Abstract
Newcastle disease virus (NDV), an avian paramyxovirus, initiates infection with attachment of the viral hemagglutinin-neuraminidase (HN) protein to sialic acid-containing receptors, followed by fusion of viral and cell membranes, which is mediated by the fusion (F) protein. Like all class 1 viral fusion proteins, the paramyxovirus F protein is thought to undergo dramatic conformational changes upon activation. How the F protein accomplishes extensive conformational rearrangements is unclear. Since several viral fusion proteins undergo disulfide bond rearrangement during entry, we asked if similar rearrangements occur in NDV proteins during entry. We found that inhibitors of cell surface thiol/disulfide isomerase activity—5′5-dithio-bis(2-nitrobenzoic acid) (DTNB), bacitracin, and anti-protein disulfide isomerase antibody—inhibited cell-cell fusion and virus entry but had no effect on cell viability, glycoprotein surface expression, or HN protein attachment or neuraminidase activities. These inhibitors altered the conformation of surface-expressed F protein, as detected by conformation-sensitive antibodies. Using biotin maleimide (MPB), a reagent that binds to free thiols, free thiols were detected on surface-expressed F protein, but not HN protein. The inhibitors DTNB and bacitracin blocked the detection of these free thiols. Furthermore, MPB binding inhibited cell-cell fusion. Taken together, our results suggest that one or several disulfide bonds in cell surface F protein are reduced by the protein disulfide isomerase family of isomerases and that F protein exists as a mixture of oxidized and reduced forms. In the presence of HN protein, only the reduced form may proceed to refold into additional intermediates, leading to the fusion of membranes.
Cell entry by enveloped viruses requires fusion of the viral envelope with host cell membranes, a step in infection that is mediated by viral fusion proteins. Viral fusion proteins have been categorized into two and possibly three groups based on their structures and mechanisms for mediating fusion (22, 58, 70). Class 1 fusion proteins, which fold as trimers, include paramyxovirus F proteins, influenza virus hemagglutinin (HA) proteins, and retrovirus envelope (Env) proteins. These proteins, synthesized as inactive precursors, are cleaved into two subunits, F1 and F2 in the case of paramyxoviruses. The sequence at the new amino terminus generated by this cleavage is the fusion peptide (FP), which inserts into the target membrane upon fusion activation (reviewed in references 12, 23, 49, and 70). These proteins also contain two important heptad repeat (HR) domains. The F protein HR domains are located just carboxyl terminal to the fusion peptide (HR1) and adjacent to the transmembrane (TM) domain (HR2). The HR1 and HR2 peptides have a strong affinity and form a very stable six-stranded coiled coil, with HR1 forming an interior trimer and HR2 binding in the grooves of the trimer in an antiparallel orientation (3). Inhibition of fusion with either the HR1 or HR2 peptide suggests that the HR1 and HR2 domains in the intact protein are not associated prior to F protein activation, while the two domains are complexed in the postfusion F protein (28, 59, 76).
Current models for class 1 fusion proteins propose that fusion activation, by receptor binding or acid pH (reviewed in references 9, 12, 24, and 34), results in dramatic conformational changes in these proteins. First, the FP is exposed for insertion into a target membrane, anchoring the protein in that membrane. It is then proposed that the protein proceeds to refold, forming a complex between heptad repeat domains, which pulls the target and the effector membranes together (reviewed in references 9, 26, and 60). Models for the mechanistic details of the subsequent hemifusion and pore formation are less well defined, although there may be additional conformational changes in the F protein during these stages of fusion (8, 35, 47). How fusion proteins accomplish these extensive conformational rearrangements is not clear.
Thiol/disulfide exchange in various cell entry proteins, including diphtheria toxin and fusion proteins of some animal viruses, has been shown to be necessary for the fusion of membranes (25, 73). In vaccinia virus infection, the disulfide bonds in core proteins are reduced during entry into the host cell (36). Disulfide bonds in the envelope protein in Sindbis virus are reduced during cell entry (2). Disulfide bond rearrangement is involved in forming the fusogenic complex of baculovirus gp64 (39). The surface (SU) subunit of the Env protein in Moloney murine leukemia virus has a CXXC motif that leads to isomerization of a disulfide bond between the SU and TM proteins, which is required for fusion (17, 56, 69). Recent studies of the human immunodeficiency virus type 1 (HIV-1) Env protein have shown that a plasma membrane-associated oxidoreductase, protein disulfide isomerase (PDI), or a related protein, is required for the fusion of membranes mediated by HIV-1 Env (16, 40, 61). It was proposed that, upon gp120 binding to receptors, thiol/disulfide isomerase activity cleaves disulfide bonds in Env, facilitating its refolding, which is required for membrane fusion. Down regulation of PDI has also been shown to inhibit infection by mouse polyoma virus (21).
PDI and PDI-like isomerases belong to the thioredoxin superfamily (14). These enzymes catalyze the reduction, formation, and isomerization of disulfide bonds in proteins in the endoplasmic reticulum (ER) (71). Although they are ER-resident proteins, they are also present in other cellular locations, including plasma membranes, where they associate with integral plasma membrane proteins through noncovalent interactions (27, 66). The redox function of PDI-like proteins is based on one or more catalytic domains bearing a CXXC motif (20, 37). In the ER, these isomerases act mainly as oxidases. Cysteines in their CXXC motifs are oxidized, leading to the formation of disulfide bonds in interacting proteins. At the cell surface, these isomerases predominantly act as reductases, and cysteines in their CXXC motifs are in the form of free thiols. These forms of the proteins lead to cleavage of disulfide bonds and production of free thiols in interacting proteins (54).
It is unknown if disulfide bond rearrangement has any role in paramyxovirus attachment and fusion, mediated by the HN and F proteins, respectively. We found that free thiols can be detected in surface-expressed Newcastle disease virus (NDV) F protein, but not in HN protein. Furthermore, we found that the non-membrane-permeating inhibitors of disulfide bond isomerases, 5′5-dithio(2-bis-nitrobenzoic acid) (DTNB) and bacitracin, as well as anti-PDI antibodies, inhibited cell-cell fusion and virus entry mediated by NDV glycoproteins. DTNB and bacitracin also inhibited the formation of free thiols in the F protein. These inhibitors also altered the conformation of cell surface-expressed F protein, as detected by conformation-sensitive antibodies. Furthermore, blocking the free thiols in F protein by covalent addition of a thiol-specific biotin inhibited cell-cell fusion. Our results suggest that disulfide bond reduction in cell surface NDV F protein is mediated by PDI-like enzymes and that disulfide bond isomerization may be required for conformational changes in F protein that are essential for membrane fusion.
MATERIALS AND METHODS
Cells, virus, and plasmids.
COS-7 cells, obtained from the American Type Culture Collection, were grown in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with nonessential amino acids, vitamins, penicillin, streptomycin, and 10% fetal calf serum. Stocks of NDV B1, an avirulent strain, were grown in embryonated chicken eggs and purified by standard protocols (42).
NDV F and HN genes were inserted into pSVL and pCAGGS expression vectors as previously described (43, 44).
Transfections.
Transfections were accomplished using Lipofectin or Lipofectamine (Invitrogen). For Lipofectamine transfection, a mixture of DNA (pSVL, 1 μg/35-mm plate, or pCAGGS, 0.5 μg/35-mm plate) and Lipofectamine (7 μl/35-mm plate) in OptiMEM medium (Gibco) was incubated at room temperature for 45 min and added to COS-7 cells grown in 35-mm plates and previously washed with OptiMEM. For Lipofectin transfection, DNA (pSVL, 1 μg/35-mm plate) and Lipofectin (10 μl/35-mm plate) were incubated with OptiMEM separately at room temperature for 40 min. Both DNA and Lipofectin were mixed and incubated further for 15 min and added to OptiMEM-washed cells grown in 35-mm plates. The cells were incubated for 5 h at 37°C, the OptiMEM was removed, and 2 ml of supplemented DMEM was added.
Antibodies and PDI inhibitors.
Anti-NDV antibody was raised in rabbits against UV-inactivated stocks of NDV strain AV by standard protocols as previously described (43). Rabbit anti-HR1 and anti-HR2 antibodies were raised against peptides with the HR1 and HR2 sequences from NDV F protein as described previously (10, 43). Anti-Fu1a is a mouse monoclonal antibody specific for the NDV F protein and was obtained from M. Peeples (51). Anti-AS antibody was raised against a peptide with a sequence from the NDV HN protein as previously described (45). The secondary antibodies used were anti-rabbit immunoglobulin G (IgG) coupled to horseradish peroxidase (Amersham), goat anti-rabbit IgG Alexa Fluor 488 (Molecular Probes), and anti-mouse IgG Alexa Fluor 568 (Molecular Probes).
PDI inhibitors, DTNB and bacitracin, were purchased from Sigma. Bacitracin was used with a protease inhibitor, phenylmethanesulfonyl fluoride (0.2 mg per ml of 7.5 mM bacitracin), to prevent the degradation of proteins by contaminating proteases (67). Polyclonal rabbit anti-PDI antibody was purchased from Stressgen. Anti-CD71 antibody (Santa Cruz Biotechnology) was used as a control IgG.
Cell viability.
COS-7 cell monolayers grown in 35-mm plates were incubated overnight with DTNB (2.5 mM, 5 mM, and 7.5 mM), bacitracin (2.5 mM, 5 mM, and 7.5 mM), or anti-PDI antibodies (3 μl and 6 μl per ml) or left untreated. The cells were then incubated with 2 ml of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (Promega) (0.5 mg/ml in phosphate-buffered saline [PBS]) for 3 h at 37°C. After the MTT solution was removed, the cells were treated with 2 ml acidified isopropanol (0.04 M HCl in absolute isopropanol) to solubilize blue formazan crystals. The blue color was quantified by measuring absorbance at a test wavelength of 570 nm and a reference wavelength of 650 nm as described previously (52).
Annexin V-fluorescein isothiocyanate (FITC) and propidium iodide (PI) (Biovision) double staining was performed as described previously (1). As a positive control for annexin V binding and PI staining by fluorescence microscopy, cells were treated with 0.05% Triton X-100 for 30 min on ice to allow access to the interior of the cells. Incubation with staurosporine (Sigma; 1 μM) for 6 h, which induces apoptosis (32), as well as 0.05% Triton X-100 (30 min on ice), was used as a positive control for flow cytometry analysis (fluorescence-activated cell sorting [FACS]). The addition of Triton X-100 enhanced PI staining. COS-7 cell monolayers were either incubated overnight with PDI inhibitors or left untreated. The cells were then detached for FACS or left on coverslips for fluorescence microscopy and incubated with annexin V-FITC (5 μl) and PI (5 μl) in annexin binding buffer (Biovision) for 10 min in the dark at room temperature. The cells were washed twice with PBS, fixed with 2% paraformaldehyde, and analyzed by visualizing them by microscopy or FACS.
Hemadsorption.
COS-7 cells grown in 35-mm plates were transfected with pSVL HN. The transfected cells, incubated overnight with DTNB (7.5 mM), bacitracin (7.5 mM), or anti-PDI antibodies (6 μl per ml), were then incubated with a 0.4% suspension of washed guinea pig red blood cells (RBCs) (Bio Link) in PBS for 30 min at 4°C. Unbound RBCs were removed by washing them three times with cold PBS. Bound RBCs were lysed with 0.75 ml of 50 mM NH4Cl, and the hemoglobin released was quantified by measuring absorbance at 540 nm.
Neuraminidase.
Cells grown in 35-mm plates, transfected with pSVL HN, were incubated overnight with PDI inhibitors. The cells were then analyzed for neuraminidase activity as previously described (50). Briefly, the cells were washed twice with PBS and once with 0.1 M sodium acetate (pH 6) and incubated for 4 h at 37°C with N-acetylneuraminyl lactose (Sigma; 0.25 mg in 0.5 ml sodium acetate [0.1 M, pH 6]). The supernatant was collected and incubated with 0.5 ml of 25 mM periodic acid for 30 min at 37°C; 0.4 ml of 2% sodium arsenite was then added to the tubes and mixed. The mixture was boiled with thiobarbituric acid (14.2 mg/ml, pH 9) for 15 min, followed by the addition of 4 ml acid-butanol (5% HCl in butanol). The tubes were centrifuged for 15 min, and the optical density of the top layer at 549 nm was determined.
Surface biotinylation.
COS-7 monolayers grown in 35-mm plates and transfected with pSVL HN and pSVL F were incubated with PDI inhibitors overnight and washed three times with PBS-CM (PBS with 0.1 mM CaCl2 and 1 mM MgCl2). PBS-CM containing 0.5 mg/ml sulfo-NHS-SS-biotin (Pierce) was added, and the cells were incubated for 40 minutes at 4°C. Unbound biotin was absorbed with 2 ml DMEM, and the cells were washed three times with PBS and lysed with RSB buffer (0.01 M Tris-HCl [pH 7.4], 0.01 M NaCl, 1.5 mM MgCl2) containing 1% Triton X-100, 0.5% sodium deoxycholate, 2.5 mg of N-ethyl maleimide per ml, and 0.2 mg of DNase per ml. The lysates were incubated for 1 h at room temperature with 0.3% sodium dodecyl sulfate (SDS) containing neutravidin-agarose (Pierce) that had been washed with PBS containing 0.5% Tween 20 and 5 mg/ml bovine serum albumin (BSA) and then with PBS containing 0.5% Tween 20 and 1 mg/ml BSA. The precipitates were washed three times with PBS containing 0.5% Tween 20 and 0.4% SDS, resuspended in gel sample buffer (125 mM Tris-HCl, pH 6.8, 2% SDS, and 10% glycerol) with 0.7 M β-mercaptoethanol, and resolved by polyacrylamide gel electrophoresis (PAGE) (10%). The gels were equilibrated in transfer buffer (25 mM Tris, pH 8.2, 192 mM glycine, 12.5% methanol), transferred to Immobilon-P (Millipore Corp.) membranes, and probed with anti-HR2 antibody (1:1,000) or anti-AS antibody (1:1,000) (primary) and goat anti-rabbit immunoglobulin G coupled to horseradish peroxidase (secondary). Bound antibodies were detected using ECL Western blotting detection reagent (Amersham Biosciences).
Syncytium formation.
COS-7 cells grown in 35-mm plates were cotransfected with pSVL HN (1 μg/plate) and pSVL F (1 μg/plate) using Lipofectin (Invitrogen). At 24 h posttransfection, DTNB (2.5 mM, 5 mM, and 7.5 mM), bacitracin (2.5 mM, 5 mM, and 7.5 mM), or anti-PDI antibody (6 μl per ml) was added. At 48 h posttransfection, the nuclei in 20 fusion areas were counted to determine the average size of the syncytia, as previously described (62). Values obtained after transfection with the empty vector were subtracted.
Content mixing.
Content mixing was measured by using a modification of a previously described protocol (46). Briefly, cells grown in 35-mm plates were transfected with pCAGGS-HN (0.5 μg/plate) and pCAGGS-F (0.5 μg/plate) DNAs and a plasmid encoding a tetracycline-responsive transcriptional activator, tTA (Clontech) (1 μg/plate). A separate population of cells was transfected with a plasmid encoding the β-galactosidase protein under the control of a tetracycline-responsive transcriptional activator (pB1-G) (Clontech; 1 μg/plate). After 36 h, PDI inhibitors were added to the plates, and after 40 h, the cells transfected with pB1-G were removed from the plates with trypsin and placed on top of the HN and F protein-expressing cells. The cells were further incubated with inhibitors for 6 h, washed twice with PBS, and lysed (Promega cell lysis buffer), and extracts were assayed for β-galactosidase activity (Promega protocols). Activity due to background fusion of COS-7 cells was measured after cells cotransfected with pTA and empty vector were mixed with cells transfected with pB1-G. The values obtained were subtracted from the values obtained with cells expressing HN and F proteins.
Lipid mixing.
The protocol used was similar to that previously described (29). Briefly, guinea pig RBCs (Bio Link) were washed in PBS and resuspended to give a 0.4% suspension and were incubated with 15 μg/ml of R18 (octadecyl rhodamine B chloride; Molecular Probes) for 30 min at room temperature in the dark. Three volumes of complete medium (DMEM with 10% fetal calf serum) was added, and incubation was continued for 30 min. The RBCs were then washed four times in ice-cold PBS and resuspended to give a 0.4% suspension of RBCs in PBS containing CaCl2 (0.01%). These RBCs were added to cells that had been grown on coverslips in 35-mm plates and transfected with pSVL HN and pSVL F, and were washed in PBS-CaCl2. The cells were incubated with labeled RBCs for 30 min on ice. The cells were then washed with ice-cold PBS-CaCl2 and incubated at 37°C for 40 min. After incubation, the cells were washed in cold PBS-CaCl2 and immediately visualized and photographed with a Nikon Diaphot 300 fluorescence microscope.
Virus infection.
COS-7 monolayers grown in 35-mm plates were incubated with egg-grown NDV strain B1 (multiplicity of infection, 10) at 37°C for 40 min in the presence or absence of inhibitors. The monolayers were washed three times with DMEM and further incubated at 37°C for 9 h with or without inhibitors. The cells were then washed and lysed. Proteins in lysates were resolved by SDS-PAGE and analyzed by Western blotting using a mixture of anti-NDV (1:1,250), anti-HN protein (anti-AS; 1:1,650), and anti-F protein (anti-HR2; 1:2,500) antibodies described above.
For analysis of infection by immunofluorescence (IF), COS-7 cells were grown in 35-mm plates containing glass coverslips and infected in the presence or absence of inhibitors as described above. After 40 min, the cells were washed three times with DMEM and further incubated for 2 h with or without inhibitors. The inhibitors were removed, and surface-bound virus was neutralized by anti-NDV antibody (1:40 dilution). After 6 h of incubation at 37°C, the cells were washed twice with PBS, fixed in 1% paraformaldehyde, and incubated at 4°C in IF buffer (PBS containing 1% bovine serum albumin, 0.02% sodium azide, and 0.01% CaCl2) for 1 h. The cells were incubated with primary antibody (anti-NDV) diluted in IF buffer (1:200) at 4°C for 1 h, washed three times with IF buffer, and incubated for 1 h with anti-rabbit IgG coupled to Alexa-488 diluted in IF buffer (1:200). The cells were washed in ice-cold IF buffer. Coverslips were mounted on slides, and the cells were photographed immediately with a Nikon Diaphot 300 fluorescence microscope.
Biotinylation with MPB.
MPB [3-(N-maleimidylpropionyl) biocytin] (Molecular Probes) was used to biotinylate free thiols in cell surface proteins. Transfected cells grown in 35-mm plates were washed with PBS-CM and incubated with MPB (0.5 mM in PBS) at 25°C for 40 min in the presence or absence of DTNB (7.5 mM) or bacitracin (7.5 mM). The cells were then washed once with DMEM and twice with PBS and lysed using RSB lysis buffer as described above. The lysates were precipitated with 0.3% SDS containing neutravidin-agarose that had been washed sequentially with PBS containing 0.5% Tween 20 and 5 mg/ml BSA and PBS containing 0.5% Tween 20 and 1 mg/ml BSA. The precipitates were washed three times with PBS containing 0.5% Tween 20 and 0.4% SDS, resolved by SDS-PAGE, and analyzed by Western blotting using anti-F (anti-HR2) and anti-HN (anti-AS) antibodies as described previously.
Immunofluorescence.
COS-7 cells were grown in 35-mm plates containing glass coverslips and transfected with pSVL HN and pSVL F as described above. After 36 h, DTNB (7.5 mM) or bacitracin (7.5 mM) was added, and the cells were incubated overnight. After 48 h, the cells were washed twice with ice-cold IF buffer and incubated for 1 h at 4°C in IF buffer with or without inhibitors. The cells were then incubated at 4°C for 1 h with IF buffer containing antibody (diluted 1:200) and inhibitors. The cells were washed three times with ice-cold IF buffer and incubated for 1 h on ice with IF buffer containing Alexa 488-labeled anti-rabbit IgG or Alexa 570-labeled anti-mouse IgG (diluted 1:200), as well as inhibitors. The cells were washed with ice-cold IF buffer, fixed with 2% paraformaldehyde, and mounted for microscopy.
Quantification of surface immunofluorescence was accomplished by determining the mean fluorescence intensities for cells, using Adobe Photoshop as described previously (64). Individual cells were outlined manually using the lasso tool in immunofluorescence pictures opened in Photoshop. The mean fluorescences of selected areas were determined with the histogram submenu. Background fluorescence values were obtained from empty-vector-transfected cells and were subtracted from the values obtained from cells expressing the HN and F proteins.
RESULTS
PDI inhibitors did not affect cell viability or viral glycoprotein surface expression.
To explore the role of disulfide bond rearrangement in the functions of NDV glycoproteins, we determined the effects of inhibitors of disulfide bond exchange—DTNB, bacitracin, and anti-PDI antibodies—on virus attachment and membrane fusion.
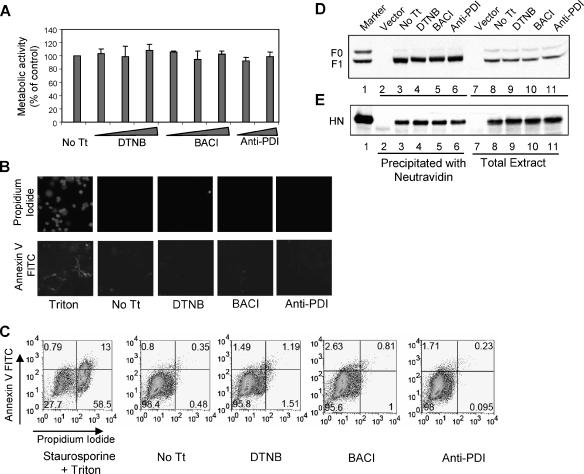
For these studies, it was first necessary to determine the effects of these compounds on cell viability and on glycoprotein surface expression. Cell viability was determined using the compound MTT, which is metabolized to form blue formazan crystals in live cells (52). COS-7 cells, treated overnight with increasing concentrations of DTNB, bacitracin, and anti-PDI antibodies, were incubated with MTT, and the development of blue color in these cells was compared to that in untreated controls. As shown in Fig. 1A, there were no differences in viability between untreated cells and cells subjected to prolonged incubation with the inhibitors.
FIG. 1.
Effects of PDI inhibitors on cell viability and protein expression. (A) Cells incubated overnight with DTNB (2.5 mM, 5 mM, or 7.5 mM), bacitracin (BACI) (2.5 mM, 5 mM, or 7.5 mM), or anti-PDI antibody (3 μl or 6 μl/ml) were incubated with MTT for 3 h at 37°C. The blue metabolic product of MTT, measured by optical density at 570 nm, is represented as a percentage of the value obtained for untreated cells (No Tt). The results shown are the averages of three independent experiments, and the error bars indicate the standard deviations. (B and C) Cells were treated with Triton X-100 (0.05%) (B) or with Triton X-100 and staurosporine (1 μM) (C) as a positive control or with DTNB (7.5 mM), bacitracin (BACI) (7.5 mM), or anti-PDI antibody (6 μl/ml) or were left untreated (No Tt). The cells were stained with annexin V-FITC and PI and were analyzed for double staining by immunofluorescence (B) and FACS (C). The numbers in each quadrant indicate percentages of total cells. (D and E) Cells, transfected with empty vector (lanes 2 and 7) or HN and F protein cDNAs (lanes 3 to 6 and 8 to 11), were incubated with DTNB (7.5 mM) (lanes 4 and 9), bacitracin (BACI) (7.5 mM) (lanes 5 and 10), or anti-PDI antibody (6 μl/ml) (lanes 6 and 11) or without inhibitor (No Tt) (lanes 3 and 8). Surface F protein (D) or HN protein (E), biotinylated using sulfo-NHS-SS-biotin, were precipitated with neutravidin-agarose (lanes 3 to 6), and total F or HN protein in the extracts (lanes 8 to 11) was resolved by SDS-PAGE and analyzed by Western blotting using anti-F protein antibody (anti-HR2) or anti-HN protein antibody (anti-AS). The amount of the total extract loaded represents one-third of the amount of extract used to precipitate biotinylated surface proteins. Lane 1 shows infected-cell extract used as a marker.
The effects of inhibitors on cell viability were further explored using annexin V-FITC and PI double staining. Annexin V binds to cell surfaces, and propidium iodide stains nuclei only in cells undergoing apoptosis (77). Annexin V-FITC and PI double staining of control and inhibitor-treated cells was determined by both fluorescence microscopy and FACS, and the results are shown in Fig. 1B and C, respectively. These results confirmed that PDI inhibitors do not affect cell viability.
To determine the effects of PDI inhibitors on total expression of HN and F glycoproteins or on their cell surface expression, intact COS-7 cells expressing the NDV HN and F proteins were either treated with PDI inhibitors overnight or left untreated, and the surface proteins were biotinylated with sulfo-NHS-SS-biotin. Incubation with inhibitors had no effect on total expression of F protein (Fig. 1D, lanes 8 to 11) or HN protein (Fig. 1E, lanes 8 to 11). Biotinylated surface proteins in these lysates were precipitated with neutravidin, and F and HN proteins in the precipitates are shown in Fig. 1B and C, lanes 3 to 6, respectively. Clearly, cell surface expression of both F protein and HN protein was unaffected by prolonged incubation with inhibitors.
PDI inhibitors did not affect receptor binding or neuraminidase activity of HN protein.
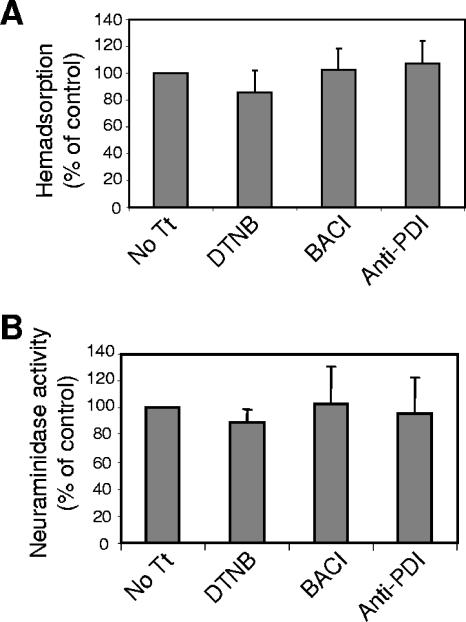
The effects of inhibitors on the receptor binding activity of NDV HN protein expressed on cell surfaces were quantified by measuring the ability of these cells to bind RBCs. COS-7 cells expressing NDV HN protein were treated overnight with DTNB, bacitracin, or anti-PDI antibodies and then incubated with RBCs. Bound RBCs were quantified as described in Materials and Methods. As shown in Fig. 2A, PDI inhibitors did not affect the binding of RBCs to surface-expressed HN protein.
FIG. 2.
Effects of PDI inhibitors on HN protein activities. (A) Effects of overnight incubation in DTNB (7.5 mM), bacitracin (BACI) (7.5 mM), or anti-PDI antibody (6 μl/ml) on the attachment activity of cell surface HN protein were analyzed by binding of RBCs. Cells were incubated with RBCs for 30 min at 4°C, bound RBCs were lysed, and released hemoglobin was quantified as described in Materials and Methods. RBC binding to inhibitor-treated cells is expressed as percentages of the binding obtained using untreated cells (No Tt). The results are averages of three independent experiments, and the error bars indicate standard deviations. (B) Effects of overnight incubation in DTNB (7.5 mM), bacitracin (BACI) (7.5 mM), or anti-PDI antibody (6 μl/ml) on the neuraminidase activity of cell surface HN protein were analyzed as described in Materials and Methods. The results are expressed as percentages of values obtained for untreated cells (No Tt) and are averages of three independent experiments.
To determine if PDI inhibitors affected the neuraminidase activity of HN protein expressed on cell surfaces, cells expressing NDV HN protein were treated overnight with PDI inhibitors or left untreated. Cell surface neuraminidase activity was quantified as described in Materials and Methods. Figure 2B shows that PDI inhibitors did not affect the neuraminidase activity of cell surface HN protein. These results also verify that the levels of HN protein expressed on the surfaces of inhibitor-treated cells were similar to levels on untreated cells.
PDI inhibitors inhibited cell-cell fusion.
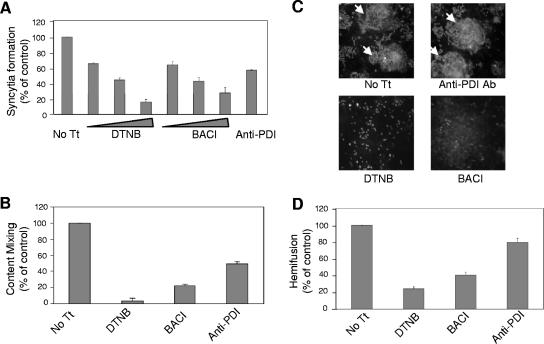
We next asked if PDI inhibitors blocked F protein-directed cell-cell fusion. Cell-cell fusion was measured in three separate assays: syncytium formation, content mixing between effector and target cells, and hemifusion between HN and F protein-expressing cells and red blood cells. As shown in Fig. 3A, increasing concentrations of both DTNB and bacitracin increasingly blocked syncytium formation. At the highest concentrations, DTNB (7.5 mM) inhibited syncytium formation by 83% and bacitracin (7.5 mM) by 71%. Anti-PDI antibodies inhibited syncytium formation by 42%.
FIG. 3.
Effects of PDI inhibitors on cell-cell fusion. (A) Syncytium formation in the presence of DTNB (2.5 mM, 5 mM, or 7.5 mM), bacitracin (BACI) (2.5 mM, 5 mM, or 7.5 mM), or anti-PDI antibody (6 μl/ml) was quantified as described in Materials and Methods. Inhibitors were added 24 h posttransfection, and at 48 h posttransfection, the average size of syncytia was determined. The results are expressed as percentages of the values obtained for untreated cells (No Tt) and are the averages of three independent experiments, with the error bars indicating standard deviations. (B) Effects of DTNB (7.5 mM), bacitracin (BACI) (7.5 mM), or anti-PDI antibody (6 μl/ml) on content mixing were assayed using a β-galactosidase reporter assay. Effector cells cotransfected with HN and F protein cDNAs, as well as pTA, were treated with inhibitors or left untreated. Treated or untreated target cells, transfected with pB1-G only, were overlaid onto HN and F protein-expressing cells. Content mixing between effector and target cells was quantified by measuring β-galactosidase in cell extracts and is shown as a percentage of the activity detected in untreated cells (No Tt). The results are the averages of three independent experiments, and the error bars indicate standard deviations. (C and D) The effects of DTNB (7.5 mM), bacitracin (BACI) (7.5 mM), or anti-PDI antibody (6 μl/ml) on lipid mixing were assayed in COS-7 cells transfected with HN and F protein cDNAs. At 24 h posttransfection, the inhibitors were added to the cells, and at 48 h posttransfection, the cells were incubated with R18-labeled RBCs on ice, followed by incubation at 37°C for 40 min. The cells were visualized with a fluorescence microscope for dye transfer (C). Cells with positive dye transfer are indicated by arrows. The average numbers of cells with positive dye transfer per field were determined and are represented as percentages of the values determined for untreated cells (No Tt) (D). The results are the averages of three different experiments, and the error bars represent standard deviations.
The effects of PDI inhibitors on pore formation were determined by measuring the extent of content mixing between effector and target cells using a reporter assay as described in Materials and Methods. As showed in Fig. 3B, DTNB inhibited content mixing by 95%, bacitracin by 77%, and anti-PDI antibodies by 51%.
The effects of PDI inhibitors on hemifusion were determined by assessing their effects on lipid mixing between HN and F protein-expressing cells and RBCs labeled with the fluorescence-labeled lipid R18. The extent of lipid dye transfer from RBC membranes to COS-7 cell membranes, which occurs during the initial stages of fusion, was determined by fluorescence microscopy, and representative results are shown in Fig. 3C. Fluorescent-dye transfer occurred between RBCs and untreated cells or cells treated with anti-PDI antibodies, while DTNB and bacitracin significantly decreased the number of cells with positive dye transfer. The number of cells with positive dye transfer per field was expressed as a percentage of the number of positive, untreated cells. As shown in Fig. 3D, DTNB inhibited hemifusion by 75%, bacitracin by 60%, and anti-PDI antibodies by 20%. These results suggested that PDI inhibitors inhibited cell-cell fusion and that the inhibition occurred prior to the onset of fusion, before hemifusion.
PDI inhibitors inhibited virus entry.
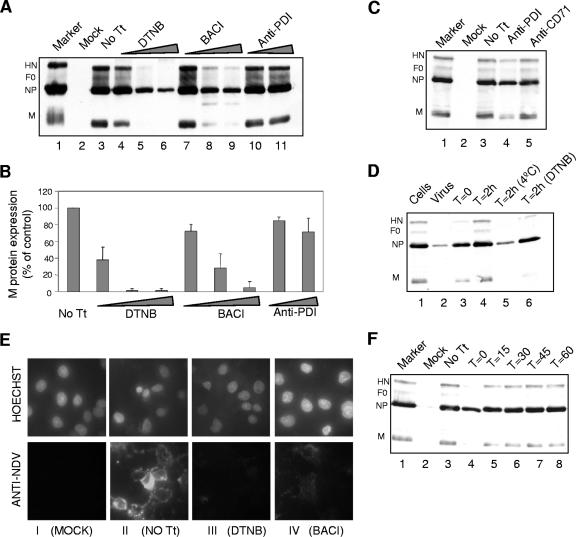
To determine if PDI inhibitors can inhibit virus entry, COS-7 cells were infected with egg-grown NDV strain B1 in the presence or absence of inhibitors. The extents of infection were measured by the levels of newly made proteins in infected cells. NDV protein expression at 9 h in untreated cells is shown in Fig. 4A, lane 3. Incubation with increasing concentrations of DTNB (lanes 4 to 6) and bacitracin (lanes 7 to 9) significantly decreased NDV protein expression compared to untreated cells (lane 3), while anti-PDI antibody (lanes 10 and 11) was not as effective. Levels of NDV protein expression were quantified using M protein (Fig. 4B). Higher concentrations of DTNB and bacitracin inhibited M protein expression by 95%, and anti-PDI antibody inhibited it by only 20%.
FIG. 4.
Effects of PDI inhibitors on virus entry. (A and B) COS-7 cells, incubated with DTNB (2.5 mM, 5 mM, or 7.5 mM) (lanes 4 to 6), bacitracin (BACI) (2.5 mM, 5 mM, or 7.5 mM) (lanes 7 to 9), or anti-PDI antibody (3 μl or 6 μl/ml) (lanes 10 and 11), were infected with NDV strain B1 (multiplicity of infection, 10) in the presence (lanes 4 to 11) or absence (lane 3; No Tt) of inhibitors. The cells were further incubated with or without inhibitors for 9 h at 37°C. Proteins in the resulting cell lysates were resolved by SDS-PAGE (10%) and analyzed for NDV protein expression by Western blotting using a mixture of anti-NDV, anti-HN, and anti-F protein antibodies as described in Materials and Methods (A). Lane 2 shows mock-infected-cell extract, and lane 1 shows infected-cell extract used as a marker. The M protein bands were quantified by densitometry, and values are expressed as percentages of the values for cells infected in the absence of inhibitors (B). The results are averages of three different experiments. (C) Cells were incubated overnight with anti-PDI antibody (lane 4) or anti-CD71 antibody (lane 5) or left untreated (lane 3), infected with NDV strain B1 in the presence or absence of antibodies, and analyzed for NDV protein expression as described above. (D) Lane 1 shows cells preincubated with anti-NDV antibody, washed, and then infected with virus (negative control). Lane 2 shows cells infected with virus preincubated with antibody (positive control). Lanes 3 and 4 show infected cells with antibody added at the time of infection and 2 h after infection, respectively. Lane 5 shows cells to which anti-NDV antibody was added after 2 h of virus adsorption at 4°C. Lane 6 shows cells infected in the presence of DTNB and incubated with anti-NDV antibody after 2 h. (E) Cells plated on coverslips were incubated with inhibitors and infected with NDV as described above. After 2 h of incubation, the inhibitors were removed and bound virus was neutralized by anti-NDV antibody. The cells were further incubated for 6 h, and surface-expressed NDV proteins were detected by immunofluorescence using anti-NDV antibody. The top panels show nuclei of cells stained with Hoechst stain, and the bottom panels show cells incubated with anti-NDV antibody. The first column (I) shows mock-infected cells, the second column (II) shows cells infected without any inhibitor treatment (No Tt), the third column (III) shows cells infected in the presence of DTNB (7.5 mM), and the fourth column (IV) shows cells infected in the presence of bacitracin (BACI) (7.5 mM). (F) DTNB (7.5 mM) was added either at the time of infection (as described above) (lane 4) or 15 min (lane 5), 30 min (lane 6), 45 min (lane 7), or 60 min (lane 8) after the infection of cells.
To determine if the slight inhibition of virus infection by anti-PDI antibody was a specific effect of the antibody, cells were infected in the presence of anti-CD71 antibody, a rabbit polyclonal IgG. As shown in Fig. 4C, anti-CD71 antibody did not affect the NDV protein expression (lane 5), while anti-PDI antibody slightly decreased the protein expression (lane 4).
To ensure that the inhibition of infection by PDI inhibitors was not due to effects on viral replication after virus entry, cells were incubated with the inhibitors for only 2 h after virus adsorption. To prevent the entry of bound virus after the removal of inhibitors, anti-NDV antibody was added after the inhibitors were removed. The efficiency of anti-NDV antibody in neutralizing virus already bound to the target cells is shown in Fig. 4D. Infected cells were incubated at 4°C for 2 h (lane 5). At 4°C, virus particles bind to cells but are unable to proceed to membrane fusion. After 2 h, the cells were washed, and anti-NDV antibody was added. The cells were further incubated and analyzed for NDV protein expression. The protein expression was decreased and was comparable to lane 2, which shows cell lysates infected with virus preincubated with anti-NDV antibody. This result indicates that anti-NDV antibody can neutralize NDV bound to cells.
Figure 4E shows that removal of inhibitors at 2 h postinfection still inhibited virus infection, results consistent with effects on virus entry. Cells were infected for 2 h in the presence of inhibitors, the inhibitors were removed, and anti-NDV antibody was added. The cells were incubated for 6 h and then analyzed for NDV protein expression by cell surface immunofluorescence (Fig. 4E). Cells infected in the presence of DTNB (panel E, III) and bacitracin (panel E, IV) showed significantly decreased surface expression of NDV proteins compared to cells infected without any inhibitor (panel E, II). These results are consistent with an inhibition of virus entry.
To further confirm that PDI inhibitors inhibit a very early step in infection, DTNB was added at different time points after the start of infection. As shown in Fig. 4F, adding the inhibitor as early as 15 min (lane 5) and 30 min (lane 6) after infection resulted in significant levels of virus infection, while adding the inhibitor with the virus blocked infection (lane 4). These results are consistent with effects of DTNB on virus entry.
Free thiols were detected in F protein but not in HN protein.
Cell surface PDI molecules are thought to primarily reduce disulfide bonds of surface-expressed proteins (38, 41). Thus, it seemed likely that these inhibitors blocked fusion by preventing the formation of free thiols. It follows that either surface-expressed HN or F protein may have free thiols. To detect free thiols on cell surface proteins, we used MPB, a non-membrane-permeating, thiol-specific biotin that binds to and biotinylates only free-thiol-containing proteins.
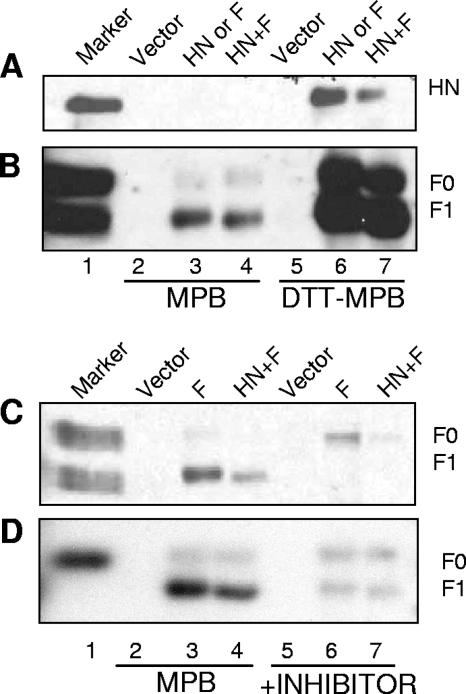
COS-7 cells expressing either HN or F or coexpressing both proteins were incubated with MPB. MPB-labeled proteins present in lysates of these cells were precipitated under stringent conditions with neutravidin and resolved by SDS-PAGE, and HN and F proteins in the polyacrylamide gels were detected by Western analysis. SDS was included in the precipitation with neutravidin to eliminate any association of host proteins with either viral protein. As a positive control, cells cultured in parallel were incubated with the reducing agent dithiothreitol (DTT) in order to generate free thiols. Figure 5A shows detection of HN protein in the precipitates. HN protein expressed alone or with F protein was not biotinylated by MPB (lanes 3 and 4), while HN protein expressed on cell surfaces treated with DTT prior to incubation with MPB was biotinylated (lanes 6 and 7). This result indicates that HN protein at the cell surface does not have free thiols. Figure 5B shows that F protein was biotinylated by MPB when expressed alone or with HN protein (lanes 3 and 4). This result suggests that surface-expressed F protein has free thiols. The slightly lower levels of HN protein in lane 7 or F protein in lane 4 can be accounted for by a slight reduction in the recovery of total HN or F protein when coexpressed (data not shown), as has been previously noted (5).
FIG. 5.
Biotinylation using a thiol-specific biotin, MPB. (A and B) COS-7 monolayers were incubated with MPB (0.5 mM in PBS) at 25°C for 40 min. The proteins in the resulting cell lysates were precipitated with neutravidin as described in Materials and Methods, and precipitated proteins were resolved by SDS-PAGE (10%) and analyzed by Western blotting using anti-HN protein antibody (A) and anti-F protein antibody (B). Lane 1 contains infected-cell extract used as a marker, lanes 2 and 5 contain empty-vector-transfected cell extracts, lanes 3 and 6 contain HN (A) or F (B) protein-transfected cell extracts, and lanes 4 and 7 contain HN and F protein-cotransfected cell extracts. Lanes 2 to 4 show cells incubated with MPB, and lanes 5 to 7 are cells incubated with DTT prior to biotinylation with MPB. (C and D) Cells transfected with empty vector (lanes 2 and 5), F protein cDNA (lanes 3 and 6), or HN and F protein cDNAs (lanes 4 and 7) were incubated with MPB in the presence (lanes 5 to 7) or absence (lanes 2 to 4) of DTNB (C) or bacitracin (D). Proteins in the resulting cell lysates were precipitated with neutravidin. Precipitated proteins were resolved by SDS-PAGE (10%) and analyzed by Western blotting using anti-F protein antibody.
PDI inhibitors blocked detection of free thiols in F protein.
To determine if PDI inhibitors block the formation of free thiols in surface-expressed F protein, F protein-expressing cells were incubated with MPB in the presence of DTNB or bacitracin. Figure 5C shows that both DTNB (Fig. 5C, lanes 6 and 7) and bacitracin (Fig. 5D, lanes 6 and 7) inhibited the biotinylation of F protein by MPB. These results indicate that the PDI inhibitors effectively blocked the formation of free thiols in the F protein, which correlated with their ability to block membrane fusion.
MPB binding to F protein blocked cell-cell fusion.
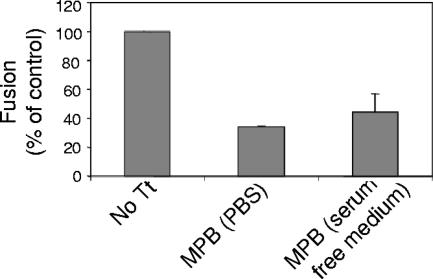
Using the content-mixing assay, we also investigated whether MPB binding to free thiols in F protein could block cell-cell fusion. Effector cells were overlaid with target cells in the presence or absence of MPB, and the extent of fusion between the two cell populations was determined by measuring the levels of β-galactosidase activity as described in Materials and Methods. Figure 6 shows that MPB binding inhibited fusion by 60 to 70%, indicating that blocking the free thiols in the F protein inhibited cell-cell fusion. This result also shows that F protein with free thiols is a biologically functional form of the protein.
FIG. 6.
Effect of MPB on cell-cell fusion. The effect of MPB on cell-cell fusion was assayed using a β-galactosidase reporter assay. Cells, cotransfected with HN and F protein cDNAs, as well as pTA, were overlaid with target cells transfected with pB1-G in the presence of MPB (0.5 mM in PBS or serum-free medium) for 1 h at 25°C. The cells were further incubated with supplemented DMEM for 5 h at 37°C. The cells were washed, and content mixing between HN and F protein-expressing cells and target cells was quantified by measuring β-galactosidase as described in Materials and Methods and is shown as percentages of the activities detected in cell mixtures treated with PBS without MPB (No Tt). The results are the averages of three independent experiments, and the error bars indicate standard deviations.
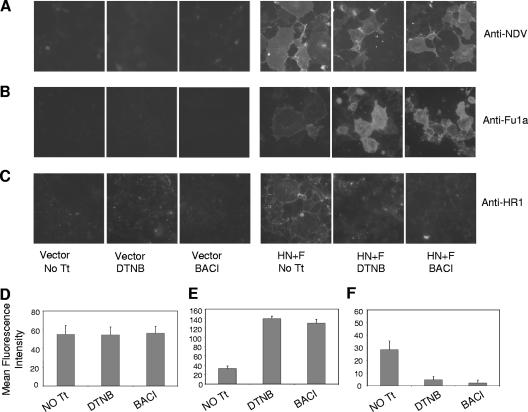
DTNB and bacitracin altered antibody binding to surface F protein.
If disulfide bond reduction has a role in conformational changes associated with the onset of fusion, then inhibition of the formation of free thiols in F protein may alter the conformation of F protein on the cell surfaces. Since antibodies are often used to detect conformation differences in proteins, two different conformation-sensitive anti-F protein antibodies, anti-Fu1a (51) and anti HR1 (43) antibodies, were used to determine if incubation with disulfide bond isomerase inhibitors affected the binding of either of these antibodies to F protein. Anti-NDV antibody was used as a control, and as shown in Fig. 7A, anti-NDV antibody binding to cells was unaffected by overnight incubation in DTNB or bacitracin. By contrast, incubation with inhibitors significantly increased the binding of anti-Fu1a (Fig. 7B). Furthermore, incubation with inhibitors eliminated the binding of anti-HR1 antibody (Fig. 7C). Mean fluorescence intensities were quantified for the cells shown in panels A, B, and C and are shown in panels D (using anti-NDV antibody), E (using anti-Fu1a antibody), and F (using HR1 antibody). These results indicate that inhibitors of disulfide bond isomerases altered the conformation of the F protein expressed on cell surfaces.
FIG. 7.
Binding of different conformation-sensitive antibodies to surface-expressed F protein in the presence of inhibitors. (A, B, and C) Cell surface immunofluorescence using anti-NDV antibody, anti-Fu1a antibody, and anti-HR1 antibody, respectively. Column 1 shows cells transfected with empty vector without any treatment (No Tt). Columns 2 and 3 show cells transfected with empty vector and treated with DTNB (7.5 mM) and bacitracin (BACI) (7.5 mM), respectively. Column 4 shows cells transfected with HN and F protein cDNAs without any treatment (No Tt), and columns 5 and 6 show cells transfected with HN and F protein cDNAs and treated with DTNB (7.5 mM) and bacitracin (BACI) (7.5 mM), respectively. (D, E, and F) Quantification of the fluorescence intensities of the cells shown in panels A, B, and C, respectively. The results shown are averages of the mean fluorescences of 10 different cells and are normalized for background fluorescence by subtracting the mean fluorescence of cells transfected with vector alone.
DISCUSSION
Fusion proteins are proposed to undergo extensive conformational changes with the activation of fusion. Evidence for these changes includes alterations in antibody reactivities upon activation (11, 13, 18, 30, 33, 68), alterations in protease sensitivity (31, 48, 57, 68), changes in oligomeric status (36, 68), and differential binding of peptides with sequences from one of the HR domains (59). Structural analyses of the prefusion and postfusion forms of influenza virus HA (6, 63, 65, 72) are quite consistent with these proposed conformational shifts. In addition, significant conformational differences have been observed between two crystallized forms of the paramyxovirus F protein. One form, exemplified by the structures of the parainfluenza virus 3 F protein (74) and the NDV F protein (7), contains a six-stranded coiled coil composed of the two HR domains of the proteins. Although these crystals were derived from uncleaved forms of the F proteins, they are proposed to be characteristic of the postfusion form of the paramyxovirus F protein (75). Another F protein structure (75), derived from a crystal formed by the ectodomain of an uncleaved simian virus 5 F protein fused at the carboxyl terminus of the HR2 domain with the yeast GCN4 sequence, was significantly different. The GCN4 sequence forces the HR2 domain to trimerize, preventing the formation of the HR1-HR2 complex. It has been proposed that this alternative structure represents the conformation of the prefusion F protein (60, 75). Based on these two different structures, a model for the conformational changes in the paramyxovirus F protein, changes that involve significant rearrangement of the molecule, has been presented (60, 75).
Disulfide bond isomerization in the HIV Env protein is proposed to accompany and facilitate conformational shifts in that molecule during fusion. Evidence for this idea includes observations that inhibitors, such as DTNB, bacitracin, and anti-PDI antibody, which block the activities of PDI and related isomerases, inhibit HIV entry and HIV cell-cell fusion (16, 40, 61). Second, free thiols have been detected in the gp120 subunit of the Env protein (4, 19, 40). These free thiols are reported to form upon CD4 binding (4, 19, 40) or upon binding to CXCR4 (4). Third, other retroviruses, including murine leukemia virus, have, within the Env sequence, CXXC motifs, which are used during fusion to isomerize the disulfide bond between the TM and SU subunits (17, 56, 69). It has been suggested that HIV relies on host isomerases while other retroviruses have evolved to encode their own isomerase activities (61, 69).
Additionally, supporting the idea that disulfide bond isomerization may be involved in membrane fusion is a theoretical analysis of disulfide bonds in proteins involved in cell entry (73). This study described some unusual disulfide bonds present in these proteins. These bonds, called cross-strand disulfides (CSDs), link cysteine residues in adjacent strands in the same β-sheet and have high potential energy stored in them (41, 73). CSDs are identified by higher dihedral strain energy in these bonds due to torsional strain produced by linking adjacent strands (25, 41, 73). Also, CSDs are more readily cleaved than other disulfide bonds and are more susceptible to reducing agents, thioredoxin or PDI (41, 73). These bonds were found at higher frequency in proteins involved in cell entry, including bacterial toxins and the viral fusion proteins, HIV gp120, influenza virus HA, and NDV F protein. CSD in the NDV fusion protein links Cys338 and Cys347, which raises the possibility that cleavage of this disulfide bond may be involved in cell entry by NDV (73).
The results presented here are consistent with the idea that thiol/disulfide exchange in NDV F protein, mediated by a PDI or PDI-like isomerases, is required for virus entry or cell-cell fusion mediated by F protein. Inhibitors of PDI, DTNB and bacitracin, inhibited cell-cell fusion. The results showing an inhibition of virus protein expression are consistent with effects on virus entry. Furthermore, inhibition of cell-cell fusion occurred at an early stage in the onset of fusion, since these inhibitors blocked hemifusion, a result consistent with effects on fusion activation. These results are less likely to be due to nonspecific effects of these inhibitors, since the inhibitors had no effect on cell viability, glycoprotein surface expression, or the attachment and neuraminidase activities of the HN protein.
Anti-PDI antibody was less effective in inhibiting cell-cell fusion, as well as virus entry. DTNB and bacitracin are not specific inhibitors of PDI. DTNB is a nonspecific inhibitor of free thiols (15), and bacitracin binds to the CXXC motif in catalytic domains that are present in all of the members of the thioredoxin family (53). The lower efficiency of anti-PDI antibody in fusion inhibition suggests that isomerization of disulfide bonds in the F protein may be due to one or several other thiol-reactive proteins that are found in cells (14, 71). Indeed, it has been shown that down regulation of PDI using small interfering RNA had only a small effect on infection or cell fusion mediated by HIV-1 (55), suggesting that other thiol-active enzymes at the cell surfaces are involved in reduction of the HIV envelope glycoprotein.
Since cell surface PDI and related proteins usually serve to reduce bonds in interacting proteins, it seemed likely that the inhibitors blocked the reduction of one or more disulfide bonds in F protein and that surface F protein may have free thiols. Indeed, the results showed that MPB, a membrane-impermeable thiol-specific reagent, binds to the F protein. This binding was inhibited by DTNB and bacitracin, suggesting that the free thiols in F protein are due to reduction by a PDI-like isomerase. The result that inhibitors blocked both fusion and detection of the free thiols is consistent with the idea that only F protein with free thiols can proceed to direct fusion. Interestingly, MPB also inhibited fusion. This result may indicate that completion of the fusion process requires reoxidation of the free thiols in F protein, perhaps as the protein refolds during the onset of fusion. Alternatively, addition of MPB to the F protein may interfere with subsequent conformational changes necessary for fusion. Inhibition of fusion by MPB indicates that F protein with free thiols is not an aberrant, biologically irrelevant form, nor is it in a postfusion conformation.
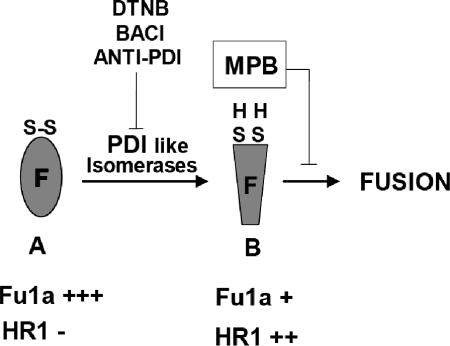
The results described above are consistent with the model shown in Fig. 8. Form A shows F protein in a prefusion form with a full complement of intramolecular disulfide bonds. Cleavage of disulfide bonds by PDI-like isomerase produces free thiols in the F protein, as shown in form B. Inhibitors of PDI-like enzymes inhibit the production of these free thiols so that the prefusion form of F protein is the predominant form in the presence of inhibitors. Binding of MPB to the free thiols in F protein in form B inhibits fusion. This binding may inhibit further conformational changes in the F protein, preventing the refolding required for fusion (60, 75).
FIG. 8.
Predicted model for the role of PDI in fusion mediated by NDV F protein. Form A represents the F protein in prefusion metastable conformation with intact disulfide bonds. Disulfide bonds are cleaved by PDI-like isomerase, resulting in free thiols in the F protein (form B). PDI inhibitors block the production of free thiols. MPB binds to the free thiols shown in form B, and the binding of MPB prevents further conformational changes required for fusion.
The existence of these predicted conformational forms is supported by changes in reactivity to the conformation-sensitive anti-F protein antibodies, anti-Fu1a and anti-HR1 antibodies, after incubation in inhibitors of disulfide bond isomerases. Fu1a antibody binding is significantly enhanced in the presence of DTNB and bacitracin. By contrast, reactivity to antibodies specific for the HR1 domain was lost in the presence of these inhibitors. Both results are consistent with the suggestion that a significant population of F protein on cell surfaces contains free thiols and that the reduction alters the conformation of the protein, decreasing reactivity to anti-Fu1a and increasing the accessibility of the HR1 domain.
Free thiols were detected on both F protein expressed alone and F protein coexpressed with HN protein. This result may indicate that binding of the attachment protein to receptors is not required for the formation of free thiols in the F protein. Rather, the results are consistent with the possibility that F protein may exist as a mixture of oxidized and reduced forms on cell surfaces. In the presence of HN protein, only the reduced form may proceed to refold into additional intermediates. The result that covalent addition of MPB blocks fusion provides additional evidence for further conformational changes that are required for fusion.
Acknowledgments
We thank M. Peeples for antibodies.
This work was supported by grant AI 30572 from the National Institutes of Health.
Footnotes
Published ahead of print on 6 December 2006.
REFERENCES
- 1.Abdel-Latif, L., B. K. Murray, R. L. Renberg, K. L. O'Neill, H. Porter, J. B. Jensen, and F. B. Johnson. 2006. Cell death in bovine parvovirus-infected embryonic bovine tracheal cells is mediated by necrosis rather than apoptosis. J. Gen. Virol. 87:2539-2548. [DOI] [PubMed] [Google Scholar]
- 2.Abell, B. A., and D. T. Brown. 1993. Sindbis virus membrane fusion is mediated by reduction of glycoprotein disulfide bridges at the cell surface. J. Virol. 67:5496-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 4.Barbouche, R., R. Miquelis, I. M. Jones, and E. Fenouillet. 2003. Protein-disulfide isomerase-mediated reduction of two disulfide bonds of HIV envelope glycoprotein 120 occurs post-CXCR4 binding and is required for fusion. J. Biol. Chem. 278:3131-3136. [DOI] [PubMed] [Google Scholar]
- 5.Bousse, T., T. Takimoto, K. G. Murti, and A. Portner. 1997. Elevated expression of the human parainfluenza virus type 1 F gene downregulates HN expression. Virology 232:44-52. [DOI] [PubMed] [Google Scholar]
- 6.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9:255-266. [DOI] [PubMed] [Google Scholar]
- 8.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 95:3425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colman, P. M., and M. C. Lawrence. 2003. The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell. Biol. 4:309-319. [DOI] [PubMed] [Google Scholar]
- 10.Dolganiuc, V., L. McGinnes, E. J. Luna, and T. G. Morrison. 2003. Role of the cytoplasmic domain of the Newcastle disease virus fusion protein in association with lipid rafts. J. Virol. 77:12968-12979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Earl, P. L., C. C. Broder, D. Long, S. A. Lee, J. Peterson, S. Chakrabarti, R. W. Doms, and B. Moss. 1994. Native oligomeric human immunodeficiency virus type 1 envelope glycoprotein elicits diverse monoclonal antibody reactivities. J. Virol. 68:3015-3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earp, L. J., S. E. Delos, H. E. Park, and J. M. White. 2005. The many mechanisms of viral membrane fusion proteins. Curr. Top. Microbiol. Immunol. 285:25-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, J., E. Mann, and D. T. Brown. 1983. Conformational changes in Sindbis virus envelope proteins accompanying exposure to low pH. J. Virol. 45:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellgaard, L., and L. W. Ruddock. 2005. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 6:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feener, E. P., W. C. Shen, and H. J. Ryser. 1990. Cleavage of disulfide bonds in endocytosed macromolecules. A processing not associated with lysosomes or endosomes. J. Biol. Chem. 265:18780-18785. [PubMed] [Google Scholar]
- 16.Fenouillet, E., R. Barbouche, J. Courageot, and R. Miquelis. 2001. The catalytic activity of protein disulfide isomerase is involved in human immunodeficiency virus envelope-mediated membrane fusion after CD4 cell binding. J. Infect. Dis. 183:744-752. [DOI] [PubMed] [Google Scholar]
- 17.Ferrari, D. M., and H. D. Soling. 1999. The protein disulphide-isomerase family: unravelling a string of folds. Biochem. J. 339:1-10. [PMC free article] [PubMed] [Google Scholar]
- 18.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2002. Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J. Virol. 76:12123-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallina, A., T. M. Hanley, R. Mandel, M. Trahey, C. C. Broder, G. A. Viglianti, and H. J. Ryser. 2002. Inhibitors of protein-disulfide isomerase prevent cleavage of disulfide bonds in receptor-bound glycoprotein 120 and prevent HIV-1 entry. J. Biol. Chem. 277:50579-50588. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert, H. F. 1997. Protein disulfide isomerase and assisted protein folding. J. Biol. Chem. 272:29399-29402. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, J., W. Ou, J. Silver, and T. Benjamin. 2006. Downregulation of protein disulfide isomerase inhibits infection by the mouse polyomavirus. J. Virol. 80:10868-10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heinz, F. X., and S. L. Allison. 2001. The machinery for flavivirus fusion with host cell membranes. Curr. Opin. Microbiol. 4:450-455. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez, L. D., R. J. Peters, S. E. Delos, J. A. Young, D. A. Agard, and J. M. White. 1997. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ALSV envelope glycoprotein. J. Cell Biol. 139:1455-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogg, P. J. 2003. Disulfide bonds as switches for protein function. Trends Biochem. Sci. 28:210-214. [DOI] [PubMed] [Google Scholar]
- 26.Jardetzky, T. S., and R. A. Lamb. 2004. Virology: a class act. Nature 427:307-308. [DOI] [PubMed] [Google Scholar]
- 27.Jordan, P. A., and J. M. Gibbins. 2006. Extracellular disulfide exchange and the regulation of cellular function. Antioxid. Redox. Signal 8:312-324. [DOI] [PubMed] [Google Scholar]
- 28.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 29.Kemble, G. W., Y. I. Henis, and J. M. White. 1993. GPI- and transmembrane-anchored influenza hemagglutinin differ in structure and receptor binding activity. J. Cell Biol. 122:1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kielian, M., and A. Helenius. 1985. pH-induced alterations in the fusogenic spike protein of Semliki Forest virus. J. Cell Biol. 101:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kielian, M., S. Jungerwirth, K. U. Sayad, and S. DeCandido. 1990. Biosynthesis, maturation, and acid activation of the Semliki Forest virus fusion protein. J. Virol. 64:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knerr, I., M. Schnare, K. Hermann, S. Kausler, M. Lehner, T. Vogler, W. Rascher, and U. Meissner. 2 November 2006. Fusiogenic endogenous-retroviral syncytin-1 exerts anti-apoptotic functions in staurosporine-challenged CHO cells. Apoptosis. doi: 10.1007/s10495-006-0329-9. [DOI] [PubMed]
- 33.Kostolansky, F., G. Russ, V. Mucha, and B. Styk. 1988. Changes in the influenza virus haemagglutinin at acid pH detected by monoclonal antibodies to glycopolypeptides HA1 and HA2. Arch. Virol. 101:13-24. [DOI] [PubMed] [Google Scholar]
- 34.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., X. Han, A. L. Lai, J. H. Bushweller, D. S. Cafiso, and L. K. Tamm. 2005. Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion. J. Virol. 79:12065-12076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Locker, J. K., and G. Griffiths. 1999. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J. Cell Biol. 144:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luz, J. M., and W. J. Lennarz. 1996. Protein disulfide isomerase: a multifunctional protein of the endoplasmic reticulum. EXS 77:97-117. [DOI] [PubMed] [Google Scholar]
- 38.Mandel, R., H. J. Ryser, F. Ghani, M. Wu, and D. Peak. 1993. Inhibition of a reductive function of the plasma membrane by bacitracin and antibodies against protein disulfide-isomerase. Proc. Natl. Acad. Sci. USA 90:4112-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Markovic, I., H. Pulyaeva, A. Sokoloff, and L. V. Chernomordik. 1998. Membrane fusion mediated by baculovirus gp64 involves assembly of stable gp64 trimers into multiprotein aggregates. J. Cell Biol. 143:1155-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Markovic, I., T. S. Stantchev, K. H. Fields, L. J. Tiffany, M. Tomic, C. D. Weiss, C. C. Broder, K. Strebel, and K. A. Clouse. 2004. Thiol/disulfide exchange is a prerequisite for CXCR4-tropic HIV-1 envelope-mediated T-cell fusion during viral entry. Blood 103:1586-1594. [DOI] [PubMed] [Google Scholar]
- 41.Matthias, L. J., and P. J. Hogg. 2003. Redox control on the cell surface: implications for HIV-1 entry. Antioxid. Redox. Signal 5:133-138. [DOI] [PubMed] [Google Scholar]
- 42.Mc Ginnes, L. W., J. Reiter, H. Pantua, and T. G. Morrison. 2006. Newcastle disease virus propagation, quantification and storage, 15F.2.3-15F.2.11. In R. Coica, T. Kowalik, J. Quarles, B. Steveman, and R. Taylor (ed.), Current protocols in microbiology, vol. 1. John Wiley and Sons, Inc., Hoboken, NJ. [DOI] [PubMed] [Google Scholar]
- 43.McGinnes, L. W., K. Gravel, and T. G. Morrison. 2002. Newcastle disease virus HN protein alters the conformation of the F protein at cell surfaces. J. Virol. 76:12622-12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McGinnes, L. W., and T. G. Morrison. 2006. Inhibition of receptor binding stabilizes Newcastle disease virus HN and F protein-containing complexes. J. Virol. 80:2894-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGinnes, L. W., and T. G. Morrison. 1994. The role of the individual cysteine residues in the formation of the mature, antigenic HN protein of Newcastle disease virus. Virology 200:470-483. [DOI] [PubMed] [Google Scholar]
- 46.McGinnes, L. W., T. Sergel, H. Chen, L. Hamo, S. Schwertz, D. Li, and T. G. Morrison. 2001. Mutational analysis of the membrane proximal heptad repeat of the Newcastle disease virus fusion protein. Virology 289:343-352. [DOI] [PubMed] [Google Scholar]
- 47.Melikyan, G. B., S. Lin, M. G. Roth, and F. S. Cohen. 1999. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell 10:1821-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meyer, W. J., S. Gidwitz, V. K. Ayers, R. J. Schoepp, and R. E. Johnston. 1992. Conformational alteration of Sindbis virion glycoproteins induced by heat, reducing agents, or low pH. J. Virol. 66:3504-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrison, T. G. 2003. Structure and function of a paramyxovirus fusion protein. Biochim. Biophys. Acta 1614:73-84. [DOI] [PubMed] [Google Scholar]
- 50.Morrison, T. G., and L. W. McGinnes. 1989. Avian cells expressing the Newcastle disease virus hemagglutinin-neuraminidase protein are resistant to Newcastle disease virus infection. Virology 171:10-17. [DOI] [PubMed] [Google Scholar]
- 51.Morrison, T. G., M. E. Peeples, and L. W. McGinnes. 1987. Conformational change in a viral glycoprotein during maturation due to disulfide bond disruption. Proc. Natl. Acad. Sci. USA 84:1020-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65:55-63. [DOI] [PubMed] [Google Scholar]
- 53.Mou, Y., H. Ni, and J. A. Wilkins. 1998. The selective inhibition of beta 1 and beta 7 integrin-mediated lymphocyte adhesion by bacitracin. J. Immunol. 161:6323-6329. [PubMed] [Google Scholar]
- 54.Noiva, R. 1999. Protein disulfide isomerase: the multifunctional redox chaperone of the endoplasmic reticulum. Semin. Cell Dev. Biol. 10:481-493. [DOI] [PubMed] [Google Scholar]
- 55.Ou, W., and J. Silver. 2006. Role of protein disulfide isomerase and other thiol-reactive proteins in HIV-1 envelope protein-mediated fusion. Virology 350:406-417. [DOI] [PubMed] [Google Scholar]
- 56.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Puri, A., F. P. Booy, R. W. Doms, J. M. White, and R. Blumenthal. 1990. Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J. Virol. 64:3824-3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roche, S., S. Bressanelli, F. A. Rey, and Y. Gaudin. 2006. Crystal structure of the low-pH form of the vesicular stomatitis virus glycoprotein G. Science 313:187-191. [DOI] [PubMed] [Google Scholar]
- 59.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Russell, C. J., and L. E. Luque. 2006. The structural basis of paramyxovirus invasion. Trends Microbiol. 14:243-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryser, H. J., E. M. Levy, R. Mandel, and G. J. DiSciullo. 1994. Inhibition of human immunodeficiency virus infection by agents that interfere with thiol-disulfide interchange upon virus-receptor interaction. Proc. Natl. Acad. Sci. USA 91:4559-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sergel, T., L. W. McGinnes, and T. G. Morrison. 1993. The fusion promotion activity of the NDV HN protein does not correlate with neuraminidase activity. Virology 196:831-834. [DOI] [PubMed] [Google Scholar]
- 63.Skehel, J. J., P. M. Bayley, E. B. Brown, S. R. Martin, M. D. Waterfield, J. M. White, I. A. Wilson, and D. C. Wiley. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. USA 79:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takizawa, N., T. C. Smith, T. Nebl, J. L. Crowley, S. J. Palmieri, L. M. Lifshitz, A. G. Ehrhardt, L. M. Hoffman, M. C. Beckerle, and E. J. Luna. 2006. Supervillin modulation of focal adhesions involving TRIP6/ZRP-1. J. Cell Biol. 174:447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tamm, L. K., X. Han, Y. Li, and A. L. Lai. 2002. Structure and function of membrane fusion peptides. Biopolymers 66:249-260. [DOI] [PubMed] [Google Scholar]
- 66.Turano, C., S. Coppari, F. Altieri, and A. Ferraro. 2002. Proteins of the PDI family: unpredicted non-ER locations and functions. J. Cell Physiol. 193:154-163. [DOI] [PubMed] [Google Scholar]
- 67.Vitkovic, L., and H. L. Sadoff. 1977. Purification of the extracellular protease of Bacillus licheniformis and its inhibition by bacitracin. J. Bacteriol. 131:891-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wahlberg, J. M., and H. Garoff. 1992. Membrane fusion process of Semliki Forest virus. I: Low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 116:339-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallin, M., M. Ekstrom, and H. Garoff. 2004. Isomerization of the intersubunit disulphide-bond in Env controls retrovirus fusion. EMBO J. 23:54-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 71.Wilkinson, B., and H. F. Gilbert. 2004. Protein disulfide isomerase. Biochim. Biophys. Acta 1699:35-44. [DOI] [PubMed] [Google Scholar]
- 72.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 73.Wouters, M. A., K. K. Lau, and P. J. Hogg. 2004. Cross-strand disulphides in cell entry proteins: poised to act. Bioessays 26:73-79. [DOI] [PubMed] [Google Scholar]
- 74.Yin, H. S., R. G. Paterson, X. Wen, R. A. Lamb, and T. S. Jardetzky. 2005. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. USA 102:9288-9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yin, H. S., X. Wen, R. G. Paterson, R. A. Lamb, and T. S. Jardetzky. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Young, J. K., R. P. Hicks, G. E. Wright, and T. G. Morrison. 1997. Analysis of a peptide inhibitor of paramyxovirus (NDV) fusion using biological assays, NMR, and molecular modeling. Virology 238:291-304. [DOI] [PubMed] [Google Scholar]
- 77.Zelenkov, P., R. Baumgartner, K. Bise, M. Heide, R. Meier, S. Stocker, R. Sroka, R. Goldbrunner, and W. Stummer. 27 September 2006. Acute morphological sequelae of photodynamic therapy with 5-aminolevulinic acid in the C6 spheroid model. J Neurooncol. doi: 10.1007/s11060-006-9252-8. [DOI] [PubMed]