terminus. The fact that this motif also functions as its NLS serves to emphasize the similarity between hUL47 and the classical RNA-exporting protein HIV-1 Rev. While we as yet have no direct evidence to show that the ultimate role of UL47 nucleocytoplasmic shuttling is in the transport of RNA, our combined studies of hUL47 and bUL47 now form a strong basis for evaluating the contribution these proteins may make to RNA biogenesis in the infected cell.
Acknowledgments
We thank Roger Everett for HSV-1 expressing CFP-ICP4 and the ICP0 plasmids, Vikram Misra for BHV-1 strain P8-2, and Peter O'Hare for HSV-1 expressing GFP-VP16.
This work was funded by Marie Curie Cancer Care.
Footnotes
Published ahead of print on 13 December 2006.
REFERENCES
- 1.Attrill, H. L., S. A. Cumming, J. B. Clements, and S. V. Graham. 2002. The herpes simplex virus type 1 US11 protein binds the coterminal UL12, UL13, and UL14 RNAs and regulates UL13 expression in vivo. J. Virol. 76:8090-8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter, D. E., and V. Misra. 1991. The most abundant protein in bovine herpes 1 virions is a homologue of herpes simplex virus type 1 UL47. J. Gen. Virol. 72:3077-3084. [DOI] [PubMed] [Google Scholar]
- 3.Chen, I. H., K. S. Sciabica, and R. M. Sandri-Goldin. 2002. ICP27 interacts with the RNA export factor Aly/REF to direct herpes simplex virus type 1 intronless mRNAs to the TAP export pathway. J. Virol. 76:12877-12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cullen, B. R. 2000. Nuclear RNA export pathways. Mol. Cell. Biol. 20:4181-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly, T. J., K. S. Cook, G. S. Gray, T. E. Maione, and J. R. Rusche. 1989. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 342:816-819. [DOI] [PubMed] [Google Scholar]
- 6.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252:162-178. [DOI] [PubMed] [Google Scholar]
- 7.Dimaano, C., and K. S. Ullman. 2004. Nucleocytoplasmic transport: integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 24:3069-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, and M. A. Skinner. 1990. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 9:4145-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingwall, C., I. Ernberg, M. J. Gait, S. M. Green, S. Heaphy, J. Karn, A. D. Lowe, M. Singh, M. A. Skinner, and R. Valerio. 1989. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc. Natl. Acad. Sci. USA 86:6925-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnelly, M., and G. Elliott. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 75:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly, M., and G. Elliott. 2001. Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 75:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 15.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, U., S. Meyer, M. Teufel, C. Heckel, R. Luhrmann, and G. Rautmann. 1994. Evidence that HIV-1 Rev directly promotes the nuclear export of unspliced RNA. EMBO J. 13:4105-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hafezi, W., E. Bernard, R. Cook, and G. Elliott. 2005. Herpes simplex virus tegument protein VP22 contains an internal VP16 interaction domain and a C-terminal domain that are both required for VP22 assembly into the virus particle. J. Virol. 79:13082-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiriart, E., L. Bardouillet, E. Manet, H. Gruffat, F. Penin, R. Montserret, G. Farjot, and A. Sergeant. 2003. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J. Biol. Chem. 278:37790-37798. [DOI] [PubMed] [Google Scholar]
- 20.Kamine, J., P. Loewenstein, and M. Green. 1991. Mapping of HIV-1 Tat protein sequences required for binding to Tar RNA. Virology 182:570-577. [DOI] [PubMed] [Google Scholar]
- 21.Khoo, D., C. Perez, and I. Mohr. 2002. Characterization of RNA determinants recognized by the arginine- and proline-rich region of Us11, a herpes simplex virus type 1-encoded double-stranded RNA binding protein that prevents PKR activation. J. Virol. 76:11971-11981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kingsbury, D. W., I. M. Jones, and K. G. Murti. 1987. Assembly of influenza ribonucleoprotein in vitro using recombinant nucleoprotein. Virology 156:396-403. [DOI] [PubMed] [Google Scholar]
- 23.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koffa, M. D., J. B. Clements, E. Izaurralde, S. Wadd, S. A. Wilson, I. W. Mattaj, and S. Kuersten. 2001. Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J. 20:5769-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Boissiere, S., A. Izeta, S. Malcomber, and P. O'Hare. 2004. Compartmentalization of VP16 in cells infected with recombinant herpes simplex virus expressing VP16-green fluorescent protein fusion proteins. J. Virol. 78:8002-8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lengyel, J., C. Guy, V. Leong, S. Borge, and S. A. Rice. 2002. Mapping of functional regions in the amino-terminal portion of the herpes simplex virus ICP27 regulatory protein: importance of the leucine-rich nuclear export signal and RGG box RNA-binding domain. J. Virol. 76:11866-11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malim, M. H., L. S. Tiley, D. F. McCarn, J. R. Rusche, J. Hauber, and B. R. Cullen. 1990. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 60:675-683. [DOI] [PubMed] [Google Scholar]
- 29.McLean, G., F. Rixon, N. Langeland, L. Haarr, and H. Marsden. 1990. Identification and characterization of the virion protein products of herpes simplex virus type 1 gene UL47. J. Gen. Virol. 71:2953-2960. [DOI] [PubMed] [Google Scholar]
- 30.Mears, W. E., and S. A. Rice. 1996. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J. Virol. 70:7445-7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meredith, D. M., J. A. Lindsay, I. W. Halliburton, and G. R. Whittaker. 1991. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J. Gen. Virol. 72:2771-2775. [DOI] [PubMed] [Google Scholar]
- 32.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 33.Normand, N., H. van Leeuwen, and P. O'Hare. 2001. Particle formation by a conserved domain of the herpes simplex virus protein VP22 facilitating protein and nucleic acid delivery. J. Biol. Chem. 276:15042-15050. [DOI] [PubMed] [Google Scholar]
- 34.Pellett, P. E., J. L. McKnight, F. J. Jenkins, and B. Roizman. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. USA 82:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phelan, A., and J. B. Clements. 1997. Herpes simplex virus type 1 immediate early protein IE63 shuttles between nuclear compartments and the cytoplasm. J. Gen. Virol. 78:3327-3331. [DOI] [PubMed] [Google Scholar]
- 36.Phelan, A., J. Dunlop, and J. B. Clements. 1996. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J. Virol. 70:5255-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 71:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinard, M. F., R. Simard, and V. Bibor-Hardy. 1987. DNA-binding proteins of herpes simplex virus type 1-infected BHK cell nuclear matrices. J. Gen. Virol. 68:727-735. [DOI] [PubMed] [Google Scholar]
- 39.Richard, N., S. Iacampo, and A. Cochrane. 1994. HIV-1 Rev is capable of shuttling between the nucleus and cytoplasm. Virology 204:123-131. [DOI] [PubMed] [Google Scholar]
- 40.Roller, R. J., L. L. Monk, D. Stuart, and B. Roizman. 1996. Structure and function in the herpes simplex virus 1 RNA-binding protein U(s)11: mapping of the domain required for ribosomal and nucleolar association and RNA binding in vitro. J. Virol. 70:2842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roller, R. J., and B. Roizman. 1990. The herpes simplex virus Us11 open reading frame encodes a sequence-specific RNA-binding protein. J. Virol. 64:3463-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandri-Goldin, R. M. 2004. Viral regulation of mRNA export. J. Virol. 78:4389-4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sciortino, M. T., M. Suzuki, B. Taddeo, and B. Roizman. 2001. RNAs extracted from herpes simplex virus 1 virions: apparent selectivity of viral but not cellular RNAs packaged in virions. J. Virol. 75:8105-8116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sciortino, M. T., B. Taddeo, A. P. Poon, A. Mastino, and B. Roizman. 2002. Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc. Natl. Acad. Sci. USA 99:8318-8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sokolowski, M., J. E. Scott, R. P. Heaney, A. H. Patel, and J. B. Clements. 2003. Identification of herpes simplex virus RNAs that interact specifically with regulatory protein ICP27 in vivo. J. Biol. Chem. 278:33540-33549. [DOI] [PubMed] [Google Scholar]
- 47.Soliman, T. M., R. M. Sandri-Goldin, and S. J. Silverstein. 1997. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J. Virol. 71:9188-9197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stutz, F., A. Bachi, T. Doerks, I. C. Braun, B. Seraphin, M. Wilm, P. Bork, and E. Izaurralde. 2000. REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6:638-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tang, H., K. L. Kuhen, and F. Wong-Staal. 1999. Lentivirus replication and regulation. Annu. Rev. Genet. 33:133-170. [DOI] [PubMed] [Google Scholar]
- 50.Toth, Z., P. Lischka, and T. Stamminger. 2006. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 34:1237-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Truant, R., and B. R. Cullen. 1999. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin beta-dependent nuclear localization signals. Mol. Cell. Biol. 19:1210-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verhagen, J., M. Donnelly, and G. Elliott. 2006. Characterization of a novel transferable CRM-1-independent nuclear export signal in a herpesvirus tegument protein that shuttles between the nucleus and cytoplasm. J. Virol. 80:10021-10035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verhagen, J., I. Hutchinson, and G. Elliott. 2006. Nucleocytoplasmic shuttling of bovine herpesvirus 1 UL47 protein in infected cells. J. Virol. 80:1059-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vittone, V., E. Diefenbach, D. Triffett, M. W. Douglas, A. L. Cunningham, and R. J. Diefenbach. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566-9571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weiss, M. A., and N. Narayana. 1998. RNA recognition by arginine-rich peptide motifs. Biopolymers 48:167-180. [DOI] [PubMed] [Google Scholar]
- 56.Whittaker, G. R., M. P. Riggio, I. W. Halliburton, R. A. Killington, G. P. Allen, and D. M. Meredith. 1991. Antigenic and protein sequence homology between VP13/14, a herpes simplex virus type 1 tegument protein, and gp10, a glycoprotein of equine herpesvirus 1 and 4. J. Virol. 65:2320-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zapp, M. L., T. J. Hope, T. G. Parslow, and M. R. Green. 1991. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc. Natl. Acad. Sci. USA 88:7734-7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang, Y., and J. L. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in alpha TIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng, C., R. Brownlie, L. A. Babiuk, and S. van Drunen Littel-van den Hurk. 2004. Characterization of nuclear localization and export signals of the major tegument protein VP8 of bovine herpesvirus-1. Virology 324:327-339. [DOI] [PubMed] [Google Scholar]